Abstract
Alternative pre-mRNA splicing determines many changes in gene expression during development. Two regulators known to control splicing patterns during neuron and muscle differentiation are the polypyrimidine tract-binding protein (PTB) and its neuronal homolog nPTB. These proteins repress certain exons in early myoblasts, but upon differentiation of mature myotubes PTB/nPTB expression is reduced, leading to increased inclusion of their target exons. We show here that the repression of nPTB expression during myoblast differentiation results from its targeting by the muscle-restricted microRNA miR-133. During differentiation of C2C12 myoblasts, nPTB protein but not mRNA expression is strongly reduced, concurrent with the up-regulation of miR-133 and the induction of splicing for several PTB-repressed exons. Introduction of synthetic miR-133 into undifferentiated C2C12 cells leads to a decrease in endogenous nPTB expression. Both the miR-133 and the coexpressed miR-1/206 microRNAs are extremely conserved across animal species, and PTB proteins are predicted targets for these miRNAs in Drosophila, mice, and humans. There are two potential miR-133-responsive elements (MRE) within the nPTB 3′ untranslated region (UTR), and a luciferase reporter carrying this 3′ UTR is repressed by miR-133 in an MRE-dependent manner. Transfection of locked nucleic acid (LNA) oligonucleotides designed to block the function of miR-133 and miR-1/206 increases expression of nPTB and decreases the inclusion of PTB dependent exons. These results indicate that miR-133 directly down-regulates a key splicing factor during muscle development and establishes a role for microRNAs in the control of a developmentally dynamic splicing program.
Keywords: Locked nucleic acids, alternative splicing, miR-133, microRNAs, myogenesis, nPTB
Alternative splicing is an important means of post-transcriptional gene regulation that contributes to the diversification of protein products in different cell types. Polypyrimidine tract-binding protein (PTB/hnRNPI/PTBP1) is a well-studied splicing regulator that is expressed in a wide range of tissues (Garcia-Blanco et al. 1989; Patton et al. 1991; Ghetti et al. 1992; Wagner and Garcia-Blanco 2001; Spellman and Smith 2006). At least two additional PTB homologs exist in mammals: ROD1, which is restricted to hematopoetic cells (Yamamoto et al. 1999), and nPTB/brPTB/PTBP2, which is expressed primarily in neurons and testes and at lower levels in muscle (Ashiya and Grabowski 1997; Chan and Black 1997; Irwin et al. 1997; Kikuchi et al. 2000; Markovtsov et al. 2000; Polydorides et al. 2000; Lillevali et al. 2001). Rodents also contain a smooth muscle expressed homolog, smPTB (Gooding et al. 2003). PTB and its homologs regulate many exons that are spliced in a neuron- or muscle-specific manner, such as c-src N1, α-actinin SM, α-tropomyosin exon 2, NMDAR1 exon 5, and cardiac troponin T exon 5 (Chan and Black 1997; Gooding et al. 1998; Southby et al. 1999; Zhang et al. 1999; Charlet-B et al. 2002). During the development of embryonic muscle and repair of adult muscle injury, alternative splicing plays a critical role in the establishment of the mature pattern of muscle gene expression, including definition of the excitatory-contractile apparatus unique to a particular muscle type (Morano 2003; Agarkova and Perriard 2005). Various myopathies caused by aberrant splicing underscore its importance in myogenesis (Ver Heyen et al. 2001; Faustino and Cooper 2003; Day and Ranum 2005; Ladd et al. 2005).
In addition to alternative splicing, other post-transcriptional regulatory mechanisms such as the RNA interference (RNAi) pathway play critical roles during development. MicroRNAs (miRNAs or miRs) are a large class of noncoding RNA present in diverse organisms, including plants, nematodes, insects, and mammals (Lee et al. 1993; Reinhart et al. 2000; Lagos-Quintana et al. 2001; Lau et al. 2001; Lee and Ambros 2001). Animal microRNAs typically form imperfectly base-paired duplexes with microRNA-response elements (MREs) in target mRNAs. This interaction can inhibit translation and may ultimately also lead to the degradation of the mRNA (Olsen and Ambros 1999; Bagga et al. 2005; Lim et al. 2005). For effective repression, base-pairing between the MRE and the first 8 nucleotides (nt) of the miRNA—the “seed” region—are particularly important (Lewis et al. 2003, 2005; Lai 2004; Brennecke et al. 2005; Xie et al. 2005). However, the mechanisms leading to translational repression by the bound miRNA within the RNA-induced silencing complex (RISC) are not known (Humphreys et al. 2005; Pillai et al. 2005; Petersen et al. 2006).
In mammals, hundreds of microRNAs have been identified, many of which are tissue specific and/or temporally regulated in their expression, but whose biological functions are mostly not yet known (Lagos-Quintana et al. 2001, 2003; Mourelatos et al. 2002; Dostie et al. 2003; Berezikov et al. 2005). Targets for these miRNAs have been identified computationally, and some targets have been validated in vivo (Farh et al. 2005; O’Donnell et al. 2005; Stark et al. 2005; Zhao et al. 2005; Chen et al. 2006). Several microRNAs have been shown to regulate key events in cell differentiation, controlling the expression of important transcription factors or specialized functions of the cell (Poy et al. 2004; Yekta et al. 2004; Zhao et al. 2005; Chen et al. 2006). In this study, we describe the regulation of nPTB protein expression by the muscle-specific microRNA miR-133. The decrease in nPTB protein brought about by miR-133 alters the splicing of several mRNAs involved in muscle maturation. Thus, in addition to controlling transcription regulators and functional products of the fully differentiated cell, microRNAs also regulate mRNA processing events that lead to the mature muscle proteome.
Results
nPTB protein is present in C2C12 myoblasts but absent from differentiated myotubes
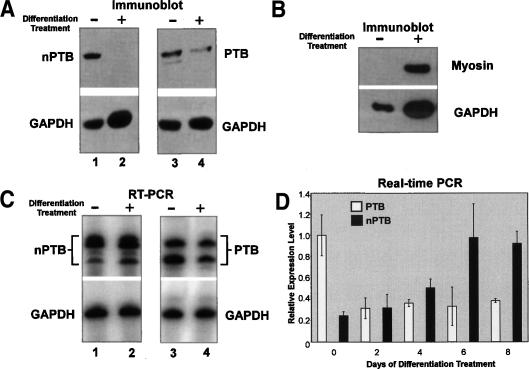
Because nPTB and PTB regulate the alternative splicing of exons known to change during muscle-cell differentiation, we examined their expression in C2C12 cells in both the proliferative growth phase and after 5 d of differentiation treatment. C2C12 cells are derived from adult mouse satellite cells and undergo fusion to form contractile myotubes when exposed at high density to differentiation medium. Myotube formation and further maturation was monitored visually and through Western blotting for myosin heavy chain protein expression, a marker for differentiated muscle (Fig. 1B). Although present in proliferating myoblasts, nPTB protein was virtually eliminated in differentiated myotubes (Fig. 1A, lanes 1,2). In contrast to nPTB protein, both the quantity of nPTB mRNA and the ratio of alternatively spliced variants remain unchanged during this period as measured by semiquantitative RT–PCR (Fig. 1C, lanes 1,2). The nPTB paralog PTB displayed a different expression pattern. PTB protein was reduced but not eliminated during differentiation, and PTB mRNA decreased in parallel with the protein (Fig. 1A,C, lanes 3,4). The changes in nPTB and PTB mRNA levels during myotube differentiation were confirmed by real-time PCR on cDNA derived from C2C12 cells. As measured relative to two separate control mRNAs, GAPDH and β-actin, nPTB mRNA levels actually increase during the differentiation process (Fig. 1D; data not shown). In contrast, after 2 d of differentiation, PTB mRNA dropped to ∼40% of the level seen in proliferating myoblasts and then remained relatively constant (Fig. 1D). These data confirm that the loss of nPTB and PTB proteins coincides with the onset of muscle differentiation, but that for nPTB a corresponding loss of mRNA does not occur.
Figure 1.
nPTB protein is nearly eliminated during differentiation of C2C12 myoblasts, while mRNA levels increase. (A, top panel) Immunoblot of total protein lysates from C2C12 cells using α-nPTB IS2 (lanes 1,2) or α-PTB-NT antibodies (lanes 3,4). Cells were cultured in either growth or differentiation medium as indicated above each lane. (Bottom panel) GAPDH loading control. (B, top panel) Immunoblot of myosin heavy chains, a mature muscle marker in the same cells used in A and B. The bottom panel is blotted for GAPDH. (C) The top panel shows RT–PCR of the nPTB (lanes 1,2) and PTB (lanes 3,4) alternatively spliced regions in mRNA from proliferating (−) and differentiating (+) cells (same samples as used in A). (Bottom panel in all lanes) Equivalent amounts of RNA were used as indicated by the RT–PCR of GAPDH. (D) mRNA quantification for PTB and nPTB relative to β-actin mRNA between days 0 and 8 of differentiation treatment as determined by real-time PCR.
nPTB contains phylogenetically conserved MREs for miR-133
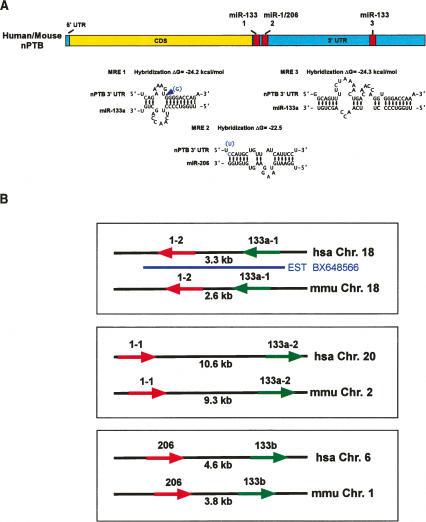
The loss of nPTB protein upon myotube formation without a similar reduction in mRNA suggested that a translational repression mechanism was controlling nPTB protein levels. MicroRNAs repress translation of mRNAs through binding to specific sequences in their RNA targets, often in the 3′ untranslated region (UTR) (Olsen and Ambros 1999). We examined whether the human and mouse nPTB mRNAs contain likely response elements (MREs) for any of the known microRNAs. MiR-133 was found to have two potential binding sites, which are conserved almost exactly between mouse and human (Fig. 2A). The same miR-133 responsive MREs were also identified in computational screens published by other groups (John et al. 2004; Kiriakidou et al. 2004; Krek et al. 2005). An important determinant for miRNA repression is the stability of the miRNA:MRE duplex within the 8-nt “seed” region at the 5′ end of the miRNA. Among all the predicted MREs in nPTB, the elements specific to miR-133 had the lowest predicted free energy of hybridization with the cognate miRNA (−24.2 and −24.3 kcal/mol) (Fig. 2A).
Figure 2.
nPTB contains conserved MREs for miR-133 and miR-1/206. (A) The MREs in both mouse and human nPTB are shown as red boxes. Untranslated regions are indicated in blue and coding sequence are in yellow. Each MRE is numbered, the predicted structure of each base-paired MRE/miRNA hybrid is diagrammed, and the predicted free energy of hybridization of the miR with the MRE is indicated. The top strand in each diagram represents the MRE sequence in the target mRNA, 5′–3′, and the bottom strand represents the indicated microRNA. Paired bases are indicated by a black oval, G:U pairs are indicated by two dots. All nucleotides in the miR and almost all in the MREs are identical between the mouse and human; where they differ, the mouse-specific nucleotides are shown in blue. (B) In mouse and human, the miR-133 (green arrows) and miR-1/206 (red arrows) microRNAs are coexpressed from three distinct loci. The chromosomes and the nucleotide distances between the pre-miR sequences are indicated. The arrows indicate the 5′–3′ direction of the mature microRNA. The miR locus on chromosome 18 is on the reverse strand. The blue underline represents a human EST sequence containing the miRNA precursors.
