Abstract
A family of p160 coactivators was initially identified based on ligand-dependent interactions with nuclear receptors and thought to function, in part, by recruiting CREB-binding protein/p300 to several classes of transcription factors. One of the p160 factors, p/CIP/AIB1, often amplified and overexpressed in breast cancer, also exhibits particularly strong interaction with CREB-binding protein/p300. In this manuscript, we report that p/CIP, which exhibits regulated transfer from cytoplasm to nucleus, is required for normal somatic growth from embryonic day 13.5 through maturity. Our data suggest that a short stature phenotype of p/CIP gene-deleted mice reflect both altered regulation of insulin-like growth factor-1 (IGF-1) gene expression in specific tissues and a cell-autonomous defect of response to IGF-1, including ineffective transcriptional activities by several classes of regulated transcription factors under specific conditions. The actions of p/CIP are therefore required for full expression of a subset of genes critical for regulating physiological patterns of somatic growth in mammals.
Nuclear receptors comprise a family of transcription factors that regulate gene expression in a ligand-dependent manner and include receptors for steroid hormones, such as estrogen and glucocorticoid, receptors for nonsteroidal ligands, such as thyroid hormone receptor and retinoic acid receptor, as well as receptors that bind diverse products of lipid metabolism (reviewed in refs. 1–5). A combination of genetic, biochemical, and functional data suggests that many factors, including the BRG (Swi/Snf) complex (6), CREB-binding protein (CBP)/p300, p160 factors, p/CAF, and the TRAP/DRIP/ARC (7, 8) complex are likely to be critical regulators for at least subsets of nuclear receptor-regulated genes (9–15). However, by the simple criteria of ligand-dependent binding and the ability to synergize on cotransfection assays, numerous additional proteins have been suggested to exert coactivator roles (4, 5).
Proteins of approximately 160 kDa molecular mass were among the first factors identified that interact with nuclear receptor in a highly ligand-dependent manner both in solution (16, 17) and on DNA (18) and could themselves associate with CBP (11, 12). Expression cloning and yeast two-hybrid screening approaches led to the identification of three related genes that encode these p160 factors, referred to as SRC-1/NCoA-1, TIF2/GRIP-1/NCoA2, and p/CIP/AIB1/ACTR/RAC3/TRAM-1 (11, 19–25). These factors bind to nuclear receptors by interactions of LXXLL motif-containing helices within the interaction domain formed by conserved residues in helix3 and helix12 of the liganded receptor (5).
Members of the p160 family of nuclear receptor coactivators contain a highly conserved N-terminal bHLH-PAS domain that is also present in members of the Per/Arnt/Sim family of transcription factors and mediates protein–protein interactions. Although several lines of evidence support the idea that p160 factors play important roles as nuclear receptor coactivators, in part by recruiting CBP/p300 and/or arginine methyltransferase (26), the extent of their role is not proven. Consistent with a potential functional redundancy, the deletion of the SRC-1 gene in mice results in only relatively subtle defects in the development of estrogen receptor-dependent tissues, including uterus and breast, that may be explained by the observed compensatory increase in GRIP-1/TIF-2/NcoA2 expression (27, 28).
In this manuscript, we report the in vivo role of p/CIP, based on analysis of p/CIP gene-deleted mice, revealing an unexpected function of p/CIP for regulating normal somatic growth. This phenotype is attributable to both alteration in the levels of the insulin-like growth factor 1 (IGF-1) as independently described (29) and a cell-autonomous defect in a program of gene transcription that results in ineffective proliferative responses to IGF-1 and other growth factors.
Materials and Methods
Generation of p/CIP Gene-Deleted Mice.
A mouse p/CIP genomic bacterial artificial chromosome (BAC) clone was obtained by using a cDNA fragment encoding nuclear receptor interaction domain. We determined the intron and exon structures of the genomic locus. A 5′-flanking 5-kb BamHI fragment immediately upstream of the nuclear receptor interaction domain and a 3′-flanking 3.5-kb HindIII fragment just downstream of CBP-binding domain were cloned into the plasmid pBluescript KSII. The fragments were sequenced and cloned into SalI–XbaI and XhoI–NotI sites of the pPGK–TKNeo-targeting vector (30). The NotI-linearized construct (20 μg) was electroporated into R1 embryonic stem (ES) cells (in 0.8 ml of electroporation buffer) at 250 V and 500 μF with a gene-pulser. ES cells were grown for 7–9 days in 150 μg/ml G418 and 2 mM gancyclovir for positive and negative selections, respectively. Homologous recombination was identified by using appropriate probes (400-bp cDNA fragment and 5-kb BamHI genomic fragment). External probes were used to identify homologous recombination in the 3′- and 5′-flanking regions. Two ES cell lines exhibiting homologous recombination were injected into C57BL/6 blastocysts that were then implanted into pseudopregnant females. Chimeric mice were backcrossed to C57BL/6 and germ-line transmission was examined. Heterozygous and homozygous mice were identified by Southern blot analysis and genomic PCR, with a pair of neomycin primers (product ≈500 bp) and a pair primers from deleted nuclear receptor interaction regions of p/CIP (P1 5′-AGTGTCCTCCTCAACATCAGG-3′ and P2 5′-CTTCTTAGGACTCAGCTGCTCC-3′, and the product is 215 bp). Two lines were generated and analyzed.
