Abstract
The Engrailed Homeodomain protein has the highest refolding and unfolding rate constants directly observed to date. Temperature jump relaxation measurements gave a refolding rate constant of 37,500 s−1 in water at 25°C, rising to 51,000 s−1 around 42°C. The unfolding rate constant was 1,100 s−1 in water at 25°C and 205,000 s−1 at 63°C. The unfolding half-life is extrapolated to be ≈7.5 ns at 100°C, which allows real-time molecular dynamics unfolding simulations to be tested on this system at a realistic temperature. Preliminary simulations did indeed conform to unfolding on this time scale. Further, similar transition states were observed in simulations at 100°C and 225°C, suggesting that high-temperature simulations provide results applicable to lower temperatures.
Keywords: molecular dynamics, fast unfolding, helix docking, residual structure
Our understanding of protein folding has increased enormously in the past decade because of developments in experimental and theoretical methods and their integration (1, 2). The observed time scale for two-state protein folding and unfolding has decreased from tens of milliseconds to tens of microseconds over the same period (3–6). Unfortunately, the most detailed and realistic simulation procedure, molecular dynamics (MD), is currently on a time scale that is generally too long for direct comparison with experiment under accessible conditions. The longest simulation performed so far is for about 1 μs on the folding of a 36-residue protein, the villin headpiece, during which time the protein collapses to an intermediate (7). Raising the temperature of simulation of folding does not circumvent the problem of incompatible time scales: the equilibrium for most proteins favors unfolding at temperatures above 40–70°C, and the folding rate constant has a maximum at the lower end of that temperature range and then decreases because of a large specific heat of activation (8). But, simulation of protein unfolding is particularly useful because unfolding does speed up with increasing temperature, and simulation at some 200°C detects complete unfolding in the nanosecond range (9) (S. L. Kazmirski, K. B. Wong, S. M. V. Freund, Y.-J. Tan, A.R.F., and V.D., unpublished observations). Although simulation of unfolding at high temperature is subject to the criticism that the conditions are unnatural, simulations do benchmark very well with detailed experimental data (for example, φ-values and NMR data) obtained at 25°C over the millisecond time scale (9–11). Also, recent studies of the detailed unfolding pathway of chymotrypsin inhibitor 2 as a function of temperature (75–225°C) demonstrate that unfolding is accelerated at high temperature, but the pathway does not change as a function of temperature (D. O. V. Alonso, Y. P. Pan, S. Ham, B. Benniun, and V.D., unpublished observations).
Simulations have reproduced particular events in protein folding. Examples are the hydrophobic collapse of ubiquitin at 62°C (12, 13) and the formation of a collapsed state on the microsecond time scale for the villin headpiece (7), but true refolding did not occur in either case. We have been searching for proteins that fold and unfold very rapidly, both to explore the “speed limit” of protein folding (14–16) and to find systems that are accessible to MD simulation at the upper temperatures of life, i.e., around 100°C. We have found in this study a suitable candidate, the Engrailed Homeodomain (En-HD) from Drosophila melanogaster. This protein has 61 residues and is mainly α-helical (17). The structure of the homeodomains, including En-HD, is an extension of the prokaryotic helix–turn–helix DNA-binding motif (18). We want to report measurements on this rapidly folding and unfolding protein, together with preliminary MD simulation of its unfolding.
Materials and Methods
Purification.
En-HD was overexpressed in C41(DE3) Escherichia coli cells (19) at 37°C and purified (20). Mass spectrometry revealed >99% homogeneity of the product with the expected Mr 7, 453.3 ± 0.5.
Equilibrium Measurements.