Mature miR-133 in mammals exists in two slightly different variants, miR-133a and miR-133b, expressed from three loci located on separate human or mouse chromosomes (Chen et al. 2006). Interestingly, each of these loci has a closely linked gene encoding the precursor (pre-miRNA) for either miR-1 or the highly similar miR-206 (Fig. 2B). The proximity of the two pre-miRNAs at each locus, expressed sequence tag (EST) expression data, Northern blot analyses from others, and the identical expression patterns of the two miR families all suggest that each miR-133 plus miR-1/206 pair is coexpressed from a common precursor transcript (Chen et al. 2006). A putative MRE matching miR-1/206 is also present in the nPTB 3′ UTR, although its seed region hybridization is not as strong as the miR-133 elements. Mir-133 and miR-1/206 are highly conserved in the fruit fly, mouse, human, and other animals (Supplementary Fig. 1A). Interestingly, the Drosophila melanogaster PTB homolog hephaestus was identified in earlier studies as a putative miR-133 and miR-1 target in Drosophila (Dansereau et al. 2002; Stark et al. 2003). Using the same target prediction protocol as for nPTB, we searched for MREs in the other PTB gene family members (see Materials and Methods). Indeed, PTB also contains MREs for miR-133 and miR-1/206 that are conserved between mouse and human (Supplementary Fig. 1B). As seen previously, this analysis also identified multiple possible MREs for these microRNAs in the fruit fly hephaestus gene (Supplementary Fig. 1B). Thus, two mammalian PTB proteins, as well as the single fruit fly homolog, are all predicted targets of these two highly conserved microRNA families.
Several groups have shown that miR-133 and miR-1/206 are specifically expressed in heart and skeletal muscle (Lee and Ambros 2001; Lagos-Quintana et al. 2002; Babak et al. 2004; Barad et al. 2004; Sempere et al. 2004; Thomson et al. 2004; Baskerville and Bartel 2005; Wienholds et al. 2005). Similarly, in D. melanogaster miR-1 is restricted to muscle cells throughout development (Biemar et al. 2005; Kwon et al. 2005; Sokol and Ambros 2005). To confirm these findings and to assess their expression during our C2C12 differentiation protocol, we assayed for miR-133 and miR-1/206 in tissues and cell lines by small RNA Northern blots. As expected, both the miR-133 and miR-1/206 families were restricted to muscle and heart tissue in the mouse (data not shown). Among the cell lines tested, we observed the presence of both microRNA families only in cell lines derived from muscle tissue, specifically the human rhabdomyosarcoma cell line RD and the mouse myoblast cell line C2C12 (Fig. 3A). In C2C12 cells, the undifferentiated myoblasts maintained in proliferative conditions do not express the microRNAs. Upon differentiation into myotubes, we observe a strong up-regulation of both miR-133 and miR-1/206 (Fig. 3A,B), in agreement with the results of Chen et al. (2006). The expression of miR-133 and miR-1/206 began as early as 2 d following the switch to differentiation conditions, increased dramatically through 4 d, and decreased thereafter (Fig. 3B). Consistent with their expression from a common precursor, miR-133 and miR-1/206 always increased and decreased in parallel. At 10 d post-treatment the cells have completed myotube formation and can be observed undergoing contractile activity.
Figure 3.
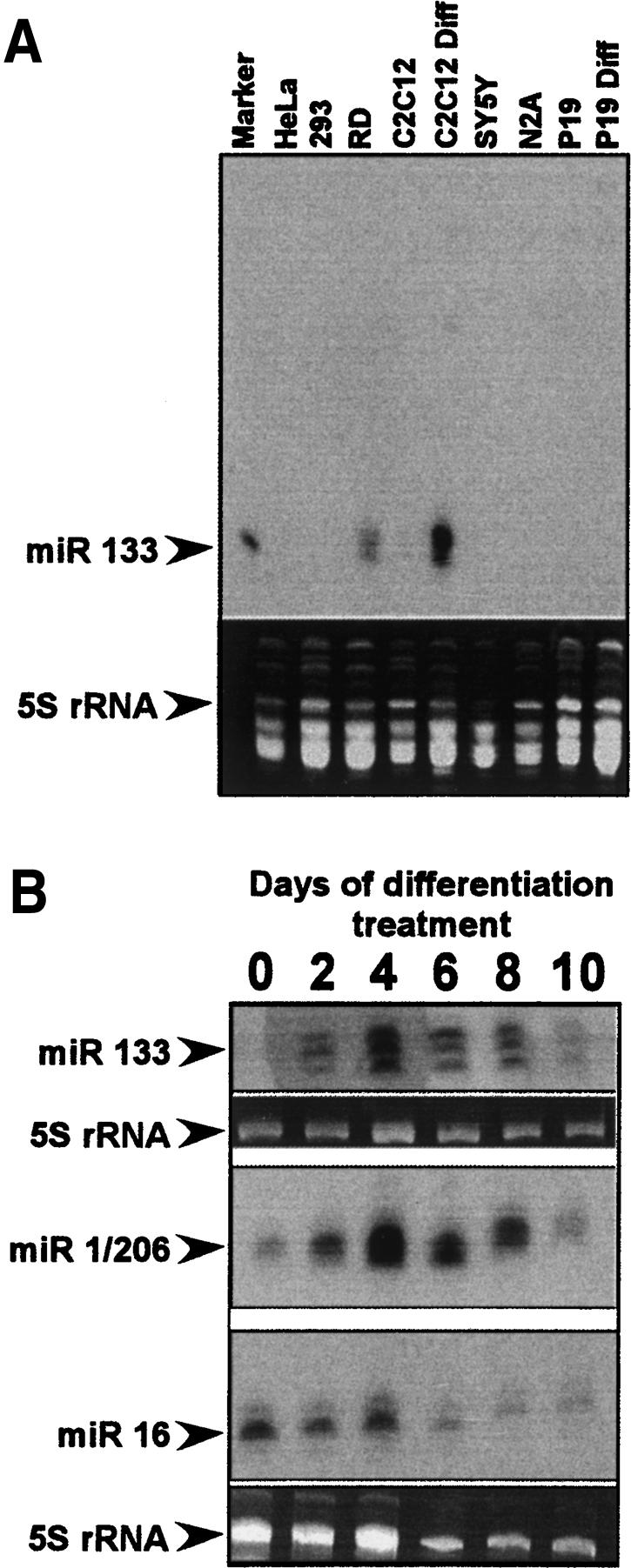
miR-133 and miR-1/206 expression is restricted to muscle-derived cell lines. Size-fractionated samples of total RNA from tissue culture cells were probed for the presence of microRNAs. Note that the probe for miR-133 cannot distinguish between miR-133a and miR-133b, and the probe for miR-1/206 cannot distinguish between miR-1 and miR-206. miR-16 is a widely expressed microRNA detected in most cell lines. 5S rRNA was stained with ethidium bromide to normalize for loading. (A) Small RNA Northern probed for miR-133 expression in cell lines. The lane marked “C2C12” contains RNA from proliferating cells, whereas “C2C12 Diff” is RNA from cells treated for 4 d in differentiation medium. Similarly, “P19” is RNA from undifferentiated cells and “P19 Diff” is RNA from retinoic acid-treated P19 cells. (B) Small RNA Northern blot of RNA from differentiating C2C12 cells probed for miR-133 and miR-1/206. Differentiation medium was added at time 0 and RNA was harvested every 48 h through day 10. Note that the miR-1/206 and miR-16 panels are probings of the same blot and are normalized to the same 5S RNA panel.
miR-133 represses nPTB mRNA translation through two conserved MREs in the 3′ UTR
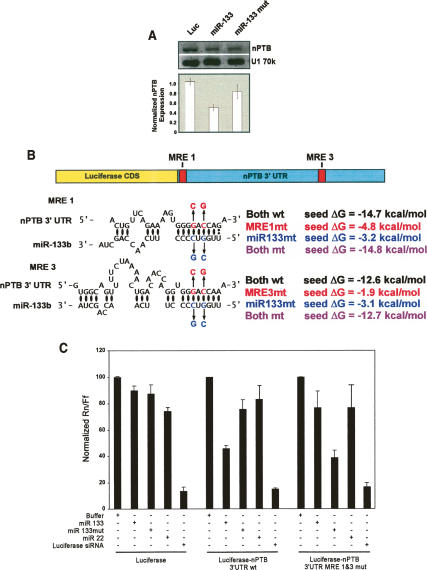
To verify the targeting of endogenous nPTB by miR-133 in C2C12 cells, we transfected proliferating myoblasts with artificial miR-133. We prepared short double-stranded RNAs (dsRNAs) resembling Dicer-processed microRNAs, in which the strand designed to be accepted into the RISC complex was identical to mature miR-133b (Doench et al. 2003; Khvorova et al. 2003; Schwarz et al. 2003). We also prepared a mutant version of miR-133 carrying point mutations that would disrupt the seed pairing with the wild-type MREs. Subconfluent myoblasts in growth medium were transfected with these miRNAs and assayed for nPTB expression. Two days after transfection of wild-type miR-133, nPTB protein was reduced by 50% compared with control cells treated with buffer or transfected with a luciferase small interfering RNA (siRNA) (Fig. 4A). This effect was dose dependent and saturated at higher amounts of miRNA (data not shown). The mutant miR-133 gave weaker repression, at about half that of the wild-type RNA. This repression may be due to the remaining base-pairing potential in the mutant affecting expression at high levels of miRNA. A miR-206 RNA gave still weaker repression (data not shown). We did not observe affects of the transfected miRNAs on the differentiation of the C2C12 cells, as reported by others (Chen et al. 2006; Kim et al. 2006). This may be due to different growth conditions, as we kept the cells well below confluency to reduce spontaneous differentiation and maintain normal nPTB expression.
Figure 4.
miR-133b represses expression of nPTB through elements in the 3′ UTR. (A) Proliferating C2C12 myoblasts were transfected with the indicated control siRNA (Luc), wild-type miR-133 (miR-133), or mutant miR-133 (miR-133 mut). At 48 h post-transfection, cells were harvested and Western blots were performed on total cell lysates are shown. Fluorescent secondary antibodies were used to quantify nPTB levels relative to a U1 70k control, which are graphed below. (B) A luciferase reporter plasmid carrying the Renilla luciferase coding sequence attached to the entire human nPTB 3′ UTR with either wild-type (wt) or mutant (mut) MREs is diagrammed. For each MRE/miR pair, the calculated free energy of hybridization (ΔG) in kilocalories per mole is shown on the right. (C) Luciferase reporters were cotransfected with the artificial miR-133 or control RNAs. The ratio of reporter (Renilla luciferase, Rn) to control plasmid (firefly luciferase, Ff) in relative luminescence units was normalized for each reporter to the buffer control and plotted as a percentage of the control value. Error bars represent the standard error for n = 3.
To test directly whether miR-133 can repress translation through binding the nPTB MREs, we constructed a luciferase reporter with the entire human nPTB 3′ UTR immediately following the Renilla luciferase coding sequence (Fig. 4B). We also constructed an equivalent luciferase reporter with the two miR-133 MREs mutated at two nucleotides each. These MRE mutations restore pairing to the mutant miR-133, with the predicted free energy of hybridization of the mutant pairing equivalent to that of the wild-type miR-133 to the wild-type MREs (Fig. 4B).
We cotransfected HEK 293 cells with each of the luciferase/nPTB 3′ UTR reporters combined with either the artificial microRNAs or a luciferase-specific siRNA and assayed luciferase expression after transient expression. These cells do not express endogenous miR-133 or miR-1/206 (Fig. 3A). Expression from the luciferase reporter lacking the nPTB 3′ UTR was unaffected by either the wild-type or the mutant miR-133, but was drastically reduced by the luciferase-specific siRNA, demonstrating that the reporter plasmids and small RNA molecules were efficiently cotransfected (Fig. 4C). In contrast, transcripts carrying the wild-type nPTB 3′ UTR exhibited a >50% reduction in luciferase expression in the presence of the wild-type miR-133b. This repression was dose dependent and saturated at higher concentrations of miR-133 (data not shown). An artificial miR-133a molecule behaved similarly (data not shown). The mutant miR-133 had a much weaker effect on the wild-type 3′ UTR, as did an unrelated microRNA, miR-22, which is not predicted to bind the nPTB 3′ UTR (Fig. 4C). Conversely, the reporter carrying the mutant MREs was repressed more strongly by the mutant miR-133. Expression from this mutant reporter was again repressed by ∼50% by the mutant miR-133, with which it could base-pair well, but was only weakly affected by the wild-type miR-133 or by miR-22 (Fig. 4C). Luciferase/nPTB constructs carrying only one mutated MRE showed similar responses to the microRNAs, but of lower magnitude (data not shown). These data indicate that miR-133 can down-regulate expression from mRNAs carrying the nPTB 3′ UTR, and that this repression is mediated through the MREs.