In Situ Hybridization, Immunohistochemistry, Radioimmunoassays of IGF-1, Fluorescence-Activated Cell Sorter (FACS) Analysis, and Analysis of Mammary Gland Development.
E15.5 mouse embryos were isolated, fixed, and sectioned, and the sections were hybridized with 35S-labeled antisense RNA probes as described (30). Immunohistochemistry were performed on 5- to 7-μm thick paraffin sections or 20-μm sections cut on a cryostat by indirect immunoperoxidase method. Serum IGF-1 levels were determined by the manufacturer's protocol (Diagnostic Systems Laboratories, Webster, TX). Annexin V staining was performed on mouse embryonic fibroblasts (MEFs) according to the manufacturer's (PharMingen) protocol. Propidium iodide- and annexin V-stained MEFs were analyzed on FACS with the cell quest program from Becton Dickinson. The fourth mammary glands from virgin and pregnant female mice were dissected out, fixed in 10% formalin overnight, and transferred and stored in 70% ethanol. The glands were defatted in acetone for 3 h with three changes of acetone. Mammary glands were stained in Harris's hematoxylin for 2 h after two passages in 100% and 95% ethanol, each for 1 h, and destained in acidic 50% ethanol. They were then dehydrated and stored in methyl salicylate.
Preparation of MEFs and Hepatocytes, Western Blot Analysis, RNase Protection Assays, and Single Cell Nuclear Injections.
The E13.5–E16.5 embryos were dissected out by C-sections of pregnant mothers, and embryonic fibroblasts were cultured in DMEM high glucose medium with 10% FCS and sodium pyruvate, after removing liver and blood from embryos and trypsin digestion. Third to sixth passages of MEFs were used in the experiments. Cultures of hepatocytes from adult mouse livers were prepared after two-step in situ perfusion with 0.5 mM solution of EGTA and collagenase type IV (Sigma) as was described (32). Kidneys were dissected from wild-type and p/CIP(−/−) mice and mechanically homogenized into single cells; whole cell extracts were prepared, and 50 μg of protein was loaded in each lane of an SDS gel. Western blots were performed as previously described. Total RNA was isolated from MEFs or individual organs of mice with RNeasy Mini kit from Qiagen (Chatsworth, CA). Total RNA (10 μg) was used in RNase protection assays with the RNP III kit from Ambion. Single cell nuclear microinjection assays were performed by using MEFs as described (31). 5-Bromodeoxyuridine (BrdU) immunofluorescence labeling and staining were performed on MEFs and isolated liver cells as described in ref. 33.
Results
p/CIP Exhibits Regulated Intracellular Localization.
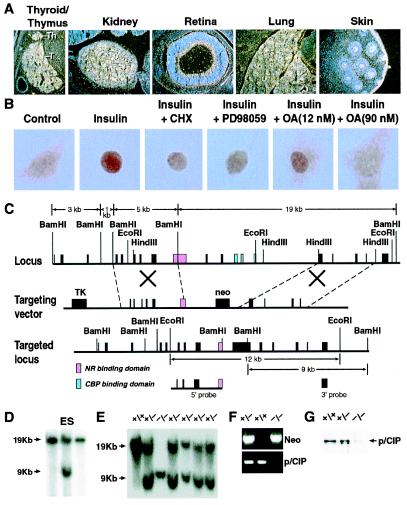
In situ hybridization and immunohistochemical analyses of members of the p160 family coactivators exhibit distinct, albeit highly overlapping patterns of expression. p/CIP was highly expressed in the thyroid gland, thymus, kidney, lung, retina, and skin (Fig. 1A). To begin to further investigate p/CIP functions as a coactivator, a yeast two-hybrid screen using the nuclear receptor and CBP interaction domains of p/CIP as bait was performed to search for interaction partners of p/CIP. Unexpectedly, we found that the cytoplasmic protein, IkBγ, interacted with p/CIP in this system and in glutathione S-transferase fusion protein-binding assays (data not shown). Based on these interactions, immunohistochemical studies were performed to determine whether p/CIP could localize to both nuclear and cytoplasm compartments. p/CIP exhibited both cytoplasmic and nuclear localization in several cell types (Fig. 1B) as confirmed by analysis of nuclear and cytoplasmic fractions using Western blotting (data not shown). However, when cells were placed in serum-free conditions, p/CIP rapidly redistributed to exhibit a predominant cytoplasmic localization; although some immunoreactive protein remained in the nucleus (Fig. 1B). Addition of insulin to the Rat-1 fibroblasts cultured under serum-free conditions caused most of the p/CIP to relocate to the nucleus; this redistribution was not altered by a mitogen-activated kinase kinase (MEK) inhibitor, PD 98059, and was not modified by treatment with cycloheximide to block protein synthesis (Fig. 1B). Intriguingly, the redistribution was sensitive to addition of high concentrations of okadaic acid, which blocks the actions of both protein phosphatase-1 (PP1) and 2A (PP2A). Low concentrations of okadaic acid, which selectively inhibit PP2A, only had a limited effect on insulin-stimulated redistribution. Therefore, it would appear that PP1-dependent dephosphorylation events are involved in the observed nuclear accumulation of p/CIP.