Experiments used 50 mM Hepes, pH 8.0, and 100 mM NaCl, except for differential scanning calorimetry, which used 50 mM sodium acetate buffer, pH 5.8. Thermal denaturation of 18.6 μM En-HD was followed at 222 nm with a Jasco (Easton, MD) J-720 spectropolarimeter equipped with a Jasco PTC-348WI temperature controller. Thermal denaturation of 360 μM En-HD also was followed by differential scanning calorimetry using a MicroCal (Northampton, MA) VP-DSC with a cell volume of 0.52 ml. In both experiments, the temperature was increased at 1° per min. Curves were fitted to the standard two-state equation. The partial heat capacity of En-HD (Fig. 1) was calculated according to Privalov (21). ΔHcal was determined by integration of the excess heat capacity in Fig. 1.
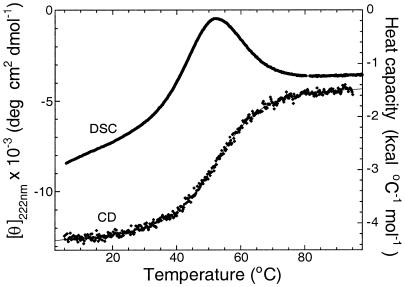
Figure 1.
Equilibrium denaturation of En-HD. Reversible thermal denaturation of En-HD as followed by circular dichroism (CD) and differential scanning calorimetry (DSC).
Kinetic Measurements.
Temperature jump measurements in a 0.8-ml cell were made using a DIA-RT T-Jump instrument (22) (Dia-Log, Düsseldorf, Germany). Protein concentrations were in the range of 15 to 300 μM in the Hepes buffer. Voltages ranging from 10 to 40 kV with 10 or 50 nF capacitors gave increases of 1–5°, as calibrated by the changes in absorbance of a pH indicator in a buffer of known pH dependence on temperature. The heating under optimal conditions was completed in 0.5 μs.
MD Unfolding Simulations.
Duplicate simulations were performed at both 100 and 225°C with encad (23) by using standard procedures (9). All atoms were explicitly present, and the protein was solvated in a box of water molecules (24, 25), by using the experimental densities for pure liquid water at these temperatures (26, 27). The transition state region for each simulation was identified by using a conformational clustering procedure (10, 11). The two primary simulations were carried out for 40 (225°C) and 70 ns (100°C).
Results
Equilibrium Measurements.
Thermal denaturation curves of En-HD followed by circular dichroism were sigmoidal, and the derived melting temperatures were coincident with those from differential scanning calorimetry data (Fig. 1), indicative of a two-state transition. Further, the calorimetric and van't Hoff denaturation enthalpies were very similar, 35.1 ± 0.4 kcal⋅mol−1 and 33 ± 3 kcal⋅mol−1, respectively, which is a necessary characteristic of a two-state transition. The thermodynamic values obtained from equilibrium measurements were independent of protein concentration, from 4 to 400 μM. Urea denaturation at 25°C gave a free energy of denaturation, ΔGD-N, of 1.85 ± 0.1 kcal⋅mol−1 in water and an m value (mD-N = ∂ΔGD-N/∂[urea]) of 0.8 ± 0.05 kcal⋅mol−1⋅M−1.
Calculation of the free energy of denaturation at different temperatures from the enthalpy and entropy of denaturation that were measured from the thermal denaturation experiments requires knowing the specific heat of denaturation (ΔCpD-N). Because changes of stability with pH in this system are small (at pH 5.8, ΔGD-N = 1.85 ± 0.1 kcal⋅mol−1; at pH 8.0, = 1.98 ± 0.1 kcal⋅mol−1, as measured from urea denaturation curves), we could not determine the specific heat by the standard procedure of measuring ΔHD-N at different values of Tm at varying values of pH. Instead, we used the average value of ΔCpD-N per residue from a set of small nondisulfide-bridged proteins for which ΔCpD-N has been calculated over a wide range of temperatures (28). The values of ΔCpD-N for En-HD is estimated to be 700 ± 200 cal·mol−1⋅K−1 through its transition region (25–75°C), in agreement with value estimated by the procedures of Myers et al. (29). This value decreases with increasing temperature, approaching 0 at around 140°C (28). Errors induced in the calculation of ΔGD-N from errors in ΔCpD-N are, in any case, greatly attenuated because they lead to compensated errors in the extrapolated components of enthalpy and entropy (30). The free energy of denaturation at pH 5.8 extrapolated to 25°C by using the estimated value of ΔCpD-N agrees well (2.08 ± 0.24 kcal⋅mol−1) with the value calculated from urea-induced denaturation.