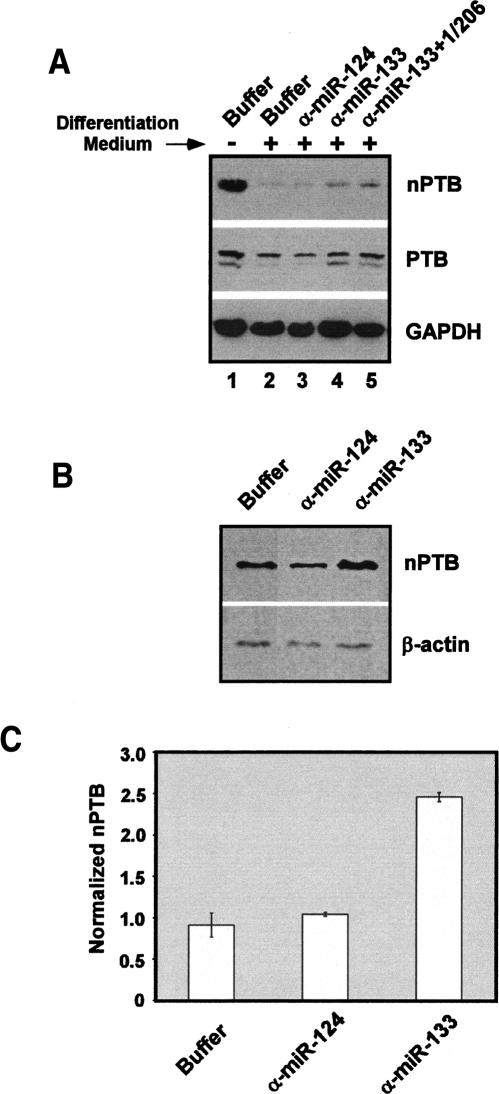
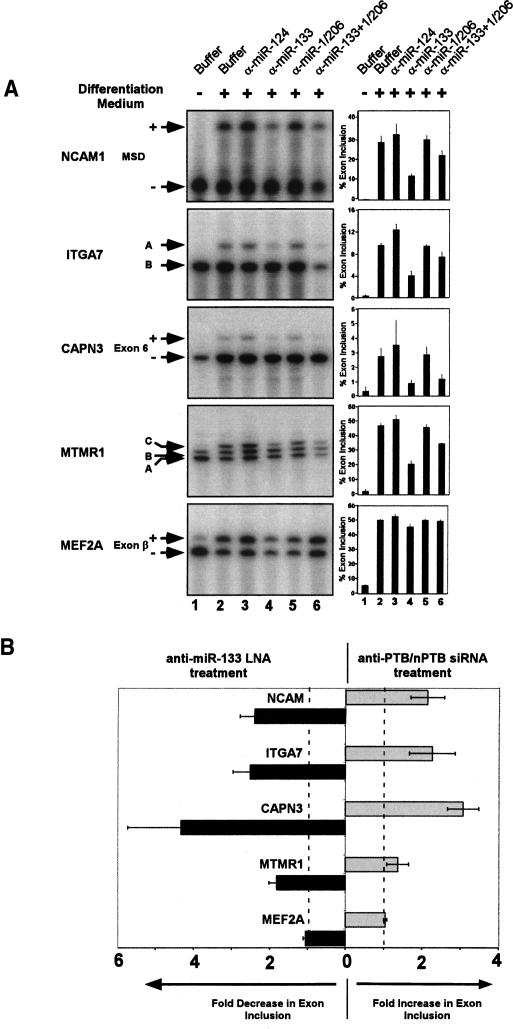
Blocking miR-133 function during muscle cell differentiation increases PTB and nPTB protein and alters splicing of muscle-specific exons
To observe directly the effect of miR-133 and miR-1/206 on nPTB in differentiating C2C12 cells, we used locked nucleic acid (LNA)-modified oligonucleotides complementary to the miRNAs to block their function (Chan et al. 2005; Fazi et al. 2005; Orom et al. 2006). As assayed by Western blot, nPTB protein was nearly eliminated after 72 h in differentiation medium (Fig. 5A, cf. lanes 1 and 2). Significantly, cells transfected with the anti-miR-133 LNA oligo exhibited a consistent doubling in nPTB expression when compared with two different loading controls (Fig. 5A [lane 4], B). In contrast, an equivalent amount of the anti-miR-124 LNA control oligo had no effect (Fig. 5A, lane 3). Treatment with a combination of three LNA oligos targeting miR-133, miR-1, and miR-206 increased nPTB levels slightly more than the miR-133 LNA alone (Fig. 5A, lane 5). Although PTB protein is not eliminated upon differentiation (Fig. 5A, lane 2), similar increases were seen in the PTB protein level upon LNA treatment (Fig. 5A, middle panel).
Figure 5.
nPTB and PTB proteins are increased in differentiating myoblasts when miR-133 is functionally blocked. (A) Undifferentiated cells (lane 1) or cells grown in differentiation medium for 72 h (lanes 2–5) were immunoblotted for nPTB (top panel), PTB (middle panel), or GAPDH (bottom panel). Note that the residual nPTB seen after 72 h is eliminated by longer culture in differentiation medium (Fig. 1A). Cells were transfected with the indicated LNA oligonucleotides and/or siRNAs before differentiation treatment. (Lane 3) An LNA-modified oligonucleotide against miR-124 was used as a control (α-miR-124). LNA oligos designed to block miR-133, miR-1, and miR-206 are labeled above the lanes (α-miR-133, α-miR-1, and α-miR-206, respectively). (B) Immunoblot showing nPTB protein levels relative to a β-actin loading control. (C) Quantification of fluorescent secondary antibody signal comparing nPTB expression against U1 70k as a loading control; based on three separate experiments.
The lifetime of the LNA effect (1–2 d) is short relative to the differentiation time course, and it was not possible to completely reverse the inhibition of nPTB expression and bring it back to predifferentiation levels. Moreover, the immunoblot chemiluminescent detection system does not give a perfectly linear response when normalized between two antibodies (data not shown). To more carefully quantify the changes in protein induced by LNA treatment, Western blots with Cy5-labeled fluorescent secondary antibodies were used. An antibody targeting a nPTB-specific peptide (nPTB-IS2) yielded a consistently linear signal over a large range of nPTB protein concentration, relative to U1 70K and β-actin control antibodies (data not shown). Using this fluorescent signal to quantify nPTB protein levels, we again observed that nPTB protein more than doubled in the anti-miR-133 LNA treated cells compared with the controls (Fig. 5C). Similar effects were observed with two different anti-miR-133 LNA oligos, one that we designed and a second that is commercially available (see Materials and Methods). At the low levels of protein seen in differentiated C2C12 cells, the observed twofold change in nPTB upon LNA treatment should be sufficient to induce changes in the splicing of PTB dependent exons (Wagner and Garcia-Blanco 2001; Wollerton et al. 2001).
To investigate the functional consequences of blocking miR-133 and up-regulating nPTB and PTB, we assayed the splicing of a number of alternative exons previously shown to change during the course of myogenesis (Hamshere et al. 1991; Ziober et al. 1993; Herasse et al. 1999; Buj-Bello et al. 2002). We confirmed the PTB/nPTB responsiveness of some of these exons as well as the known PTB-dependent exons in SRC and ACTN1 in differentiating C2C12 cells by RNAi knockdown (Fig. 6B; data not shown). Both PTB and nPTB are present in the cells and decrease with differentiation. Since PTB and nPTB often show equal repression activity on a target exon (data not shown), to observe the splicing in the absence of all PTB repression, we needed to knock down both proteins. Using RNA collected from the same cells assayed in Figure 5, we examined the splicing of PTB-dependent exons in neural cell adhesion molecule 1 (NCAM1), Integrin α 7 (ITGA7), Calpain 3 (CAPN3), Myotubularin-Related Protein 1 (MTMR1), Myocyte Enhancer Factor 2A, and other genes. All of these exons are strongly repressed in undifferentiated cells, but are induced to splice upon differentiation, ranging up to 50% inclusion for exons in MTMR1 and MEF2A (Fig. 6, lanes 1,2; data not shown). Treatment of these differentiated cells with the anti-miR-133 LNA or with all three LNA oligos (anti-miR-133, anti-miR-1, and anti-miR-206) strongly reduced the differentiation-dependent splicing for several of these exons (Fig. 6, lanes 4,6). Treatment with the control anti-miR-124 LNA or the anti-miR-1/206 LNAs alone had little effect (Fig. 6, lanes 3,5). PTB/nPTB dependent exons in NCAM1, ITGA7, CAPN3, and MTMR1 all showed similar repression in cells treated with the miR-133 LNA oligo, as expected from the increase in nPTB and PTB proteins in these cells. For these transcripts, the inclusion of the regulated exon was decreased by as much as 75% (Fig. 6). In contrast, splicing of exon β in MEF2A transcript was largely unaffected by blocking miR-133 function. This exon is presumably regulated by other splicing factors known to be up-regulated during C2C12 differentiation (Ladd et al. 2001). Thus, miR-133 has a specific effect on the splicing of a group of developmentally regulated PTB/nPTB-dependent exons in muscle cells.
Figure 6.
Alternative splicing of PTB-dependent muscle-specific exons is altered by blocking miR-133. (A) Lysates of cells from Figure 5 were processed for total mRNA, and the splicing of the indicated transcripts was assayed by RT–PCR. Lane 1 is undifferentiated cells (UD). Lanes 2–6 are cells differentiated for 72 h after transfection with the indicated LNA oligos and/or siRNAs. Polyacrylamide–urea gels with RT–PCR products incorporating 5′-32P-end-labeled primers are shown to the left, with quantification of the exon inclusion graphed to the right. The exon included and skipped products are indicated by arrows. All data are graphed as the average of three separate experiments, and the standard error is indicated by error bars. (B) Bars showing the fold increase in each exon in response to double knockdown of PTB and nPTB by RNAi extend to the right. This is measured against cells treated with anti-miR-133 LNA oligo and a control siRNA. Bars showing the fold decrease in exon inclusion as a result of anti-miR-133 LNA oligo treatment extend to the left. Error bars show the variation between three separate transfections.
Discussion
miR-133 controls alternative splicing during muscle development
We find that the muscle-specific microRNA miR-133 represses expression of the splicing factor nPTB during myoblast differentiation into myotubes. The repression of nPTB by miR-133 leads to the splicing of a group of PTB-silenced exons in mature myotubes. MicroRNAs are thought to target genes whose expression would be inappropriate in a particular tissue, as well as genes needed in the tissue, but which must be temporally controlled during development (Enright et al. 2003; Lewis et al. 2003; John et al. 2004; Farh et al. 2005; Krek et al. 2005; Stark et al. 2005). NPTB fits with the latter group, and its down-regulation along with PTB leads to important changes in splicing during muscle development. Thus, miR-133 plays a key role in controlling alternative splicing during muscle formation and defining the properties of differentiated muscle cells.
Neuronal PTB is a close homolog of PTB, expressed in adult brain, muscle, and testis. In adult brain, nPTB is specifically expressed in neurons where PTB is low. For this reason, nPTB has been studied primarily with neuronally spliced exons, where it exhibits weaker splicing repression activity than PTB (Ashiya and Grabowski 1997; Markovtsov et al. 2000; Polydorides et al. 2000). Interestingly, nPTB represses several muscle-specific exons roughly equally with PTB (Fig. 6; R. Spellman and C. Smith, pers. comm.; P. Boutz and P. Stoilov, unpubl.). We find here that the protein is also present in undifferentiated myoblasts, where it is indeed repressing muscle-specific exons. During myoblast differentiation, both PTB and nPTB levels decrease, but the mechanisms causing this loss of protein are apparently not identical. The presence of potential miRNA-binding sites within the PTB mRNA and the increase in protein expression upon blocking of miR-133 indicate that miR-133 also represses PTB expression. However, the decrease in PTB mRNA levels during C2C12 differentiation suggests that additional mechanisms of repression are also involved. The different roles of these two PTB proteins in muscle and why they need to be independently regulated are interesting questions for further study.