Figure 1.
Generation of p/CIP gene-deleted mice. (A) In situ hybridization analysis of p/CIP transcript in E15.5 embryo. Strong expression is detected in thyroid gland, thymus, kidney, retina, lung, and epidermis. (B) Cytoplasmic/nuclear localization of p/CIP. Under serum-free conditions, in Rat-1 cells, the immunoreactive p/CIP is largely cytoplasmic (control); addition of insulin (10−7 M) for 30 min causes most of p/CIP relocate to nucleus; pretreatment by cycloheximide (CHX; 10 μM) for 1 h or the mitogen-activated kinase kinase (MEK) inhibitor PD 18059 (10 μg/ml) and 12 nM okadaic acid did not alter the insulin effect; however, 90 nM okadaic acid reversed the insulin effect. (C) Strategy for homologous recombination of the p/CIP genomic locus. Probes of 5′ and 3′ are shown, and the regions targeted for deletion are nuclear receptor and CBP-binding domains. (D) Homologous recombination in ES cells. (E) Homologous recombination and proof of generation of gene-deleted mice. (F) PCR diagnostic strategy that is used to screen for genomic deletion. The primers used are from the deleted nuclear receptor binding domain. (G) Western blot analysis of kidney extract, showing the absence of p/CIP immunoreactive staining in p/CIP mice.
Generation of p/CIP Gene-Deleted Mice.
To further characterize the biological roles of p/CIP, we designed a targeting construct to delete a portion of the p/CIP genomic locus. The genomic DNA that encompasses the coding region of p/CIP is ≈30 kb, as diagrammed in Fig. 1C. A single exon encodes the entire nuclear receptor interaction domain of p/CIP, whereas the CBP-binding domain is encoded by four small exons. A targeting construct was designed to delete both the nuclear receptor interaction and CBP-binding domains, as shown in Fig. 1C.
Two ES clones exhibited homologous recombination in one allele, documented by Southern blot analysis (Fig. 1D), as used to generate chimeric mice. Mice heterozygous and homozygous for the p/CIP gene-deleted locus were obtained by appropriate breeding, and they were confirmed by the absence of wild-type locus by genomic Southern blots and the absence of p/CIP protein by Western blot analysis (Fig. 1 E and G). We detected increased expression of p300 and TIF2/GRIP1/NcoA2 in some organs, such as kidney in p/CIP(−/−) mice by immunohistochemical analysis, but not other organs, such as brain or liver (data not shown). These data suggest that other coactivators might potentially compensate for the loss of p/CIP in some organs.
Analysis of 502 pups from heterozygous mating revealed an expected Mendelian ratio of p/CIP mice [p/CIP +/+ 122, +/− 265, and (−/−) 114], which survived to adulthood and which were fertile. Standard histological analyses of E15.5 embryos and adults revealed generally normal morphology, including the kidney, liver, thyroid gland, lung, and brain. FACS analysis with CD4 and CD8 antibodies demonstrated normal thymocyte development in thymus, and a normal profile of peripheral CD4 and CD8 single positive T cells in spleen, in both wild-type and p/CIP(−/−) mice (data not shown). However, there was a facial asymmetry, characterized by a random unilateral drop of the ear that was observed in ≈10% of the homozygous mutant mice.
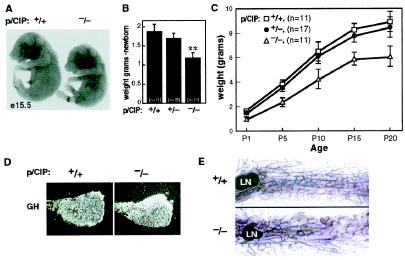
Because p/CIP is often amplified and/or overexpressed in breast cancer (20), we studied mammary gland development in female mutant mice. First, we examined mammary glands of 10-week-old virgin mice, finding no difference when comparing mutant mice and their female wild-type littermates, in the ductal branching and elongation, or in penetrance of epithelial cells into the fat pad by whole mount staining (Fig. 2E). We observed similar proliferation of epithelial cells and lobulo-alveolar development in the mammary glands at different stages of pregnancy, and similar lactation and nursing behavior between homozygous mutant mice and their wild-type littermates. On examining the onset of puberty and estrus cycles by vaginal smears, we found no significant difference between p/CIP(−/−) mice and their wild-type littermates.
Figure 2.