Kinetic Measurements.
The low values of both m and ΔGD-N led to both denatured and native states of En-HD being populated over a wide range of urea concentrations and temperatures. Those suitable characteristics of the protein have enabled us to measure directly the rate constants for unfolding and refolding of En-HD by temperature-jump experiments on the microsecond time scale. On rapidly increasing the temperature (dead time = 1–5 μs, according to experimental conditions), there was a measurable change in the equilibrium population of the states, with a relaxation rate constant kobs = ku + kf, where kf = kobs/(1 + KD-N) and ku = (KD-N kobs)/(1 + KD-N). The observed rate constants were independent of protein concentration from 10 to 300 μM and of the size of the temperature jump to the target temperature. All of the recorded traces were strictly exponential (see Fig. 2 A and B), as expected for a cooperative mechanism and generally observed for slower folding proteins.
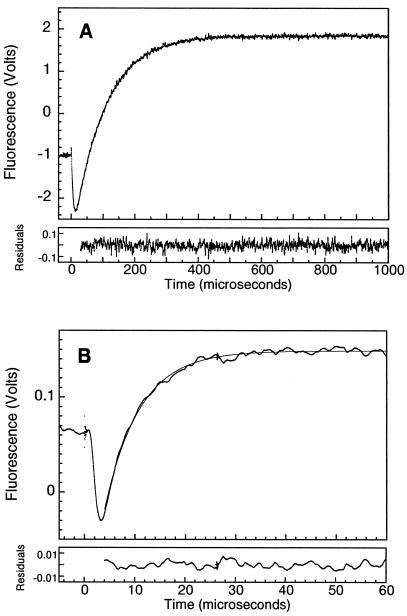
Figure 2.
Relaxation kinetics of En-HD. (A) Kinetic trace at 25°C and 2.6 M urea on a 24,000-V electrical discharge with a 50-nF capacitor (5° jump). Six traces were averaged (En-HD concentration was 49 μM). Fitting of the observed trace to a single exponential equation gives a relaxation rate of 10,300 ± 200 s−1. (B) Reaction trace at 57°C and 0 M urea on a 30,000-V electrical discharge from a 10-nF capacitor (1° jump). Seventeen traces were averaged (En-HD concentration was 300 μM). Fitting of the observed trace to a single exponential equation gives a relaxation rate of 145,000 ± 500 s−1.
Plots of of ln(kobs/T) vs. 1/T (the Eyring equation; ref. 31) were not linear for either folding and unfolding, as expected for a reaction with a change of heat capacity between the ground and transition states (8). The direct plot of logkobs vs. T (Fig. 3A) shows very high curvature and the now characteristic observation of the folding rate constant for a two-state reaction going through a maximum (8).
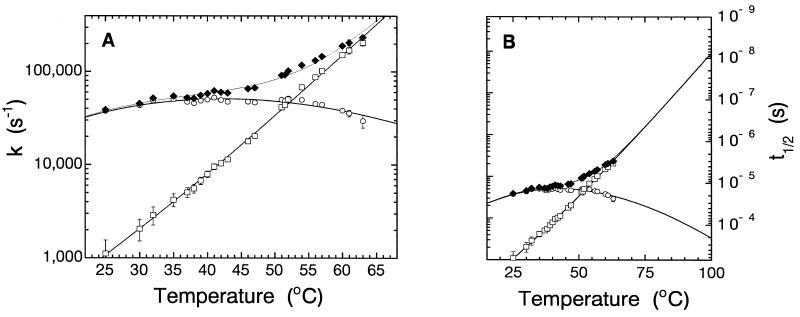
Figure 3.