The concentration of nPTB mRNA increases steadily during myoblast differentiation, even though protein expression is nearly eliminated. Other splicing regulators that are currently thought to be widely expressed based on mRNA levels may also prove more restricted in protein expression due to similar post-transcriptional repression. Interfering with miR-133 with a transfected LNA probe led to a doubling of nPTB concentration, which is sufficient to cause significant changes in splicing. Similarly, miR-133 induced a 50% decrease in luciferase expression from a reporter carrying the wild-type nPTB 3′ UTR. The magnitude of these effects is similar to those observed in other systems with these assays and may not reflect the full repressive effect of microRNAs (Fazi et al. 2005; Lim et al. 2005; Stark et al. 2005). The induction or blocking of miR function in these experiments is likely inefficient. However, given the striking absence of nPTB protein and abundant nPTB mRNA, other repression mechanisms may also be involved.
The expression of nPTB is extremely sensitive to the sequence of the 3′ UTR. Mutation of just four MRE nucleotides within the 1400-nt UTR eliminates 60% of the repressive effect of the miR-133 (Fig. 4B). Since the miR responsiveness is restored by compensating base-pair mutations in the miR, miR-133 clearly targets the predicted MREs. These experiments used the entire nPTB 3′ UTR, rather than isolated elements or fragments as is sometimes done. This should retain the context of the MREs relative to RNA secondary structure or bound proteins and perhaps yield a more meaningful measure of the miRNA response. Given the conservation of the entire UTR among vertebrates, it is likely that microRNA-mediated silencing is coupled with other mechanisms of regulating gene expression.
The physiological relevance of this nPTB repression mechanism is underscored by the conservation of this family of proteins as potential miRNA targets. MiR-133 and miR-1 are highly conserved across animal species (Stark et al. 2003). Within the large group of targets for a miRNA in an individual genome, one would expect to find a subset of conserved original target mRNAs common to all species, although few targets are known to be conserved between organisms as evolutionarily distant as insects and mammals. The Drosophila PTB homolog hephaestus was predicted previously to be a target of both miR-133 and miR-1 (Stark et al. 2003). Using the search protocol that identified MREs in nPTB, we also found strong miRNA target sites in hephaestus and in the human and mouse PTB genes. The predicted MREs in hephaestus are within the coding sequence and contain a minimum of six contiguous Watson-Crick base pairs in positions 2–7 of the seed region (Doench and Sharp 2004; Brennecke et al. 2005; Lewis et al. 2005). The MREs identified by Stark et al. (2003) are in the 3′ UTR, but contain mismatches within the seed. Many computational screens for microRNA targets have focused on 3′ UTRs to identify conservation in the absence of coding potential. Thus, it is not well studied how well MRE’s function within coding sequence, and it remains to be tested which predicted MREs in hephaestus are functional in vivo.
The functional similarity of the PTB proteins of different species is also an interesting question. Intriguingly, Drosophila hephaestus is largely restricted to neural, myogenic, and germline tissues in developing flies, an expression pattern more closely resembling that of mammalian nPTB than the more widely expressed PTB (Davis et al. 2002). It will be interesting to determine in the mouse whether nPTB is expressed during embryonic myogenesis, as well as in adult muscle satellite cells, and its relative role in these two contexts.
Genomic analyses reveal that muscle tissue is among the most enriched for tissue-specific alternative splicing (Xing and Lee 2005). Only a subset of these alternative exons will be regulated by PTB/nPTB. Although we have not found high-probability MREs in their mRNAs, we cannot rule out that other splicing regulatory proteins might also be miR-133 targets. Detailed studies have examined the role of PTB in the splicing of α-actinin, tropomyosins 1 and 2, and cardiac troponin-T (Patton et al. 1991; Mulligan et al. 1992; Gooding et al. 1998; Southby et al. 1999; Charlet-B et al. 2002), primarily in smooth or cardiac muscle. In addition to these known exons, we find that PTB and nPTB also regulate splicing of the NCAM1 muscle-specific domain exons, MTMR1 exon 2.3, ITGA7 exon A, and CAPN3 exon 6 in muscle cells. The repression of the NCAM muscle-specific exons has also been shown to depend on CU-rich elements in the flanking introns that likely serve as nPTB/PTB-binding sites (Kawahigashi et al. 1998). Importantly, where it was assayable, exons that were repressed by blocking miR-133 in differentiating cells were also derepressed by PTB/nPTB knockdown (Fig. 6B; data not shown). It is clear that the loss of nPTB/PTB expression during myoblast differentiation alters the splicing of a whole group of target transcripts. It will be important to identify the large ensemble of exons controlled by miR-133 and the PTB proteins and to understand how this regulatory system fits into the specialized biology of muscle cells.
microRNA function in embryonic and adult myogenesis
The larger roles of miR-133 and miR-1/206 in muscle development are not entirely clear. Sokol and Ambros (2005) found that miR-1 mutant embryos in Drosophila developed normal musculature and motility through the end of the first larval instar. However, during transition to the second instar the null mutants failed to grow and eventually suffered lethal muscle degeneration. During Drosophila development, a subset of myogenic precursor cells is reserved in an undifferentiated state while embryonic muscles are positioned and develop. These precursor cells are subsequently employed for the production of muscle during larval and adult development, much like vertebrate satellite cells. It is intriguing to speculate that in mammals miR-133 and miR-1/206 might also be required for later processes of adult muscle maturation and repair, rather than embryonic myogenesis. Consistent with this idea, miR-1 and miR-133 show limited expression in the mouse embryo, but are strongly induced during neonatal growth (Chen et al. 2006).
It has been reported that transfection of a double-stranded miR-133 precursor RNA reduces C2C12 differentiation, and that miR-133 may function earlier in the differentiation process than miR-1, perhaps in maintaining the undifferentiated state (Chen et al. 2006). We did not observe this with our miR-133 dsRNA, but cannot rule out such an effect. For example, the culture conditions and the sequence of the transfected microRNAs were different in the two studies. In our system, it is clear that miR-133 continues to be expressed well after the cells have entered the differentiation process and is always induced concomitantly with miR-1/206, in keeping with their linked gene loci.
To understand the fundamental roles of miR-133 and miR-1, one starting place is the targets for these RNAs that are conserved between mammalian and insect muscle. To determine whether other pairs of mammalian and Drosophila gene homologs are conserved targets of the muscle-specific microRNAs similar to the PTB family, we searched the RefSeq database for elements capable of perfect Watson-Crick base-pairing to positions 2–7 of miR-1 or miR-133 (the seed sequences). We selected mRNAs for which the fruit fly, mouse, and human homologs could be matched and which all contain a minimum of three MREs for miR-1 and/or miR-133. We identified 156 genes that fit these criteria, most of which were identified in previous target identification studies for the individual organisms (Supplementary Table 1; Enright et al. 2003; Lewis et al. 2003; Stark et al. 2003; John et al. 2004; Krek et al. 2005). Of the 156 conserved targets, there were 48 examples of a single gene in Drosophila that had expanded to a multigene family in mammals, and where at least two mammalian paralogs were predicted microRNA targets along with the fruit fly homolog. Strikingly, 34 of the 42 mammalian collagen genes are predicted targets, and many have large numbers of MREs. D. melanogaster has three collagen genes, all of which are predicted miR-133 and miR-1 targets (Supplementary Table 1). Similarly, the Drosophila target gene baboon has three mammalian homologs encoding the TGF β receptors 1, 2 and 3, all of which contain MREs for miR-133 and miR-1/206. The TGF β signaling pathway is known to inhibit myogenesis in cultured C2C12 cells (De Angelis et al. 1998). It is apparent that, as with PTB/nPTB, mammalian microRNAs must often target multiple gene homologs to achieve the same outcome of repressing a single Drosophila gene.
Among these conserved targets, there are few genes in cell fate determination pathways. Instead, most targets are structural, metabolic, or signaling molecules expected to function after the specification of cell type, such as components of the dystrophin glycoprotein complex. These results suggest that these microRNAs are controlling expression within an established cell lineage rather than the induction of that lineage (Tomczak et al. 2004). This is consistent with studies in zebrafish showing that suppression of global miRNA expression resulted largely in perturbations in morphogenesis rather than cell fate determination (Wienholds et al. 2003; Giraldez et al. 2005; Harfe et al. 2005).
A role in the refinement rather than the initiation of a differentiated state is in keeping with the known targets of PTB/nPTB, including modulating cell/cell interactions and the function of the contractile apparatus. During the development of muscle fibers, where many cells fuse to function as one, it is likely that expression of the mature contractile apparatus must be precisely controlled while the individual cells comprising a muscle fiber are appropriately organized. Other PTB/nPTB regulated exons, in genes such as NCAM1, ITGA7, CAPN3, and MTMR1, would also be expected to need similar controlled expression during this process. Thus, the targeting of this splicing factor allows the microRNA to control a larger temporal program of muscle cell properties through not just the direct translational regulation of contractile apparatus and other mRNAs, but also by altering the splicing of important mRNAs.
Materials and methods
Sequence alignments
Alignment of nPTB 3′ UTRs from multiple species was performed using the University of California at Santa Cruz (UCSC) genome browser (Kent et al. 2002; http://genome.ucsc.edu) as well as AutoAssembler (Parker 1997) and BioEdit (Hall 1998) to align mRNA sequences obtained from GenBank (http://www.ncbi.nih.gov). Initial identification of putative MREs was performed using AutoAssembler to align a library of the 5′ 8 nt of each known human and mouse microRNA with the human and mouse nPTB mRNAs from GenBank. Free energies for the hybridization of microRNA seed sequences with mRNA sequences were determined using the RNA folding and two-state hybridization servers (Mathews et al. 1999; Zuker 2003; http://www.bioinfo.rpi.edu). Clustal-W alignment of the miR-133 and miR-1/206 microRNA families from multiple species was performed using the alignment tool provided by miRBase (Griffiths-Jones 2004). Genomic data for the relative locations of microRNAs in mouse and human and relevant EST data were obtained through the UCSC genome browser. miRNA–mRNA hybridization structures were determined using RNAhybrid (Rehmsmeier et al. 2004).
Plasmid construction
To construct the luciferase/nPTB 3′ UTR wild-type reporter construct, the human nPTB 3′ UTR was amplified by PCR using a WERI-1 cDNA clone (Markovtsov et al. 2000) as the template and primers nPTBlucF (5′-GGCCTAGGAAATGGGAAGAT GAAGATTG-3′) and nPTBlucR (5′-GGCCTAGGATCCGT TAGAAAATTAATTT-3′). The PCR product was ligated into the XbaI site downstream from the Renilla luciferase coding sequence in the vector pRL-TK (Promega). Point mutations in the MREs were introduced using a two-part PCR approach. A PCR product spanning from the nPTB stop codon to the MRE was made using primers nPTBlucF and nP3m133b1mutR (5′-CTCTGCTGCCCACTTTTCTGAACAG-3′) for MRE 1 and nP3m133b2mutR (5′-CTTTGCTGCCACCTGGTTGTCAG-3′) for MRE 3. A second PCR product spanning from the MRE to the end of the 3′ UTR, overlapping the first fragment by ∼20 nt, was made using primer nPTBlucR in the reverse direction and nP3m133b1mutF (5′-GTGGGCAGCAGAGTTTGATTTTT TTTG-3′) for MRE 1 and nP3m133b2mutF (5′-GGCAGCA AAGTTTATGTGCC-3′) for MRE 3. In a second PCR reaction, equimolar amounts of the two PCR products were mixed with dNTPs, buffer, and polymerase but no primers; denatured for 5 min at 95°C; annealed for 2 min at 50°C; then extended for 15 min at 68°C to produce full-length template strands. nPTBlucF and nPTBlucR primers and additional polymerase were then added and a standard 30-cycle PCR was performed. The PCR products were gel-purified, digested with XbaI, and ligated into pRL-TK. The clone in which both MREs are mutated was made in a similar fashion, except that the reporter construct with MRE 1 mutated was used as the template for the first PCR step with the MRE 3 mutant primers.