Growth defect in p/CIP(−/−) mice. (A) Embryonic (E15.5) difference of wild-type and p/CIP(−/− mice in size. (B) Weight difference of postnatal day 1 in wild-type, heterozygous, and p/CIP(−/−) mice. **, P < 0.01. (C) Rates of growth in wild-type, heterozygous, and p/CIP(−/−) mice. (D) Expression of GH using in situ analysis on adult pituitary gland. (E) Harris hematoxylin whole-mount staining of mammary glands of 10-week-old female virgin littermates from wild-type and p/CIP(−/−) mice (lymph node, LN); no reproducible differences were observed at any developmental stages.
Growth Defects in p/CIP Gene-Deleted Mice.
Although p/CIP(−/−) mice were entirely viable, one phenotype exhibited 100% penetrance. Mice heterozygous for the p/CIP deletion were slightly smaller than their wild-type littermates (≈10%), which is at the limit of statistical significance. However, gene-deleted mice were uniformly much smaller than the age- and sex-matched littermates, whether male or female, with weight consistently ≈30–50% reduced compared to their wild-type littermates. Examination of ontogeny of growth and development revealed that the decreased size of p/CIP gene-deleted embryos was apparent on E13.5 and 50–70% of body weight of their littermates from E15.5 onward (Fig. 2 A–C). This growth impairment continued through the neonatal period, through weaning and sexual maturation, and into adulthood. Thus, somatic growth was affected both before puberty and before growth hormone (GH)-dependent control of hepatic IGF-1 modulation (34). Pituitary development exhibited both normal morphology and cellular proliferation, and all hormone-encoding genes were expressed comparably in wild-type and p/CIP(−/−) mice (Fig. 2D and data not shown).
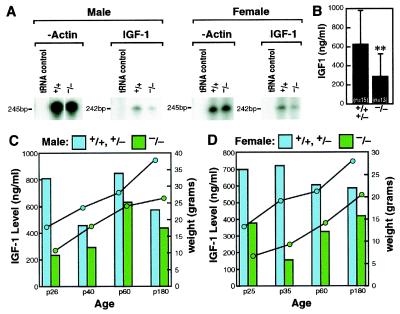
The well-known relationship of IGF-1 pathways to regulation of growth, specifically their roles during embryogenesis and postnatal life (34), raised questions of the role of the components of the IGF-1 pathway in the p/CIP(−/−) phenotype. Radioimmunoassays of sera from p/CIP(−/−) mice and wild-type littermates revealed a significant decrease, ≈30–50%, in levels of IGF-1 in p/CIP(−/−) mice, which was observed consistently from p25 to p180 in both males and females, although a diminishing difference in IGF-1 levels was found with increasing age (Fig. 3 B, C, and D), in concert with observations on mice with a different gene deletion strategy (29). Furthermore, examination of IGF-1 transcripts by using RNase protection assays revealed up to a 2- to 3-fold decrease in IGF-1 mRNA in livers in p/CIP(−/−) mice (Fig. 3A), whereas no alterations in IGF transcripts were observed in some other organs evaluated, including ovary and spleen.
Figure 3.
Decrease of IGF-1 gene expression and serum levels in p/CIP(−/−) mice. (A) RNase protection analysis on liver mRNAs by using specific probes; loading of RNA were normalized based on beta-actin levels. From 100–200% decreases of IGF-1 transcript were observed in age- and sex-matched, p25 mice. (B) IGF-1 serum levels in wild-type, heterozygous, and p/CIP(−/−) mice. **, P < 0.01. (C and D) Growth rates and serum IGF-1 levels in wild-type, heterozygous, and p/CIP(−/−) mice.
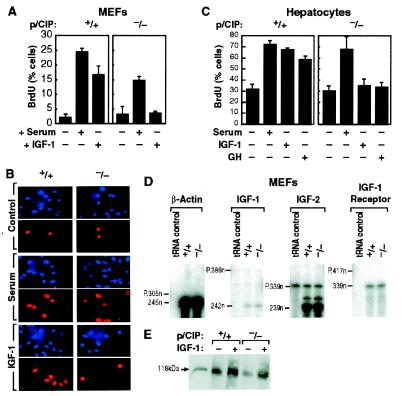
The observation that IGF-1 was affected in liver would not account for the growth phenotype because even with serum IGF-1 levels in liver-specific IGF-1 gene-deleted mice, ≈25% that of wild-type littermates, no growth defects were observed (35, 36). On the other hand, mice deleted for the suppressor of cytokine-signaling-2 (SOCS2) gene locus exhibited gigantism, with IGF-1 levels higher in certain tissues such as heart and lung, but with no alterations noted in the liver or serum (37). Therefore, we investigated the possibility that there might be a second component of p/CIP action by preparing MEF cultures and primary hepatocyte cultures from adult livers. We examined DNA synthesis in the MEFs by BrdU labeling in serum-starved cells and cells subsequently treated with serum or growth factors. Addition of serum to the starved MEFs resulted in comparable labeling in both mutant and wild-type MEFs, indicating normal DNA synthesis under this condition. Although addition of IGF-1 to wild-type MEFs resulted in significant DNA synthesis, only minimal BrdU incorporation was observed in p/CIP(−/−) MEFs (Fig. 4A), indicating a defect in the response to IGF-1 in these cells. To investigate molecular mechanisms underlying this failure to respond to IGF-1, we examined expression levels of IGF-1-signaling components. The expression levels of IGF-1, IGF-2, and IGF-1 receptor transcripts were almost equivalent in both mutant and wild-type MEFs, as demonstrated using RNase protection assays (Fig. 4D). We also examined mRNA levels of IGF-1-binding proteins IGFBP1–IGFBP6, insulin receptor, and IRS-1, IRS-2, and IRS-4 by “quantitative” PCR, with no discernable difference in any of these components between wild-type and p/CIP(−/−) MEFs (data not shown). Further, Western blot analysis suggested that IGF-1 receptor can be normally phosphorylated in response to IGF-1 in p/CIP(−/−) MEFs, although it might be expressed at very slightly lower levels than in wild-type MEFs (Fig. 4E). We also evaluated growth response in primary hepatocyte cultures, prepared from perfused livers. Hepatocytes were cultured in medium with serum overnight and then switched to defined medium without serum for 24 h. The cultures were then stimulated with serum or GH. As shown in Fig. 4C, both wild-type and p/CIP hepatocytes respond comparably to serum; however, GH caused significant BrdU incorporation in wild-type, but much less in p/CIP(−/−) hepatocytes, implicating a failure of response to a signal transducer and activator of transcription pathway-dependent growth signal.