Folding kinetics at increasing temperatures. (A) Relaxation rate constants (♦) are plotted in a logarithmic axis vs. temperature. Folding (kf, ○) and unfolding (ku, □) rate constants were extracted from kobs as described in the text, plotted in an Eyring plot of ln(kobs/T) vs. 1/T (K−1) (not shown) and fitted individually to ln (k/T) = ln (kB/h) − ΔG‡/RT to estimate the half-times of reaction up to 100°C shown in B.
The errors in the measured value of kobs were small (<3%). The errors in the values of ku and kf increase with increasing temperature difference from Tm because of errors in estimating the equilibrium constant for denaturation (see error bars in Fig. 3). The relaxation rate constant at 25°C in the absence of denaturant was 38,700 ± 1,400 s−1, where kf = 37,500 ± 1,600 s−1 and ku = 1,100 ± 450 s−1. At the thermal denaturation midpoint (52.15°C), kobs = 96,000 ± 4,000 s−1, where ku = kf = 48,000 ± 2,000 s−1. The highest value of kobs we have measured accurately for En-HD is 235,000 ± 10,000 s−1, at 63°C. The temperature of 42.3°C for the maximum refolding rate constant in Fig. 3 (kf = 51,000 ± 1,500 s−1) corresponds to the temperature where the folding enthalpy equals zero.
The position of the transition state on the reaction pathway (32), βT, may be estimated from the values of ∂(logku)/∂[urea] and ∂(logKeq)/∂[urea]. Rates up to 7.3 M urea measured either by relaxation techniques or by continuous flow mixing (unpublished observations) have been used to estimate the urea dependence of the unfolding reaction and correlate it with the urea dependence of the equilibrium constant. The measured values yield a β84 for folding of 0.9 (data not shown).
Unfolding MD Simulations.
There is some uncertainty in the extrapolated rate constant of unfolding at 100°C because of the need to estimate the specific heat of unfolding. The use of our estimate of ΔCpD-N = 700 ± 200 cal·mol−1⋅K−1 to extract the individual rate constants gave an unfolding half-life of 7.5 ns at 100°C (Fig. 3B). Underestimates or overestimates of ΔCpD-N do not change significantly the extrapolated value of unfolding half-life at 100°C; values span from 25 ns to 4.5 ns, using 460 and 930 cal·mol−1⋅K−1, respectively. A temperature-dependent ΔCpD-N—according to Privalov's calculation (28)—yields rates that also fall in this range.
The unfolding reaction of En-HD of half-life about 10 ns at 100°C is within present capabilities of MD simulation. Consequently, multiple thermal MD simulations were performed, starting from the crystal structure. The initial events in unfolding involved expansion and disruptions of helix packing. In particular, helix III moved away from the core, leading to the transition state. In addition, helix II partially unraveled and/or separated from helix I, and the extended N-terminal tail pulled away from the protein. This sequence of events was independent of temperature in multiple unfolding simulations (shown for 100 and 225°C in Fig. 4).
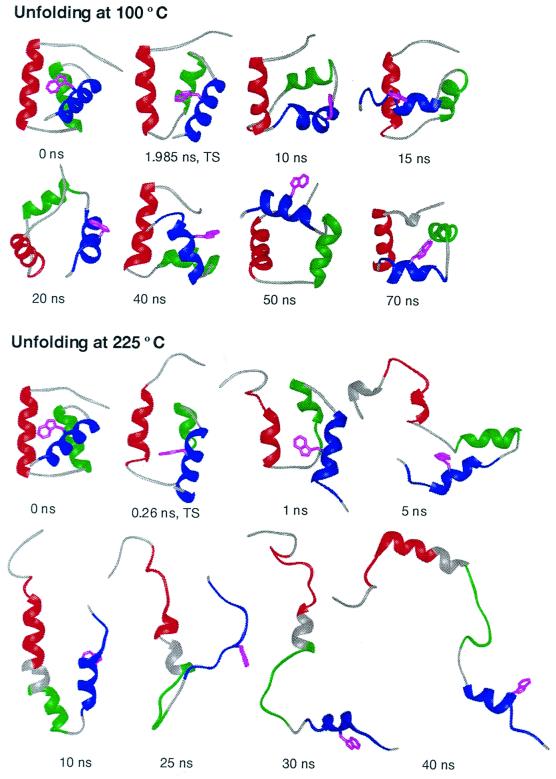
Figure 4.