Tissue culture
Cell lines were grown following standard tissue culture procedure, with guidelines provided by the American Type Culture Collection (http://www.atcc.org). C2C12 cells were grown in high-glucose DMEM (Invitrogen) with 1.5 g/L sodium bicarbonate and 1 mM sodium pyruvate, supplemented with 10% fetal bovine serum (FBS); cells were grown at 37°C in 5% CO2. For differentiation treatment, C2C12 cells were allowed to reach confluency, then switched to the same medium except with 2% donor horse serum (HS) instead of FBS.
siRNA synthesis
Artificial microRNAs and siRNAs were constructed as described previously (Amir-Ahmady et al. 2005). Briefly, a double-stranded DNA template was constructed for each sense and antisense strand of the siRNA duplex by annealing a specific oligonucleotide (listed below) with a universal, T7-promoter-containing primer (5′-TAATACGACTCACTATAGGGAGA CAGG-3′) and filling in with exo− Klenow (New England Biolabs). Each RNA strand was then synthesized using T7-RNA polymerase, the two complementary strands were mixed, and the duplex was digested with DNaseI (Roche) and RNase T1 (Ambion) to remove the template DNA and to produce the characteristic 2-nt 3′ overhang present on each strand of a Dicer-processed siRNA or miRNA (Elbashir et al. 2001). The RNA duplexes were then phenolchloroform-extracted, ethanol-precipitated, and resuspended in dsRNA annealing buffer [20 mM HEPES-KOH at pH 7.6, 50 mM KOAc, 1 mM Mg2(OAc)2, 0.2-μm filtered] to a concentration of 20 μM. Note that the miR-22-negative control dsRNA thermodynamically favors the passenger strand for RISC entry but is used here as a control for specificity of the miR-133 molecules.
Template oligodeoxynucleotides; antisense strand is equivalent to the mature miR as follows: miR-133b wild-type, sense, 5′AATTGGTCCCCTTCAACCAGCCCTGTCTC-3′; miR-133b wild-type, antisense, 5′-TAGCTGGTTGAAGGGGACCAACCT GTCTC-3′; miR-133b mutant, sense, 5′-AATTGCTGCCCTT CAACCAGCCCTGTCTC-3′; miR-133b mutant, antisense, 5′-TAGCTGGTTGAAGGGCAGCAACCTGTCTC-3′; pRL156 (luciferase siRNA), sense, 5′-AATAAATAAGAAGAGGCCGC GCCTGTCTC-3′; pRL156(luciferase siRNA), antisense, 5′-AA CGCGGCCTCTTCTTATTTACCTGTCTC-3′; miR-22 sense, 5′-TTAAGCTGCCAGTTGAAGAACTGCCTGTCTC-3′; miR- 22 antisense, 5′-TACAGTTCTTCAACTGGCAGCTTCCTGTC TC-3′.
RT–PCR
Total RNA was collected from adherent tissue culture cells using Trizol (Invitrogen) according to the manufacturer’s instructions. RNA was quantified (A260) using a Nanodrop-1000 spectrophotometer (Nanodrop Technologies). Total RNA (0.5–1 μg) was used for each sample in a 10-μL reaction with 0.25 μL of SuperScript II RT (Invitrogen). One-tenth volume of the RT reaction was used in a 25-μL PCR reaction containing 200,000–500,000 counts per minute (cpm) of 32P-labeled reverse-strand primer; PCR reactions were run in an MJ Research PTC-200 thermocycler for 25 cycles with an annealing temperature of 55°C. Half of each PCR reaction was mixed 1:1 with 95% formamide containing 5% 10 mM Tris (pH 8.0) with bromophenol blue and xylene cyanol. RT–PCR reactions were loaded onto 8% polyacrylamide, 7.5 M urea gels and electrophoresed. Subsequently, gels were dried and imaged on a Typhoon PhosphorImager (Molecular Dynamics), and bands were quantified using ImageQuant 5.1 software (Molecular Dynamics). Primers used were as follows: nPTB exon 8 forward, 5′-GCATTTGCC AAGGAGACATCC-3′; nPTB exon 11 reverse, 5′-CGCTGCACATCTCCATAAACAC-3′; PTB exon 8 forward, 5′-AAGAGCAGAGACTACACTCGA-3′; PTB exon 12 reverse, 5′-CT GCCGTCTGCCATCTGCACAA-3′; CAPN3 exon 6 forward, 5′-TTCACCAAATCCAACCACCG-3′; CAPN3 exon 6 reverse, 5′-ACTCCAAGAACCGTTCCACT-3′ (Herasse et al. 1999); NCAM MSD forward (MH1B2 F), 5′-CCCCCGCCCC GAATTCCCACTGAGTTCAAGACACAG-3′; NCAM MSD reverse (MH2BR), 5′-GCCGGCGCGGAGCTTTCTGCCCTT CCAGCTTGGGT-3′ (Hamshere et al. 1991); ITGA7 A7A/B forward:, 5′-GCTGCTCAGAGATGCATCC-3′; ITGA7 A7A/B reverse, 5′-CACCGGATGTCCATCAGGAC-3′ (Ziober et al. 1993); MTMR1 exon 2 forward, 5′-CATGTTGAATGGTGTA AACAG-3′; MTMR1 exon 5 reverse, 5′-AATTATCCCCATG GCTCTGT-3′ (Buj-Bello et al. 2002); MEF2A forward, 5′-CTT GATTGGAAATACTGGTGC;-3′ MEF2A reverse, 5′-TCGGA GTTGTCACAGACA-3′ (Buj-Bello et al. 2002).
Real-time PCR
cDNA was prepared from Trizol total RNA preps as described in the RT–PCR section, except that 5 μg RNA was used in a 20-μL reaction. Real-time PCR was performed using Bio-Rad iQ SYBR Green Supermix on a Bio-Rad iQ 5 real-time thermocycler for 35 cycles, 58°C annealing temperature. Relative mRNA levels were determined by comparing threshold cycles for PTB and nPTB with GAPDH and β-actin using the ΔCT method (Bio-Rad Laboratories 2005).
Primers were designed using Primer3 (http://fokker.wi.mit. edu/primer3), as follows: GAPDH forward, 5′-AACTTTGGCA TTGTGGAAGG-3′; GAPDH reverse, 5′-CACATTGGGGGTA GGAACAC-3′; ACTB forward, 5′-TACAGCTTCACCACCA CAGC-3′; ACTB reverse, 5′-ATGCCACAGGATTCCATACC 3′; PTB forward, 5′-TCTACCCAGTGACCCTGGAC-3′; PTB reverse, 5′-GAGCTTGGAGAAGTCGATGC-3′; nPTB forward, 5′-ACCAGGCATTTTTGGAACTG-3′; nPTB reverse, 5′-TGT GGTGCCACTAAGAGGTG-3′.
Western blotting
Total protein lysates from adherent cell lines were prepared by scraping cells off of tissue culture plates in PBS, pelleting cells, and resuspending them in RIPA buffer (150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris at pH 8.0) (Harlow and Lane 1999) with protease inhibitors (Complete protease inhibitor cocktail; Roche) and Benzonase (90%, 1:10,000, Sigma) to break down chromatin. Lysates were diluted in 2× SDS loading buffer, heated for 10 min at 95°C, and loaded onto 10% polyacrylamide Laemmli SDS PAGE gels. Gels were run under standard electrophoresis conditions, then transferred to nitrocellulose (Schleicher & Schuell Protran) in a Bio-Rad Trans-Blot Cell apparatus. For blotting with fluorophore-conjugated secondary antibodies, transfers were performed on a Novex X-Cell mini-cell transfer apparatus (Invitrogen) onto Immobilon-FL PVDF membranes (Millipore). Membranes were probed under standard conditions with α-GAPDH 6C5 (1:200,000, Research Diagnostics, Inc.), α-β-actin (1:2500, Novus Biologicals, Inc.), α-U1 70k (Sharma et al. 2005), α-nPTB IS2 (1:500) (Sharma et al. 2005), α-MHC MF20 (Developmental Hybridoma Bank, University of Iowa) (1:2000), and α-PTB antibody PTB-NT (1:3000) (Markovtsov et al. 2000). Blots were then probed with HRP-conjugated secondary antibodies (1:5000, Amersham Pharmacia) and developed using standard enhanced chemiluminescence (ECL) and Kodak BioMax XAR film or probed with ECL Plex Cy5-conjugated goat α-mouse and goat α-rabbit secondary antibodies, dried, and scanned on a Typhoon PhosphorImager (GE Healthcare) for fluorescent blots. Quantification of fluorescent signal was performed using ImageQuant 5.1 software (Molecular Dynamics).
Northern blotting
Small RNAs were isolated from cell lines of different origins as well as tissues using the mirVana miRNA Isolation kit (Ambion), according to the manufacturer’s protocol. The small RNA fraction contained RNAs <200 nt including miRNAs, and 1–2 μg of RNA were used to load each lane. RNA from different cell lines or tissues was loaded onto 15% urea-PAGE gels. The gels were stained with ethidium bromide to visualize total RNA levels, followed by transfer onto Zeta-probe membranes (Bio-Rad 162-0156). Prehybridization was carried out for 2 h at 42°C using Ultrahyb-Ultrasensitive Hybridization buffer (Ambion). Hybridization was carried out in the presence of 32P-end-labeled RNA probes (106 cpm/mL) for miR-1 (UGGAAU GUAAAGAAGUAUGUAUU) or miR-133b (UUGGUCCC CUUCAACCAGCUA) overnight at 42°C. A probe recognizing miR-16 (UAGCAGCACGUAAAUAUUGGCG), a ubiquitously expressed miRNA, was used as a control for loading. After washing, the blots were exposed to PhosphorImager screens and visualized on a Typhoon PhosphorImager using ImageQuant software. RNA probes were transcribed from double-stranded DNA oligonucleotide templates prepared as described in the siRNA synthesis section and end-labeled using T4 polynucleotide kinase (New England Biolabs).
Luciferase assays
Luciferase assays were performed in accordance with previously published protocols (Doench et al. 2003; Doench and Sharp 2004). A 1:7 mixture of the firefly luciferase control plasmid (0.1 μg) (pGL3, Promega) and a wild-type or mutant luciferase/nPTB 3′ UTR construct, 0.1 μg pUC18 plasmid as carrier DNA, and 0.2 μL of a 20 μM solution of artificial, double-stranded microRNA were transfected into 40,000 HEK 293 cells with Lipofectamine 2000 (Invitrogen) as previously described (Underwood et al. 2005), and were plated in white 96-well tissue culture-treated plates (Corning Costar). At 24 h post-transfection, cells were lysed and assayed for firefly and Renilla luciferase activity using the Dual Glo Luciferase Assay System (Promega) according to the manufacturer’s instructions and measured on the Molecular Devices Analyst AD microplate reader. Data were normalized to the control samples (each construct, without microRNA added), and the results of three independent transfections were combined to determine standard error. Data were processed and plotted using Microsoft Excel.
LNA oligonucleotides and siRNA transfections
LNA oligos were obtained from Integrated DNA Technologies. All LNA oligos were synthesized with 5′ C6 amino modifications. Standard DNA bases are shown in lowercase, and LNA-modified bases are shown in uppercase, as follows: anti-miR 133 LNA, 5′-acaGctGgtTgaAggGgaCcaA; anti-miR-1 LNA, 5′-taCAtaCttCttTaCAttCCa; anti-miR-206 LNA, 5′-cCacA caCttCctTacAttCca; anti-miR-124 LNA, 5′-tggCatTcaCcgCgt GccTtaA.