Figure 4.
Growth defects in p/CIP(−/−) cells. (A) MEFs from wild-type and p/CIP(−/−) littermates embryos (E13.5–E16.5) were starved for 24 h, 10% serum or 100 ng/ml human recombinant IGF-1 (GIBCO/BRL) was added for 14 h, and cells were then pulse-labeled with BrdU for 2 h. MEFs were then immunostained with anti-BrdU antibody, and labeled cells were counted as percentage of total cells. (B) A representative experiment of BrdUrd labeling of MEFs. (C) Growth responses of primary hepatocytes to growth stimuli. Hepatocyte cultures were maintained in 10% serum overnight and then switched to serum-free medium for 24 h. IGF-1 (100 ng/ml), 10 μg/ml GH, or 10% serum was added to cultures for 14 h. BrdU labeling was performed as described in A. (D) RNase protection analysis of IGF-1, IGF-2, and IGF-1 receptor transcripts from MEFs. Total RNA (10 μg) was used in each experiment. (E) Tyrosine phosphorylation of IGF-1 receptor in response to IGF-1 stimulation in MEFs. Starved MEFs were stimulated with IGF-1 for 10 min as described in 4A; equal amount of whole cell extracts were immunoprecipitated with anti-IGF-1 receptor IgG. The precipitates were then subjected to SDS/PAGE and Western blot analysis by using anti-phosphotyrosine IgG.
Role of p/CIP in Regulating Transcriptional Responses.
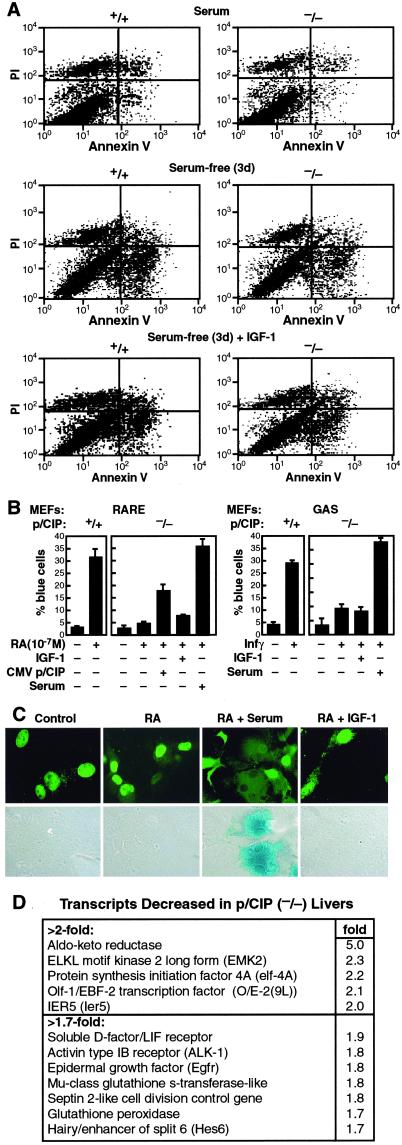
IGF-1 is well established to also serve as a survival factor, acting in part via protein kinase B (AKT) kinase (38). This event inhibits expression of the FAS ligand gene as a result of phosphorylation of the forkhead protein, FKHRL1, preventing its nuclear translocation (38). We therefore investigated whether there was an increase in apoptosis in p/CIP(−/−) MEFs, using Annexin V staining and FACS analysis. As shown in Fig. 5A Top, both wild-type and p/CIP(−/−) MEFs contained very few preapoptotic cells (low right, LR), when they were maintained in normal 10% serum medium. Serum starvation for 72 h resulted in significant numbers of both dead cells [PI only staining, upper left (UL)] and preapoptotic cells [annexin V only staining, lower right (LR)], but comparably in both wild-type and p/CIP(−/−) MEFs (Fig. 5A Top and Middle). Addition of IGF-1 to starved MEFs for 24 h resulted in comparable rescues for preapoptotic cells in both wild-type and p/CIP(−/−) MEFs (Fig. 5A Bottom). These results demonstrated that the survival pathway of IGF-1 appeared to be intact in p/CIP(−/−) MEFs, whereas the proliferative response was impaired.