Time progression of the thermal denaturation of En-HD at neutral pH from MD simulations at 100 and 225°C. The early steps in unfolding to the transition state and the disruption of helix packing are illustrated at 100°C. The unfolding at 100°C is on the proper time scale, as estimated from experiment. However, as the process is accelerated by temperature, the conformational heterogeneity of the denatured state is better sampled at 225°C. The crystal structure (17) is given as the 0-ns conformation. All structures are colored according to the placement of native helical structure: helix I, residues 10–22, in red; helix II, residues 28–38, in green; and helix III, residues 42–55, in blue. Helical structure, as determined using the method of Kabsch and Sander (43), is illustrated by ribbons. Trp-48, the fluorescence probe, is shown in magenta. This residue becomes exposed to solvent at the transition state and remains exposed thereafter at both temperatures. Note the similarity between the transition states at different temperatures: the displayed transition state structures have an α-carbon rms deviation of 3.8 Å, which is lower than their rms deviations from the starting structure (3.9 and 5.3 Å for 100 and 225°C, respectively).
The simulated transition state ensemble was native like and very similar for the two different temperatures (Fig. 4). In fact, the transition states are more similar to one another than to their starting structure (Fig. 4 legend). There was a 14 and 20% increase in the solvent-accessible surface area of the 100 and 225°C transition states, respectively, compared with the native state. The difference between these values is mostly because of the N-terminal tail. The value of βT is 0.9, implying that the transition state is native like with a 10% increase in solvent exposure, consistent with the simulations.
Further expansion occurred after the transition state in the simulations, resulting in almost complete separation of the helices. At this point, the interhelical orientations were very fluid, and packing interactions were disrupted and those that formed were transient (see the 100°C structures in Fig. 4). After further expansion, the helices underwent cycles of unfolding and refolding, which is better seen in the 225°C simulation, where sampling of the denatured state was improved (Fig. 4).
Discussion
Temperature Jump Kinetics.
We previously applied temperature jump kinetics to protein folding by rapidly heating a cold-denatured solution of a protein and so measuring the increase in population of native protein (33). Here, we have used the more conventional procedure of jumping through a thermal unfolding transition and measuring the rate of relaxation to a new equilibrium in which the unfolded state is more populated. The relaxation rate constant thus obtained is the sum of those for the forward and reverse reactions. We calculated the individual rate constants from the relaxation rate constant and the measured equilibrium constant under the same conditions. It is worth noting that although the classical Eigen–de Maeyer temperature jump apparatus (22) is slow compared with modern laser-based machines, being limited to the microsecond time range, it has exceptionally good signal-to-noise ratio, is very stable, and has exceptionally reproducible traces. Further, unlike more modern equipment, the whole cell is rapidly heated and so the signal does not have to be processed to allow for thermal diffusion as happens when just a small part of the sample is rapidly heated by a focused laser flash. As a consequence, we can analyze the nature of the relaxation traces with high precision. The relaxation processes for folding and unfolding fit extremely well to simple, single, first-order curves, without any detectable hints of stretching, inhomogeneity, or multiple processes (Fig. 2). This behavior ranges from >200,000 s−1 at high temperature, which represents predominantly the rate constant for unfolding, to 38,700 s−1 at 25°C, which is predominantly the folding rate constant. The refolding rate constant is 37,500 s−1 in water at 25°C, with a maximum of 51,000 s−1 around 42°C. The unfolding rate constant was 1,100 s−1 in water at 25°C and 205,000 s−1 at 63°C. These are the highest rate constants ever directly measured for protein folding and unfolding. The rate constant for folding is approaching within an order of magnitude the value predicted by Eaton and colleagues for the diffusional collapse of a polypeptide chain (15).