A second antisense oligonucleotide targeting the miR-133 microRNAs was obtained from Exiqon (miRCURY LNA knockdown oligo mmu-miR-133a).
Transfections were performed in 12-well plates, with 200,000 C2C12 cells per well, 2 μg of pUC18 plasmid as carrier, 4 μL of Lipofectamine2000 (Invitrogen), and 80 nM LNA oligo for each target microRNA. Anti-miR 124 (240 nM) was used to equal the total LNA oligo concentration in the anti-133/1/206 triple-cotransfection. The anti-PTB siRNA was described previously (Amir-Ahmady et al. 2005). Twenty-six nanomolar siRNA was used in each transfection. All transfections were performed as previously described by Underwood et al. (2005).
miR133a/b and miR1/206 target prediction
The search for the potential mir133a/b and miR206/1 targets was performed on the human and mouse RefSeq mRNAs. To avoid potential sequencing errors and vector sequences that are present in the RefSeq mRNAs, we retrieved the coordinates of their alignment to the respective genomes from the UCSC genome assembly RefGene tables (Karolchik et al. 2004) and assembled the mRNAs using the aligned genomic sequence. The sequences of the microRNAs were obtained from miRBase (Griffiths-Jones 2004). The microRNA sequences were trimmed to the 5′-most 8 nt. RNAhybrid (Rehmsmeier et al. 2004) was used to find potential miRNA seeds. The seeds discovered by RNAhybrid were further filtered to remove any G∷U pairs in nucleotides 2–7 of the duplex. All data are stored in the PostgreSQL relational database. D. melanogaster microRNA:target predictions were performed in a similar way using RefSeq mRNAs, and the human/mouse homologs of each predicted Drosophila target were determined using the GeneKey database (Kirov et al. 2005). Gene ontology assignments were based on analysis of published work available through PubMed via the NCBI Web site.
Acknowledgments
We thank Chonghui Cheng, Calvin Jan, and members of the Black laboratory for helpful discussions; Stephan Kirov for the use of the GeneKey database; and Abigail Silver, Daniel Boutz, and Calvin Jan for critical reading of the manuscript. This work was supported by a grant from the NIH (RO1 GM49662) to D.L.B., and a Warsaw Family Fellowship at UCLA to P.B. D.L.B. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1500707
References
- Agarkova I., Perriard J.C., Perriard J.C. The M-band: An elastic web that crosslinks thick filaments in the center of the sarcomere. Trends Cell Biol. 2005;15:477–485. doi: 10.1016/j.tcb.2005.07.001. [DOI] [PubMed] [Google Scholar]
- Amir-Ahmady B., Boutz P.L., Markovtsov V., Phillips M.L., Black D.L., Boutz P.L., Markovtsov V., Phillips M.L., Black D.L., Markovtsov V., Phillips M.L., Black D.L., Phillips M.L., Black D.L., Black D.L. Exon repression by polypyrimidine tract binding protein. RNA. 2005;11:699–716. doi: 10.1261/rna.2250405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashiya M., Grabowski P.J., Grabowski P.J. A neuron-specific splicing switch mediated by an array of pre-mRNA repressor sites: Evidence of a regulatory role for the polypyrimidine tract binding protein and a brain-specific PTB counterpart. RNA. 1997;3:996–1015. [PMC free article] [PubMed] [Google Scholar]
- Babak T., Zhang W., Morris Q., Blencowe B.J., Hughes T.R., Zhang W., Morris Q., Blencowe B.J., Hughes T.R., Morris Q., Blencowe B.J., Hughes T.R., Blencowe B.J., Hughes T.R., Hughes T.R. Probing microRNAs with microarrays: Tissue specificity and functional inference. RNA. 2004;10:1813–1819. doi: 10.1261/rna.7119904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagga S., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Bracht J., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Hunter S., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Massirer K., Holtz J., Eachus R., Pasquinelli A.E., Holtz J., Eachus R., Pasquinelli A.E., Eachus R., Pasquinelli A.E., Pasquinelli A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- Barad O., Meiri E., Avniel A., Aharonov R., Barzilai A., Bentwich I., Einav U., Gilad S., Hurban P., Karov Y., Meiri E., Avniel A., Aharonov R., Barzilai A., Bentwich I., Einav U., Gilad S., Hurban P., Karov Y., Avniel A., Aharonov R., Barzilai A., Bentwich I., Einav U., Gilad S., Hurban P., Karov Y., Aharonov R., Barzilai A., Bentwich I., Einav U., Gilad S., Hurban P., Karov Y., Barzilai A., Bentwich I., Einav U., Gilad S., Hurban P., Karov Y., Bentwich I., Einav U., Gilad S., Hurban P., Karov Y., Einav U., Gilad S., Hurban P., Karov Y., Gilad S., Hurban P., Karov Y., Hurban P., Karov Y., Karov Y., et al. MicroRNA expression detected by oligonucleotide microarrays: System establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baskerville S., Bartel D.P., Bartel D.P. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E., Guryev V., de van Belt J., Wienholds E., Plasterk R.H., Cuppen E., Guryev V., de van Belt J., Wienholds E., Plasterk R.H., Cuppen E., de van Belt J., Wienholds E., Plasterk R.H., Cuppen E., Wienholds E., Plasterk R.H., Cuppen E., Plasterk R.H., Cuppen E., Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Biemar F., Zinzen R., Ronshaugen M., Sementchenko V., Manak J.R., Levine M.S., Zinzen R., Ronshaugen M., Sementchenko V., Manak J.R., Levine M.S., Ronshaugen M., Sementchenko V., Manak J.R., Levine M.S., Sementchenko V., Manak J.R., Levine M.S., Manak J.R., Levine M.S., Levine M.S. Spatial regulation of microRNA gene expression in the Drosophila embryo. Proc. Natl. Acad. Sci. 2005;102:15907–15911. doi: 10.1073/pnas.0507817102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bio-Rad Laboratories, Inc. Real-time PCR applications guide. Bio-Rad Laboratories, Inc.; Hercules, CA: 2005. [Google Scholar]
- Brennecke J., Stark A., Russell R.B., Cohen S.M., Stark A., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buj-Bello A., Furling D., Tronchere H., Laporte J., Lerouge T., Butler-Browne G.S., Mandel J.L., Furling D., Tronchere H., Laporte J., Lerouge T., Butler-Browne G.S., Mandel J.L., Tronchere H., Laporte J., Lerouge T., Butler-Browne G.S., Mandel J.L., Laporte J., Lerouge T., Butler-Browne G.S., Mandel J.L., Lerouge T., Butler-Browne G.S., Mandel J.L., Butler-Browne G.S., Mandel J.L., Mandel J.L. Muscle-specific alternative splicing of myotubularin-related 1 gene is impaired in DM1 muscle cells. Hum. Mol. Genet. 2002;11:2297–2307. doi: 10.1093/hmg/11.19.2297. [DOI] [PubMed] [Google Scholar]
- Chan R.C., Black D.L., Black D.L. The polypyrimidine tract binding protein binds upstream of neural cell-specific c-src exon N1 to repress the splicing of the intron downstream. Mol. Cell. Biol. 1997;17:4667–4676. doi: 10.1128/mcb.17.8.4667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J.A., Krichevsky A.M., Kosik K.S., Krichevsky A.M., Kosik K.S., Kosik K.S. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Charlet-B N., Logan P., Singh G., Cooper T.A., Logan P., Singh G., Cooper T.A., Singh G., Cooper T.A., Cooper T.A. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell. 2002;9:649–658. doi: 10.1016/s1097-2765(02)00479-3. [DOI] [PubMed] [Google Scholar]
- Chen J.F., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z., Mandel E.M., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z., Thomson J.M., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z., Wu Q., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z., Callis T.E., Hammond S.M., Conlon F.L., Wang D.Z., Hammond S.M., Conlon F.L., Wang D.Z., Conlon F.L., Wang D.Z., Wang D.Z. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansereau D.A., Lunke M.D., Finkielsztein A., Russell M.A., Brook W.J., Lunke M.D., Finkielsztein A., Russell M.A., Brook W.J., Finkielsztein A., Russell M.A., Brook W.J., Russell M.A., Brook W.J., Brook W.J. Hephaestus encodes a polypyrimidine tract binding protein that regulates Notch signalling during wing development in Drosophila melanogaster. Development. 2002;129:5553–5566. doi: 10.1242/dev.00153. [DOI] [PubMed] [Google Scholar]
- Davis M.B., Sun W., Standiford D.M., Sun W., Standiford D.M., Standiford D.M. Lineage-specific expression of polypyrimidine tract binding protein (PTB) in Drosophila embryos. Mech. Dev. 2002;111:143–147. doi: 10.1016/s0925-4773(01)00595-0. [DOI] [PubMed] [Google Scholar]
- Day J.W., Ranum L.P., Ranum L.P. RNA pathogenesis of the myotonic dystrophies. Neuromuscul. Disord. 2005;15:5–16. doi: 10.1016/j.nmd.2004.09.012. [DOI] [PubMed] [Google Scholar]
- De Angelis L., Borghi S., Melchionna R., Berghella L., Baccarani-Contri M., Parise F., Ferrari S., Cossu G., Borghi S., Melchionna R., Berghella L., Baccarani-Contri M., Parise F., Ferrari S., Cossu G., Melchionna R., Berghella L., Baccarani-Contri M., Parise F., Ferrari S., Cossu G., Berghella L., Baccarani-Contri M., Parise F., Ferrari S., Cossu G., Baccarani-Contri M., Parise F., Ferrari S., Cossu G., Parise F., Ferrari S., Cossu G., Ferrari S., Cossu G., Cossu G. Inhibition of myogenesis by transforming growth factor β is density-dependent and related to the translocation of transcription factor MEF2 to the cytoplasm. Proc. Natl. Acad. Sci. 1998;95:12358–12363. doi: 10.1073/pnas.95.21.12358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Sharp P.A., Sharp P.A. Specificity of microRNA target selection in translational repression. Genes & Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doench J.G., Petersen C.P., Sharp P.A., Petersen C.P., Sharp P.A., Sharp P.A. siRNAs can function as miRNAs. Genes & Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostie J., Mourelatos Z., Yang M., Sharma A., Dreyfuss G., Mourelatos Z., Yang M., Sharma A., Dreyfuss G., Yang M., Sharma A., Dreyfuss G., Sharma A., Dreyfuss G., Dreyfuss G. Numerous microRNPs in neuronal cells containing novel microRNAs. RNA. 2003;9:180–186. doi: 10.1261/rna.2141503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir S.M., Lendeckel W., Tuschl T., Lendeckel W., Tuschl T., Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enright A.J., John B., Gaul U., Tuschl T., Sander C., Marks D.S., John B., Gaul U., Tuschl T., Sander C., Marks D.S., Gaul U., Tuschl T., Sander C., Marks D.S., Tuschl T., Sander C., Marks D.S., Sander C., Marks D.S., Marks D.S. MicroRNA targets in Drosophila. Genome Biol. 2003;5:R1. doi: 10.1186/gb-2003-5-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farh K.K., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Grimson A., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Jan C., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Lewis B.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Johnston W.K., Lim L.P., Burge C.B., Bartel D.P., Lim L.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. The widespread impact of mammalian microRNAs on mRNA repression and evolution. Science. 2005;310:1817–1821. doi: 10.1126/science.1121158. [DOI] [PubMed] [Google Scholar]
- Faustino N.A., Cooper T.A., Cooper T.A. Pre-mRNA splicing and human disease. Genes & Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- Fazi F., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., Rosa A., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., Fatica A., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., Gelmetti V., De Marchis M.L., Nervi C., Bozzoni I., De Marchis M.L., Nervi C., Bozzoni I., Nervi C., Bozzoni I., Bozzoni I. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPα regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco M.A., Jamison S.F., Sharp P.A., Jamison S.F., Sharp P.A., Sharp P.A. Identification and purification of a 62,000-dalton protein that binds specifically to the polypyrimidine tract of introns. Genes & Dev. 1989;3:1874–1886. doi: 10.1101/gad.3.12a.1874. [DOI] [PubMed] [Google Scholar]