Figure 5.
Role of p/CIP in regulated gene transcription and identification of gene targets. (A) Analysis of apoptosis using annexin V staining (using reagents and the protocol of PharMingen) of MEFs from wild-type and p/CIP(−/−) mice. MEFs were starved in serum-free conditions for 72 h and were then stimulated with 100 ng/ml IGF-1 for 24 h. (B) Results of retinoic acid (RA) and IFN-γ responsive element reporters in p/CIP(+/+) and p/CIP(−/−) MEFs, cultured under serum-free conditions, or in addition of 10% serum, or IGF-1 (100 ng/ml). (C) Photographs of retinoic acid response on an RARE/β-gal reporter in p/CIP(−/−) MEFs. (D) Affymetrix array analysis using total RNA from livers of age- and sex-matched p/CIP(+/+) and (−/−) littermates was performed in three separate experiments; only targets differing in all three experiments are shown.
We next tested whether the transcriptional activity of several types of DNA-binding transcription factors were maintained in the p/CIP(−/−) MEFs. As shown in Fig. 5B, we found that MEFs from both wild-type and p/CIP(−/−) mice exhibited normal transcriptional activities in response to the retinoic acid and IFN-γ by using the single cell nuclear microinjection assay (31), when cultured in the presence of serum. However, in serum-free conditions, we found that the retinoic acid IFN-γ and responses were markedly impaired in the p/CIP(−/−) MEFs (Fig. 5B). In the presence of IGF-1, we found that there was only a minimal response of the retinoic acid receptor to its ligand, and the response to an IFN-γ was ≈50% of that of wild-type MEFs (Fig. 5B). Therefore, p/CIP regulates a function(s) that in nonproliferative cells resulted in impaired transcriptional activities by several types of regulated transcription factors.
To begin to define the molecular mechanism for the cell-autonomous growth defects, we initiated studies of patterns of gene expressions in the livers of age- and sex-matched littermates using the Affymetrix U74 gene chips. Only the genes that were down-regulated in the p/CIP(−/−) livers in all three pairs of littermates were selected (Fig. 5D). IGF-1 met this requirement, and two other genes that were decreased more than 2-fold were aldo–keto reductase and ELKL motif kinase 2 (EMK2).
Discussion
A large number of potential coactivators have been implicated in ligand-dependent transcriptional activation of nuclear receptors. One family of potential coactivators, the p160 family, contains three known related members, of which p/CIP is unique in exhibiting regulated translocation between nuclear and cytoplasmic compartments. The previous discovery of p/CIP amplification/overexpression in human breast cancers (20) suggests a potential role in growth. In this manuscript, we present genetic evidence that p/CIP is a required component for normal patterns of somatic growth. Unexpectedly, the defects in p/CIP gene-deleted mice reflect alternation in a growth factor and cell-autonomous responses.
First, we have found evidence that p/CIP deletion causes a decrease in serum IGF-1 levels, in agreement with independent studies of a knockout mouse in which a different region of p/CIP was deleted (29). We detected a decrease of IGF-1 transcripts in the adult livers of both sexes of p/CIP(−/−) mice. However, this difference in IGF-1 levels in liver and serum does not account for the growth defects in the knockout mice because only 25% of normal serum IGF-1 levels are sufficient for normal somatic growth, as shown in liver-specific knockout mice of IGF-1 (34, 35). This result implies that additional genes are modulated by p/CIP. Intriguingly, there is a second, cell-autonomous defect in p/CIP(−/−) mice, reflecting its physiological role in regulating a small number of growth-related genes. The consequence of this defect is to permit normal responses both in regulated gene expression and in proliferation in the presence of serum; however, cell proliferation responses under serum-free conditions in both MEFs and hepatocytes of mutant mice in response to GH and IGF-1 are concomitantly decreased. These data suggest that the proliferative response to IGF-1 and GH are both sensitive to p/CIP levels, but this requirement is clearly compensated for in serum-treated cells. Furthermore, responses to retinoic acid and IFN-γ are selectively inhibited under serum-free conditions, unmasking a key role of p/CIP in regulating gene transcription under particular conditions. These results correlate with that of single cell nuclear microinjections in Rat-1 cells analyzed by using anti-p/CIP IgG (25). These findings are particularly interesting in light of the report that alterations in genes regulated by the signal transducer and activator of transcription pathway are common in breast cancers (39).