The only technique other than temperature jump that has been used in this time regime is line shape analysis of one-dimensional NMR spectra (5). The highest determined value for the refolding rate constant of the G46A/G48A double mutant of λ6–85 repressor was 3,000 s−1 at 37°C in urea solution (5). A value of 30,000 s−1 in water at 30°C was extrapolated from moderate urea concentrations (34). Refolding rate constants up to 22,000 s−1, at 46°C, have been determined for the peripheral subunit-binding domain protein (6) by using the same approach.
Bringing Together Theory and Experiment.
We estimate the half-life for unfolding to be in the region of 10 ns at 100°C, which is easily in the current time realm of MD simulation. MD simulations do, indeed, find unfolding on this time scale. The protein unfolded by means of expansion of the core and movement of helix III away from the core, regardless of the simulation temperature, to yield the transition state. The transition states at 100 and 225°C are similar; in fact, they are more similar to one another than they are to their starting structure. Also, the simulated transition states are consistent with available experimental data, such as the βT value. After the transition state, further expansion and “undocking” of the helices occurred at both temperatures. At 100°C, various arrangements of the helices were sampled in the 70-ns simulation, but the helices remained largely intact although lacking in well-formed tertiary packing interactions.
Better sampling of the denatured state was obtained in the 225°C simulation. In this case, all three helices unfolded and refolded many times over the simulation. This was particularly true for helices I and III, whereas helix II was less stable. Helices I and III contained the highest helix contents over time, and after 40 ns both helices had recovered two turns of helix. These helices are unusually stable, and we note that it is not just overestimation of the helix content by the force field. We have measured experimentally the equilibrium constants for formation of helical structure in peptide fragments encompassing the helical regions in the native structure of En-HD (unpublished observations). Helices I and III were found to be exceptionally stable. The high stability of these fragments may well underestimate the amount of residual helical content in the denatured state. It has been noted before in simulations of barnase that tertiary interactions within the denatured state not only stabilize helical structure but also facilitate or trigger its formation (9, 35). Similarly, NMR measurements on the denatured state of barstar show more helix than is found in the fragments (33, 36). Simulation and experiment thus suggest that folding involves the docking of essentially preformed helices. A similar mechanism may be operative for monomeric λ repressor (34).
It has been suggested (37, 38) and found by Serrano and coworkers (39–41), that overstabilization of native structure, so that it is highly populated in the denatured state, is wasteful of stabilization energy, as can be seen in the low ΔGD-N of En-HD. But, the fluctuating residual helical structure in the denatured state does assure that the protein is poised for rapid productive docking of the helices. However, a consequence of the significant uncoupling of the formation of secondary and tertiary structure is that unfolding is also very fast.
The collaboration between the Seattle and Cambridge laboratories in simulation and experiment rests on benchmarking the simulation with experimental observables, usually with φ-values or NMR. Here, the commonalities are unfolding on the same and unusual time scale and the finding of high helical content in the denatured state. The combined approaches of experiment and unfolding simulations suggest that this protein achieves such rapid folding as a consequence of decoupling of the formation of secondary and tertiary structure. In the folding direction, the process appears to be primarily the docking of preformed helices and, in the unfolding reaction, mainly the breaking of tertiary interactions. The estimated half-life for unfolding of approximately 10 ns at 100°C has allowed for real-time unfolding MD simulation at experimentally accessible temperatures. The half-life of refolding of 18 μs should be accessible in the near future to direct real-time MD refolding simulations. We expect that the continued discovery of such fast folding proteins and the development of computational power will lead to a more general detailed understanding of protein folding at atomic resolution.