- Ghetti A., Pinol-Roma S., Michael W.M., Morandi C., Dreyfuss G., Pinol-Roma S., Michael W.M., Morandi C., Dreyfuss G., Michael W.M., Morandi C., Dreyfuss G., Morandi C., Dreyfuss G., Dreyfuss G. hnRNP I, the polypyrimidine tract-binding protein: Distinct nuclear localization and association with hnRNAs. Nucleic Acids Res. 1992;20:3671–3678. doi: 10.1093/nar/20.14.3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez A.J., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Cinalli R.M., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Glasner M.E., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Enright A.J., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Thomson J.M., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Baskerville S., Hammond S.M., Bartel D.P., Schier A.F., Hammond S.M., Bartel D.P., Schier A.F., Bartel D.P., Schier A.F., Schier A.F. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Gooding C., Roberts G.C., Smith C.W., Roberts G.C., Smith C.W., Smith C.W. Role of an inhibitory pyrimidine element and polypyrimidine tract binding protein in repression of a regulated α-tropomyosin exon. RNA. 1998;4:85–100. [PMC free article] [PubMed] [Google Scholar]
- Gooding C., Kemp P., Smith C.W., Kemp P., Smith C.W., Smith C.W. A novel polypyrimidine tract-binding protein paralog expressed in smooth muscle cells. J. Biol. Chem. 2003;278:15201–15207. doi: 10.1074/jbc.M210131200. [DOI] [PubMed] [Google Scholar]
- Griffiths-Jones S. The microRNA Registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall T. 1998. [Google Scholar]
- Hamshere M., Dickson G., Eperon I., Dickson G., Eperon I., Eperon I. The muscle specific domain of mouse N-CAM: Structure and alternative splicing patterns. Nucleic Acids Res. 1991;19:4709–4716. doi: 10.1093/nar/19.17.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfe B.D., McManus M.T., Mansfield J.H., Hornstein E., Tabin C.J., McManus M.T., Mansfield J.H., Hornstein E., Tabin C.J., Mansfield J.H., Hornstein E., Tabin C.J., Hornstein E., Tabin C.J., Tabin C.J. The RNaseIII enzyme Dicer is required for morphogenesis but not patterning of the vertebrate limb. Proc. Natl. Acad. Sci. 2005;102:10898–10903. doi: 10.1073/pnas.0504834102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow E., Lane D., Lane D. Using antibodies: A laboratory manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1999. [Google Scholar]
- Herasse M., Ono Y., Fougerousse F., Kimura E., Stockholm D., Beley C., Montarras D., Pinset C., Sorimachi H., Suzuki K., Ono Y., Fougerousse F., Kimura E., Stockholm D., Beley C., Montarras D., Pinset C., Sorimachi H., Suzuki K., Fougerousse F., Kimura E., Stockholm D., Beley C., Montarras D., Pinset C., Sorimachi H., Suzuki K., Kimura E., Stockholm D., Beley C., Montarras D., Pinset C., Sorimachi H., Suzuki K., Stockholm D., Beley C., Montarras D., Pinset C., Sorimachi H., Suzuki K., Beley C., Montarras D., Pinset C., Sorimachi H., Suzuki K., Montarras D., Pinset C., Sorimachi H., Suzuki K., Pinset C., Sorimachi H., Suzuki K., Sorimachi H., Suzuki K., Suzuki K., et al. Expression and functional characteristics of calpain 3 isoforms generated through tissue-specific transcriptional and posttranscriptional events. Mol. Cell. Biol. 1999;19:4047–4055. doi: 10.1128/mcb.19.6.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys D.T., Westman B.J., Martin D.I., Preiss T., Westman B.J., Martin D.I., Preiss T., Martin D.I., Preiss T., Preiss T. MicroRNAs control translation initiation by inhibiting eukaryotic initiation factor 4E/cap and poly(A) tail function. Proc. Natl. Acad. Sci. 2005;102:16961–16966. doi: 10.1073/pnas.0506482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin N., Baekelandt V., Goritchenko L., Benowitz L.I., Baekelandt V., Goritchenko L., Benowitz L.I., Goritchenko L., Benowitz L.I., Benowitz L.I. Identification of two proteins that bind to a pyrimidine-rich sequence in the 3′-untranslated region of GAP-43 mRNA. Nucleic Acids Res. 1997;25:1281–1288. doi: 10.1093/nar/25.6.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S., Aravin A., Tuschl T., Sander C., Marks D.S., Tuschl T., Sander C., Marks D.S., Sander C., Marks D.S., Marks D.S. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J., Sugnet C.W., Haussler D., Kent W.J., Haussler D., Kent W.J., Kent W.J. The UCSC table browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahigashi H., Harada Y., Asano A., Nakamura M., Harada Y., Asano A., Nakamura M., Asano A., Nakamura M., Nakamura M. A cis-acting regulatory element that affects the alternative splicing of a muscle-specific exon in the mouse NCAM gene. Biochim. Biophys. Acta. 1998;1397:305–315. doi: 10.1016/s0167-4781(98)00021-9. [DOI] [PubMed] [Google Scholar]
- Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D., Pringle T.H., Zahler A.M., Haussler D., Zahler A.M., Haussler D., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khvorova A., Reynolds A., Jayasena S.D., Reynolds A., Jayasena S.D., Jayasena S.D. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209–216. doi: 10.1016/s0092-8674(03)00801-8. [DOI] [PubMed] [Google Scholar]
- Kikuchi T., Ichikawa M., Arai J., Tateiwa H., Fu L., Higuchi K., Yoshimura N., Ichikawa M., Arai J., Tateiwa H., Fu L., Higuchi K., Yoshimura N., Arai J., Tateiwa H., Fu L., Higuchi K., Yoshimura N., Tateiwa H., Fu L., Higuchi K., Yoshimura N., Fu L., Higuchi K., Yoshimura N., Higuchi K., Yoshimura N., Yoshimura N. Molecular cloning and characterization of a new neuron-specific homologue of rat polypyrimidine tract binding protein. J. Biochem. 2000;128:811–821. doi: 10.1093/oxfordjournals.jbchem.a022819. [DOI] [PubMed] [Google Scholar]
- Kim H.K., Lee Y.S., Sivaprasad U., Malhotra A., Dutta A., Lee Y.S., Sivaprasad U., Malhotra A., Dutta A., Sivaprasad U., Malhotra A., Dutta A., Malhotra A., Dutta A., Dutta A. Muscle-specific microRNA miR-206 promotes muscle differentiation. J. Cell Biol. 2006;174:677–687. doi: 10.1083/jcb.200603008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiriakidou M., Nelson P.T., Kouranov A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A., Nelson P.T., Kouranov A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A., Kouranov A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A., Fitziev P., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A., Bouyioukos C., Mourelatos Z., Hatzigeorgiou A., Mourelatos Z., Hatzigeorgiou A., Hatzigeorgiou A. A combined computational-experimental approach predicts human microRNA targets. Genes & Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov S.A., Peng X., Baker E., Schmoyer D., Zhang B., Snoddy J., Peng X., Baker E., Schmoyer D., Zhang B., Snoddy J., Baker E., Schmoyer D., Zhang B., Snoddy J., Schmoyer D., Zhang B., Snoddy J., Zhang B., Snoddy J., Snoddy J. GeneKeyDB: A lightweight, gene-centric, relational database to support data mining environments. BMC Bioinformatics. 2005;6:72. doi: 10.1186/1471-2105-6-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krek A., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Grun D., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Poy M.N., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Wolf R., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Rosenberg L., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., Epstein E.J., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., MacMenamin P., da Piedade I., Gunsalus K.C., Stoffel M., da Piedade I., Gunsalus K.C., Stoffel M., Gunsalus K.C., Stoffel M., Stoffel M., et al. Combinatorial microRNA target predictions. Nat. Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- Kwon C., Han Z., Olson E.N., Srivastava D., Han Z., Olson E.N., Srivastava D., Olson E.N., Srivastava D., Srivastava D. MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc. Natl. Acad. Sci. 2005;102:18986–18991. doi: 10.1073/pnas.0509535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd A.N., Charlet N., Cooper T.A., Charlet N., Cooper T.A., Cooper T.A. The CELF family of RNA binding proteins is implicated in cell-specific and developmentally regulated alternative splicing. Mol. Cell. Biol. 2001;21:1285–1296. doi: 10.1128/MCB.21.4.1285-1296.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd A.N., Taffet G., Hartley C., Kearney D.L., Cooper T.A., Taffet G., Hartley C., Kearney D.L., Cooper T.A., Hartley C., Kearney D.L., Cooper T.A., Kearney D.L., Cooper T.A., Cooper T.A. Cardiac tissue-specific repression of CELF activity disrupts alternative splicing and causes cardiomyopathy. Mol. Cell. Biol. 2005;25:6267–6278. doi: 10.1128/MCB.25.14.6267-6278.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Lendeckel W., Tuschl T., Rauhut R., Lendeckel W., Tuschl T., Lendeckel W., Tuschl T., Tuschl T. Identification of novel genes coding for small expressed RNAs. Science. 2001;294:853–858. doi: 10.1126/science.1064921. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Rauhut R., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Yalcin A., Meyer J., Lendeckel W., Tuschl T., Meyer J., Lendeckel W., Tuschl T., Lendeckel W., Tuschl T., Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr. Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M., Rauhut R., Meyer J., Borkhardt A., Tuschl T., Rauhut R., Meyer J., Borkhardt A., Tuschl T., Meyer J., Borkhardt A., Tuschl T., Borkhardt A., Tuschl T., Tuschl T. New microRNAs from mouse and human. RNA. 2003;9:175–179. doi: 10.1261/rna.2146903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai E.C. Predicting and validating microRNA targets. Genome Biol. 2004;5:115. doi: 10.1186/gb-2004-5-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau N.C., Lim L.P., Weinstein E.G., Bartel D.P., Lim L.P., Weinstein E.G., Bartel D.P., Weinstein E.G., Bartel D.P., Bartel D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Ambros V., Ambros V. An extensive class of small RNAs in Caenorhabditis elegans. Science. 2001;294:862–864. doi: 10.1126/science.1065329. [DOI] [PubMed] [Google Scholar]
- Lee R.C., Feinbaum R.L., Ambros V., Feinbaum R.L., Ambros V., Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Shih I.H., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Jones-Rhoades M.W., Bartel D.P., Burge C.B., Bartel D.P., Burge C.B., Burge C.B. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., Bartel D.P., Burge C.B., Bartel D.P., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lillevali K., Kulla A., Ord T., Kulla A., Ord T., Ord T. Comparative expression analysis of the genes encoding polypyrimidine tract binding protein (PTB) and its neural homologue (brPTB) in prenatal and postnatal mouse brain. Mech. Dev. 2001;101:217–220. doi: 10.1016/s0925-4773(00)00566-9. [DOI] [PubMed] [Google Scholar]
- Lim L.P., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Lau N.C., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Garrett-Engele P., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Grimson A., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Schelter J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Castle J., Bartel D.P., Linsley P.S., Johnson J.M., Bartel D.P., Linsley P.S., Johnson J.M., Linsley P.S., Johnson J.M., Johnson J.M. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Markovtsov V., Nikolic J.M., Goldman J.A., Turck C.W., Chou M.Y., Black D.L., Nikolic J.M., Goldman J.A., Turck C.W., Chou M.Y., Black D.L., Goldman J.A., Turck C.W., Chou M.Y., Black D.L., Turck C.W., Chou M.Y., Black D.L., Chou M.Y., Black D.L., Black D.L. Cooperative assembly of an hnRNP complex induced by a tissue-specific homolog of polypyrimidine tract binding protein. Mol. Cell. Biol. 2000;20:7463–7479. doi: 10.1128/mcb.20.20.7463-7479.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews D.H., Sabina J., Zuker M., Turner D.H., Sabina J., Zuker M., Turner D.H., Zuker M., Turner D.H., Turner D.H. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- Morano I. Tuning smooth muscle contraction by molecular motors. J. Mol. Med. 2003;81:481–487. doi: 10.1007/s00109-003-0451-x. [DOI] [PubMed] [Google Scholar]