We conclude that p/CIP is not required for regulated gene expression of most nuclear receptor target genes, although expression of aldo–keto reductase, a well known target of liganded nuclear receptor (40), appears decreased in p/CIP(−/−) mice. However, p/CIP is clearly required for a small set of genes that regulate physiological patterns of somatic growth, both during embryogenesis and into adulthood. For example, the homolog of EMK2, EMK1, is involved in the control of somatic growth (41). Mice in which the EMK1 gene is deleted exhibit dwarfism from E18.5 and onward (41). We suggest that this reflects quantitative decreases in expression of a series of target genes that combinatorially affect somatic growth, and that the absence of p/CIP diminished transcriptional response to specific factors under particular conditions in nonproliferating cells.
Acknowledgments
We thank Drs. Greg Erickson, Mercedes Ricote, and Tiffany Seagroves for discussion; Dr. A. F. Parlow and the National Hormone and Pituitary Program for GH; H. Taylor for mouse maintenance; M. Fisher for help in preparing the manuscript; and P. Meyer for preparation of the figures. Z.W. is supported by National Research Service Award Fellowship 5 F32 DK09669, and this work was supported by grants to D.W.R. (National Institutes of Health 1 R01 DK54802–01A1), C.K.G. (National Institutes of Health), R.R. (National Institutes of Health CA58110), and M.G.R. (National Institutes of Health, Association for the Cure of Cancer of the Prostate, and California Cancer Research Program).
Abbreviations
- CBP
CREB-binding protein
- CIP
CBP-interacting protein
- IGF
insulin- like growth factor
- EMK
ELKL motif kinase
- GH
growth hormone
- FACS
fluorescence-activated cell sorter
- MEF
mouse embryonic fibroblast
- ES
embryonic stem
- BrdU
5-bromodeoxyuridine
Footnotes
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.260463097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.260463097
References
- 1.Beato M, Herrlich P, Schutz G. Cell. 1995;83:851–857. doi: 10.1016/0092-8674(95)90201-5. [DOI] [PubMed] [Google Scholar]
- 2.Chambon P. Recent Prog Horm Res. 1995;50:317–332. doi: 10.1016/b978-0-12-571150-0.50019-6. [DOI] [PubMed] [Google Scholar]
- 3.Mangelsdorf D J, Evans R M. Cell. 1995;83:841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 4.McKenna N J, Lanz R B, O'Malley B W. Endocr Rev. 1999;20:321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 5.Glass C K, Rosenfeld M G. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 6.Fryer C J, Archer T K. Nature (London) 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 7.Fondell J D, Ge H, Roeder R G. Proc Natl Acad Sci USA. 1996;93:8329–8333. doi: 10.1073/pnas.93.16.8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rachez C, Suldan Z, Ward J, Chang C P, Burakov D, Erdjument-Bromage H, Tempst P, Freedman L P. Genes Dev. 1998;12:1787–1800. doi: 10.1101/gad.12.12.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chakravarti D, LaMorte V J, Nelson M C, Nakajima T, Schulman I, Juguilon G, Montminy M, Evans R M. Nature (London) 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 10.Hanstein B R, Eckner J, DiRenzo S, Halachmi H, Liu B, Searcy R, Kurokawa H, Brown M. Proc Natl Acad Sci USA. 1996;93:11540–11545. doi: 10.1073/pnas.93.21.11540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kamei Y, Xu L, Heinzel T, Torchia J, Kurokawa R, Gloss B, Lin S C, Heyman R A, Rose D W, Glass C K, Rosenfeld M G. Cell. 1996;85:403–414. doi: 10.1016/s0092-8674(00)81118-6. [DOI] [PubMed] [Google Scholar]
- 12.Yao T P, Ku G, Zhou N, Scully R, Livingston D M. Proc Natl Acad Sci USA. 1996;93:10626–10631. doi: 10.1073/pnas.93.20.10626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kawasaki H, Eckner R, Yao T P, Taira K, Chiu R, Livingston D M, Yokoyama K K. Nature (London) 1998;393:284–289. doi: 10.1038/30538. [DOI] [PubMed] [Google Scholar]
- 14.Yao T P, Oh S P, Fuchs M, Zhou N D, Ch'ng L E, Newsome D, Bronson R T, Li E, Livingston D M, Eckner R. Cell. 1998;93:361–372. doi: 10.1016/s0092-8674(00)81165-4. [DOI] [PubMed] [Google Scholar]