Acknowledgments
We thank R. T. Sauer for providing the plasmid pSEA100 encoding En-HD and S. Poget for the mass spectrometry measurements. U.M. is supported by an Ikertzaileen prestakuntza grant from the Government of the Basque Country. V.D. is grateful for financial support from the Office of Naval Research (N00014-95-1-0484, N00014-96-1-1093, N00014-98-1-0477) and the National Institutes of Health (GM 50789). University of California, San Francisco, midasplus was used to make Fig. 4 (42).
Abbreviations
- MD
molecular dynamics
- En-HD
Engrailed Homeodomain
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250473497.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250473497
References
- 1.Chamberlain A K, Marqusee S. Adv Protein Chem. 2000;53:283–328. doi: 10.1016/s0065-3233(00)53006-x. [DOI] [PubMed] [Google Scholar]
- 2.Fersht A R. Structure and Mechanism in Protein Science. New York: Freeman; 1998. [Google Scholar]
- 3.Jackson S E, Fersht A R. Biochemistry. 1991;30:10428–10435. doi: 10.1021/bi00107a010. [DOI] [PubMed] [Google Scholar]
- 4.Schindler T, Herrler M, Marahiel M A, Schmid F X. Nat Struct Biol. 1995;2:663–673. doi: 10.1038/nsb0895-663. [DOI] [PubMed] [Google Scholar]
- 5.Huang G S, Oas T G. Proc Natl Acad Sci USA. 1995;92:6878–6882. doi: 10.1073/pnas.92.15.6878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spector S, Raleigh D P. J Mol Biol. 1999;293:763–768. doi: 10.1006/jmbi.1999.3189. [DOI] [PubMed] [Google Scholar]
- 7.Duan Y, Kollman P A. Science. 1998;282:740–744. doi: 10.1126/science.282.5389.740. [DOI] [PubMed] [Google Scholar]
- 8.Oliveberg M, Tan Y J, Fersht A R. Proc Natl Acad Sci USA. 1995;92:8926–8929. doi: 10.1073/pnas.92.19.8926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong K B, Clarke J, Bond C J, Neira J L, Freund S M V, Fersht A R, Daggett V. J Mol Biol. 2000;296:1257–1282. doi: 10.1006/jmbi.2000.3523. [DOI] [PubMed] [Google Scholar]
- 10.Li A, Daggett V. Proc Natl Acad Sci USA. 1994;91:10430–10434. doi: 10.1073/pnas.91.22.10430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li A, Daggett V. J Mol Biol. 1996;257:412–429. doi: 10.1006/jmbi.1996.0172. [DOI] [PubMed] [Google Scholar]
- 12.Alonso D O, Daggett V. J Mol Biol. 1995;247:501–520. doi: 10.1006/jmbi.1994.0156. [DOI] [PubMed] [Google Scholar]
- 13.Alonso D O, Daggett V. Protein Sci. 1998;7:860–874. doi: 10.1002/pro.5560070404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bieri O, Wirz J, Hellrung B, Schutkowski M, Drewello M, Kiefhaber T. Proc Natl Acad Sci USA. 1999;96:9597–9601. doi: 10.1073/pnas.96.17.9597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hagen S J, Hofrichter J, Szabo A, Eaton W A. Proc Natl Acad Sci USA. 1996;93:11615–11617. doi: 10.1073/pnas.93.21.11615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eaton W A, Munoz V, Hagen S J, Jas G S, Lapidus L J, Henry E R, Hofrichter J. Annu Rev Biophys Biomol Struct. 2000;29:327–359. doi: 10.1146/annurev.biophys.29.1.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clarke N D, Kissinger C R, Desjarlais J, Gilliland G L, Pabo C O. Protein Sci. 1994;3:1779–1787. doi: 10.1002/pro.5560031018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harrison S C, Aggarwal A K. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 19.Miroux B, Walker J E. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 20.Ades S E, Sauer R T. Biochemistry. 1994;33:9187–9194. doi: 10.1021/bi00197a022. [DOI] [PubMed] [Google Scholar]