- Mourelatos Z., Dostie J., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G., Dostie J., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G., Abel L., Rappsilber J., Mann M., Dreyfuss G., Rappsilber J., Mann M., Dreyfuss G., Mann M., Dreyfuss G., Dreyfuss G. miRNPs: A novel class of ribonucleoproteins containing numerous microRNAs. Genes & Dev. 2002;16:720–728. doi: 10.1101/gad.974702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan G.J., Guo W., Wormsley S., Helfman D.M., Guo W., Wormsley S., Helfman D.M., Wormsley S., Helfman D.M., Helfman D.M. Polypyrimidine tract binding protein interacts with sequences involved in alternative splicing of β-tropomyosin pre-mRNA. J. Biol. Chem. 1992;267:25480–25487. [PubMed] [Google Scholar]
- O’Donnell K.A., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T., Wentzel E.A., Zeller K.I., Dang C.V., Mendell J.T., Zeller K.I., Dang C.V., Mendell J.T., Dang C.V., Mendell J.T., Mendell J.T. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Olsen P.H., Ambros V., Ambros V. The lin-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev. Biol. 1999;216:671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- Orom U.A., Kauppinen S., Lund A.H., Kauppinen S., Lund A.H., Lund A.H. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene. 2006;372:137–141. doi: 10.1016/j.gene.2005.12.031. [DOI] [PubMed] [Google Scholar]
- Parker S.R. AutoAssembler sequence assembly software. Methods Mol. Biol. 1997;70:107–117. doi: 10.1385/0-89603-358-9:107. [DOI] [PubMed] [Google Scholar]
- Patton J.G., Mayer S.A., Tempst P., Nadal-Ginard B., Mayer S.A., Tempst P., Nadal-Ginard B., Tempst P., Nadal-Ginard B., Nadal-Ginard B. Characterization and molecular cloning of polypyrimidine tract-binding protein: A component of a complex necessary for pre-mRNA splicing. Genes & Dev. 1991;5:1237–1251. doi: 10.1101/gad.5.7.1237. [DOI] [PubMed] [Google Scholar]
- Petersen C.P., Bordeleau M.E., Pelletier J., Sharp P.A., Bordeleau M.E., Pelletier J., Sharp P.A., Pelletier J., Sharp P.A., Sharp P.A. Short RNAs repress translation after initiation in mammalian cells. Mol. Cell. 2006;21:533–542. doi: 10.1016/j.molcel.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Zoller T., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Cougot N., Basyuk E., Bertrand E., Filipowicz W., Basyuk E., Bertrand E., Filipowicz W., Bertrand E., Filipowicz W., Filipowicz W. Inhibition of translational initiation by Let-7 microRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- Polydorides A.D., Okano H.J., Yang Y.Y., Stefani G., Darnell R.B., Okano H.J., Yang Y.Y., Stefani G., Darnell R.B., Yang Y.Y., Stefani G., Darnell R.B., Stefani G., Darnell R.B., Darnell R.B. A brain-enriched polypyrimidine tract-binding protein antagonizes the ability of Nova to regulate neuron-specific alternative splicing. Proc. Natl. Acad. Sci. 2000;97:6350–6355. doi: 10.1073/pnas.110128397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Tuschl T., Rajewsky N., Rorsman P., Rajewsky N., Rorsman P., Rorsman P., et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Rehmsmeier M., Steffen P., Höchsmann M., Giegerich R., Steffen P., Höchsmann M., Giegerich R., Höchsmann M., Giegerich R., Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart B.J., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Slack F.J., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Basson M., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Pasquinelli A.E., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Bettinger J.C., Rougvie A.E., Horvitz H.R., Ruvkun G., Rougvie A.E., Horvitz H.R., Ruvkun G., Horvitz H.R., Ruvkun G., Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- Schwarz D.S., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D., Hutvagner G., Du T., Xu Z., Aronin N., Zamore P.D., Du T., Xu Z., Aronin N., Zamore P.D., Xu Z., Aronin N., Zamore P.D., Aronin N., Zamore P.D., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Sempere L.F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V., Moss E., Dmitrovsky E., Ambros V., Dmitrovsky E., Ambros V., Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S., Falick A.M., Black D.L., Falick A.M., Black D.L., Black D.L. Polypyrimidine tract binding protein blocks the 5′ splice site-dependent assembly of U2AF and the prespliceosomal E complex. Mol. Cell. 2005;19:485–496. doi: 10.1016/j.molcel.2005.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokol N.S., Ambros V., Ambros V. Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes & Dev. 2005;19:2343–2354. doi: 10.1101/gad.1356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southby J., Gooding C., Smith C.W., Gooding C., Smith C.W., Smith C.W. Polypyrimidine tract binding protein functions as a repressor to regulate alternative splicing of α-actinin mutally exclusive exons. Mol. Cell. Biol. 1999;19:2699–2711. doi: 10.1128/mcb.19.4.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman R., Smith C.W., Smith C.W. Novel modes of splicing repression by PTB. Trends Biochem. Sci. 2006;31:73–76. doi: 10.1016/j.tibs.2005.12.003. [DOI] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Russell R.B., Cohen S.M., Brennecke J., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Identification of Drosophila microRNA targets. PLoS Biol. 2003;1:E60. doi: 10.1371/journal.pbio.0000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark A., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Brennecke J., Bushati N., Russell R.B., Cohen S.M., Bushati N., Russell R.B., Cohen S.M., Russell R.B., Cohen S.M., Cohen S.M. Animal microRNAs confer robustness to gene expression and have a significant impact on 3′UTR evolution. Cell. 2005;123:1133–1146. doi: 10.1016/j.cell.2005.11.023. [DOI] [PubMed] [Google Scholar]
- Thomson J.M., Parker J., Perou C.M., Hammond S.M., Parker J., Perou C.M., Hammond S.M., Perou C.M., Hammond S.M., Hammond S.M. A custom microarray platform for analysis of microRNA gene expression. Nat. Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- Tomczak K.K., Marinescu V.D., Ramoni M.F., Sanoudou D., Montanaro F., Han M., Kunkel L.M., Kohane I.S., Beggs A.H., Marinescu V.D., Ramoni M.F., Sanoudou D., Montanaro F., Han M., Kunkel L.M., Kohane I.S., Beggs A.H., Ramoni M.F., Sanoudou D., Montanaro F., Han M., Kunkel L.M., Kohane I.S., Beggs A.H., Sanoudou D., Montanaro F., Han M., Kunkel L.M., Kohane I.S., Beggs A.H., Montanaro F., Han M., Kunkel L.M., Kohane I.S., Beggs A.H., Han M., Kunkel L.M., Kohane I.S., Beggs A.H., Kunkel L.M., Kohane I.S., Beggs A.H., Kohane I.S., Beggs A.H., Beggs A.H. Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J. 2004;18:403–405. doi: 10.1096/fj.03-0568fje. [DOI] [PubMed] [Google Scholar]
- Underwood J.G., Boutz P.L., Dougherty J.D., Stoilov P., Black D.L., Boutz P.L., Dougherty J.D., Stoilov P., Black D.L., Dougherty J.D., Stoilov P., Black D.L., Stoilov P., Black D.L., Black D.L. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol. Cell. Biol. 2005;25:10005–10016. doi: 10.1128/MCB.25.22.10005-10016.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ver Heyen M., Heymans S., Antoons G., Reed T., Periasamy M., Awede B., Lebacq J., Vangheluwe P., Dewerchin M., Collen D., Heymans S., Antoons G., Reed T., Periasamy M., Awede B., Lebacq J., Vangheluwe P., Dewerchin M., Collen D., Antoons G., Reed T., Periasamy M., Awede B., Lebacq J., Vangheluwe P., Dewerchin M., Collen D., Reed T., Periasamy M., Awede B., Lebacq J., Vangheluwe P., Dewerchin M., Collen D., Periasamy M., Awede B., Lebacq J., Vangheluwe P., Dewerchin M., Collen D., Awede B., Lebacq J., Vangheluwe P., Dewerchin M., Collen D., Lebacq J., Vangheluwe P., Dewerchin M., Collen D., Vangheluwe P., Dewerchin M., Collen D., Dewerchin M., Collen D., Collen D., et al. Replacement of the muscle-specific sarcoplasmic reticulum Ca(2+)-ATPase isoform SERCA2a by the nonmuscle SERCA2b homologue causes mild concentric hypertrophy and impairs contraction–relaxation of the heart. Circ. Res. 2001;89:838–846. doi: 10.1161/hh2101.098466. [DOI] [PubMed] [Google Scholar]
- Wagner E.J., Garcia-Blanco M.A., Garcia-Blanco M.A. Polypyrimidine tract binding protein antagonizes exon definition. Mol. Cell. Biol. 2001;21:3281–3288. doi: 10.1128/MCB.21.10.3281-3288.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E., Koudijs M.J., van Eeden F.J., Cuppen E., Plasterk R.H., Koudijs M.J., van Eeden F.J., Cuppen E., Plasterk R.H., van Eeden F.J., Cuppen E., Plasterk R.H., Cuppen E., Plasterk R.H., Plasterk R.H. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat. Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H., Kloosterman W.P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H., Berezikov E., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H., de Bruijn E., Horvitz H.R., Kauppinen S., Plasterk R.H., Horvitz H.R., Kauppinen S., Plasterk R.H., Kauppinen S., Plasterk R.H., Plasterk R.H. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
- Wollerton M.C., Gooding C., Robinson F., Brown E.C., Jackson R.J., Smith C.W., Gooding C., Robinson F., Brown E.C., Jackson R.J., Smith C.W., Robinson F., Brown E.C., Jackson R.J., Smith C.W., Brown E.C., Jackson R.J., Smith C.W., Jackson R.J., Smith C.W., Smith C.W. Differential alternative splicing activity of isoforms of polypyrimidine tract binding protein (PTB) RNA. 2001;7:819–832. doi: 10.1017/s1355838201010214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lu J., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Kulbokas E.J., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Golub T.R., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Mootha V., Lindblad-Toh K., Lander E.S., Kellis M., Lindblad-Toh K., Lander E.S., Kellis M., Lander E.S., Kellis M., Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing Y., Lee C.J., Lee C.J. Protein modularity of alternatively spliced exons is associated with tissue-specific regulation of alternative splicing. PLoS Genet. 2005;1:e34. doi: 10.1371/journal.pgen.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Tsukahara K., Kanaoka Y., Jinno S., Okayama H., Tsukahara K., Kanaoka Y., Jinno S., Okayama H., Kanaoka Y., Jinno S., Okayama H., Jinno S., Okayama H., Okayama H. Isolation of a mammalian homologue of a fission yeast differentiation regulator. Mol. Cell. Biol. 1999;19:3829–3841. doi: 10.1128/mcb.19.5.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yekta S., Shih I.H., Bartel D.P., Shih I.H., Bartel D.P., Bartel D.P. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594–596. doi: 10.1126/science.1097434. [DOI] [PubMed] [Google Scholar]
- Zhang L., Liu W., Grabowski P.J., Liu W., Grabowski P.J., Grabowski P.J. Coordinate repression of a trio of neuron-specific splicing events by the splicing regulator PTB. RNA. 1999;5:117–130. doi: 10.1017/s1355838299981530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Samal E., Srivastava D., Samal E., Srivastava D., Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- Ziober B.L., Vu M.P., Waleh N., Crawford J., Lin C.S., Kramer R.H., Vu M.P., Waleh N., Crawford J., Lin C.S., Kramer R.H., Waleh N., Crawford J., Lin C.S., Kramer R.H., Crawford J., Lin C.S., Kramer R.H., Lin C.S., Kramer R.H., Kramer R.H. Alternative extracellular and cytoplasmic domains of the integrin α 7 subunit are differentially expressed during development. J. Biol. Chem. 1993;268:26773–26783. [PubMed] [Google Scholar]
- Zuker M. Mfold Web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]