- 15.Kraus W L, Kadonaga J T. Genes Dev. 1998;12:331–342. doi: 10.1101/gad.12.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavailles V, Dauvois S, Danielian P S, Parker M G. Proc Natl Acad Sci USA. 1994;91:10009–10013. doi: 10.1073/pnas.91.21.10009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Halachmi S, Marden E, Martin G, MacKay H, Abbondanza C, Brown M. Science. 1994;264:1455–1458. doi: 10.1126/science.8197458. [DOI] [PubMed] [Google Scholar]
- 18.Kurokawa R, Soderstrom M, Horlein A, Halachmi S, Brown M, Rosenfeld M G, Glass C K. Nature (London) 1995;377:451–454. doi: 10.1038/377451a0. [DOI] [PubMed] [Google Scholar]
- 19.Onate S A, Tsai S Y, Tsai M J, O'Malley B W. Science. 1995;270:1354–1357. doi: 10.1126/science.270.5240.1354. [DOI] [PubMed] [Google Scholar]
- 20.Anzick S L, Kononen J, Walker R L, Azorsa D O, Tanner M M, Guan X Y, Sauter G, Kallioniemi O P, Trent J M, Meltzer P S. Science. 1997;277:965–968. doi: 10.1126/science.277.5328.965. [DOI] [PubMed] [Google Scholar]
- 21.Chen H, Lin R J, Schiltz R L, Chakravarti D, Nash A, Nagy L, Privalsky M L, Nakatani Y, Evans R M. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- 22.Hong H, Kohli K, Garabedian M J, Stallcup M R. Mol Cell Biol. 1997;17:2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li H, Gomes P J, Chen J D. Proc Natl Acad Sci USA. 1997;94:8479–8484. doi: 10.1073/pnas.94.16.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeshita A, Cardona G R, Koibuchi N, Suen C S, Chin W W. J Biol Chem. 1997;272:27629–27634. doi: 10.1074/jbc.272.44.27629. [DOI] [PubMed] [Google Scholar]
- 25.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 26.Chen D, Ma H, Hong H, Koh S S, Huang S M, Schurter B T, Aswad D W, Stallcup M R. Science. 1999;5423:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 27.Xu J, Qiu Y, DeMayo F J, Tsai S Y, Tsai M J, O'Malley B W. Science. 1998;279:1922–1925. doi: 10.1126/science.279.5358.1922. [DOI] [PubMed] [Google Scholar]
- 28.Qi C, Zhu Y, Pan J, Yeldandi A V, Rao M S, Maeda N, Subbarao V, Pulikuri S, Hashimoto T, Reddy J K. Proc Natl Acad Sci USA. 1999;96:1585–1590. doi: 10.1073/pnas.96.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu J, Liao L, Ning G, Yoshida-Komiya H, Deng C, O'Malley B W. Proc Natl Acad Sci USA. 2000;97:6379–6384. doi: 10.1073/pnas.120166297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bermingham J R, Jr, Scherer S S, O'Connell S, Arroyo E, Kalla K A, Powell F L, Rosenfeld M G. Genes Dev. 1996;10:1751–1762. doi: 10.1101/gad.10.14.1751. [DOI] [PubMed] [Google Scholar]
- 31.Jepsen K, Hermanson O, Onami T M, Gleiberman A S, McEvilly R J, Kurokawa R, Kumar V, Liu F, Seto E, Hedrick S M, et al. Cell. 2000;102:753–763. doi: 10.1016/s0092-8674(00)00064-7. [DOI] [PubMed] [Google Scholar]
- 32.Gleiberman A S, Kudryavtseva E I, Sharovskaya Y, Abelev G I. Mol Biol Med. 1989;6:95–107. [PubMed] [Google Scholar]
- 33.Kolch W, Philipp A, Mischak H, Dutil E M, Mullen T M, Feramisco J R, Meinkoth J L, Rose D W. Oncogene. 1996;13:1305–1314. [PubMed] [Google Scholar]
- 34.Efstratiadis A. Int J Dev Biol. 1998;42:955–976. [PubMed] [Google Scholar]
- 35.Yakar S, Liu J L, Stannard B, Butler A, Accili D, Sauer B, LeRoith D. Proc Natl Acad Sci USA. 1999;96:7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sjogren K, Liu J L, Blad K, Skrtic S, Vidal O, Wallenius V, LeRoith D, Tornell J, Isaksson O G, Jansson J O, Ohlsson C. Proc Natl Acad Sci USA. 1999;96:7088–7092. doi: 10.1073/pnas.96.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Metcalf D, Greenhalgh C J, Viney E, Willson T A, Starr R, Nicola N A, Hilton D J, Alexander W S. Nature (London) 2000;405:1069–1073. doi: 10.1038/35016611. [DOI] [PubMed] [Google Scholar]
- 38.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 39.Perou C M, Sorlie T, Eisen M B, van de Rijn M, Jeffrey S S, Rees C A, Pollack J R, Ross D T, Johnsen H, Akslen L A, et al. Nature (London) 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 40.Nishi N, Shoji H, Miyanaka H, Nakamura T. Endocrinology. 2000;141:3194–3199. doi: 10.1210/endo.141.9.7685. [DOI] [PubMed] [Google Scholar]
- 41.Bessone S, Vidal F, Le Bouc Y, Epelbaum J, Bluet-Pajot M T, Darmon M. Dev Biol. 1999;214:87–101. doi: 10.1006/dbio.1999.9379. [DOI] [PubMed] [Google Scholar]
- 42.Darmon M. Dev Biol. 1999;214:87–101. doi: 10.1006/dbio.1999.9379. [DOI] [PubMed] [Google Scholar]