- 21.Privalov P L, Potekhin S A. Methods Enzymol. 1986;131:4–51. doi: 10.1016/0076-6879(86)31033-4. [DOI] [PubMed] [Google Scholar]
- 22.Rigler R, Rabl C-B, Jovin T M. Rev Sci Instrum. 1974;45:580–588. [Google Scholar]
- 23.Levitt M. encad, Energy Calculations and Dynamics. Palo Alto, CA: Molecular Applications Group; 1990. [Google Scholar]
- 24.Levitt M, Hirshberg M, Sharon R, Daggett V. Comp Phys Commun. 1995;91:215–231. [Google Scholar]
- 25.Levitt M, Hirshberg M, Sharon R, Laidig K E, Daggett V. J Phys Chem. 1997;101:5051–5061. [Google Scholar]
- 26.Kell G S. J Chem Eng Data. 1967;12:66–68. [Google Scholar]
- 27.Haar L, Gallagher J S, Kell G S. NBS/NRC Steam Tables. Washington, DC: Hemisphere Publications; 1984. [Google Scholar]
- 28.Makhatadze G I, Privalov P L. Adv Protein Chem. 1995;47:307–425. doi: 10.1016/s0065-3233(08)60548-3. [DOI] [PubMed] [Google Scholar]
- 29.Myers J K, Pace C N, Scholtz J M. Protein Sci. 1995;4:2138–2148. doi: 10.1002/pro.5560041020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matouschek A, Matthews J M, Johnson C M, Fersht A R. Protein Eng. 1994;7:1089–1095. doi: 10.1093/protein/7.9.1089. [DOI] [PubMed] [Google Scholar]
- 31.Eyring H. Chem Rev. 1935;17:65–77. [Google Scholar]
- 32.Matouschek A, Otzen D E, Itzhaki L S, Jackson S E, Fersht A R. Biochemistry. 1995;34:13656–13662. doi: 10.1021/bi00041a047. [DOI] [PubMed] [Google Scholar]
- 33.Nölting B, Golbik R, Neira J L, Soler-Gonzalez A S, Schreiber G, Fersht A R. Proc Natl Acad Sci USA. 1997;94:826–830. doi: 10.1073/pnas.94.3.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burton R E, Myers J K, Oas T G. Biochemistry. 1998;37:5337–5343. doi: 10.1021/bi980245c. [DOI] [PubMed] [Google Scholar]
- 35.Bond C J, Wong K B, Clarke J, Fersht A R, Daggett V. Proc Natl Acad Sci USA. 1997;94:13409–13413. doi: 10.1073/pnas.94.25.13409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wong K B, Freund S M V, Fersht A R. J Mol Biol. 1996;259:805–818. doi: 10.1006/jmbi.1996.0359. [DOI] [PubMed] [Google Scholar]
- 37.Fersht A R. Proc Natl Acad Sci USA. 1995;92:10869–10873. doi: 10.1073/pnas.92.24.10869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fersht A R, Shakhnovich E I. Curr Biol. 1998;8:R478–R479. doi: 10.1016/s0960-9822(98)70310-0. [DOI] [PubMed] [Google Scholar]
- 39.Muñoz V, Serrano L. Folding Des. 1996;1:R71–R77. doi: 10.1016/S1359-0278(96)00036-3. [DOI] [PubMed] [Google Scholar]
- 40.López-Hernández E, Cronet P, Serrano L, Muñoz V. J Mol Biol. 1997;266:610–620. doi: 10.1006/jmbi.1996.0793. [DOI] [PubMed] [Google Scholar]
- 41.Viguera A R, Villegas V, Avilés F X, Serrano L. Folding Des. 1997;2:23–33. doi: 10.1016/S1359-0278(97)00003-5. [DOI] [PubMed] [Google Scholar]
- 42.Ferrin T E, Huang C C, Jarvis L E, Langridge R. J Mol Graphics. 1988;6:13–27. [Google Scholar]
- 43.Kabsch W, Sander C. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]