Abstract
The mechanism and the site of substrate (i.e., aglycone) recognition and specificity were investigated in maize β-glucosidase (Glu1) by x-ray crystallography by using crystals of a catalytically inactive mutant (Glu1E191D) in complex with the natural substrate 2-O-β-d-glucopyranosyl-4-hydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOAGlc), the free aglycone DIMBOA, and competitive inhibitor para-hydroxy-S-mandelonitrile β-glucoside (dhurrin). The structures of these complexes and of the free enzyme were solved at 2.1-, 2.1-, 2.0-, and 2.2-Å resolution, respectively. The structural data from the complexes allowed us to visualize an intact substrate, free aglycone, or a competitive inhibitor in the slot-like active site of a β-glucosidase. These data show that the aglycone moiety of the substrate is sandwiched between W378 on one side and F198, F205, and F466 on the other. Thus, specific conformations of these four hydrophobic amino acids and the shape of the aglycone-binding site they form determine aglycone recognition and substrate specificity in Glu1. In addition to these four residues, A467 interacts with the 7-methoxy group of DIMBOA. All residues but W378 are variable among β-glucosidases that differ in substrate specificity, supporting the conclusion that these sites are the basis of aglycone recognition and binding (i.e., substrate specificity) in β-glucosidases. The data also provide a plausible explanation for the competitive binding of dhurrin to maize β-glucosidases with high affinity without being hydrolyzed.
Glycoside hydrolases catalyze the selective hydrolysis of glycosidic bonds in oligosaccharides, polysaccharides, and their conjugates. They occur in all domains of living organisms (eubacteria, archaea, and eukarya). A nomenclature system classifying these enzymes into 82 families (1, 2) is now widely used and continuously updated at http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html.
β-Glucosidases constitute a major group among glycoside hydrolases. They have been the focus of much research recently because of their important roles in a variety of fundamental biological and biotechnological processes. For example, plant β-glucosidases have been implicated in defense against pests (3–5), lignification (6), and cell wall catabolism (7). They belong to families 1 and 3 of the glycoside hydrolases and hydrolyze either O-linked β-glycosidic bonds (β-d-glucoside glucohydrolase, EC 3.2.1.21), or S-linked β-glycosidic bonds (myrosinase, or β-d-thioglucoside glucohydrolase, EC 3.2.3.1).
In maize, β-glucosidase [2-O-β-d-glucopyranosyl-4-hydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOAGlc-hydrolase)] has two known isozymes, Glu1 and Glu2, which share 90% sequence identity (8). Similarly, sorghum also has two β-glucosidase (dhurrinase) isozymes, Dhr1 and Dhr2; they share ≈70% sequence identity with each other and with each of the two maize isozymes (ref. 9; S. Vicitphan and A.E., unpublished observations). The catalytically active form of both maize and sorghum β-glucosidases is a 120-kDa homodimer or its multimers (10, 11), which has now been confirmed by x-ray crystallography (M.C., M.C., V.Z., W. P. Burmeister, D.R.B., B.H., and A.E., unpublished data, and this study). The primary structures of maize and sorghum β-glucosidases contain the highly conserved peptide motifs TFNEP and ITENG, which make up part of a crater-shaped active site in all family 1 β-glycosidases (12–16). All family 1 β-glycosidases are “retaining,” in that the anomeric configuration of the glycone is the same in the product (β-d-glucose) as in the substrate (a β-d-glucoside). The hydrolysis of the β-glycosidic bond involves two steps (glycosylation and deglycosylation) and requires participation of an acid/base catalyst and a nucleophile, which are glutamic acids E191 and E406 (numbering of ZMGlu1, Fig. 1) and occur in the motifs TFNEP and I/VTENG, respectively, in all β-O-glucosidases. One of the most fundamental questions about β-glucosidase-catalyzed reactions is: What determines substrate specificity, including the site and mechanism of aglycone binding? Significant progress has been made in understanding the mechanism of catalysis and in defining the roles of the two glutamates within the active site in catalysis (17–20). However, there is little or no information as to how β-glucosidases recognize and interact with their substrates, specifically the aglycone moiety, which is the basis of subtle substrate specificity differences among β-glucosidases.
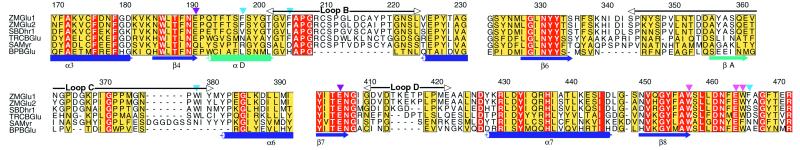
Figure 1.
Sequence alignment of selected regions from family 1 β-glycosidases, showing residues relevant for catalysis and substrate specificity. ZMGlu1, maize (Zea mays L.) Glu1; ZMGlu2, maize Glu2; SBDhr1, sorghum (Sorghum bicolor) Dhr1; TRCBGlu, white clover (Trifolium repens L.) linamarase; SAMyr, white mustard (Sinapis alba) myrosinase; BPBGlu, Bacillus polymyxa β-glucosidase. The alignment shows secondary structure elements (purple; α3, -6, and -7, and β4, -6 to -8) of the (β/α)8 barrel structure, secondary structure elements contained in loops (green; α/βA-D), the two catalytic glutamates (magenta arrowheads), highly conserved residues involved in glucose binding (pink arrowheads) and residues involved in aglycone binding (light blue arrowheads). The residue numbering is that of ZMGlu1. Red boxes denote the sites of perfect sequence identity among family 1 β-glucosidases in all three domains of living organisms, and the yellow boxes denote the sites of high sequence similarity. The figure was produced with alscript (34).
The maize β-glucosidase isozymes Glu1 and Glu2 and their sorghum homologues Dhr1 and Dhr2 constitute an ideal model system to address questions related to substrate specificity, because they represent extremes in substrate specificity. For example, Glu1 and Glu2 hydrolyze a broad spectrum of artificial and natural substrates in addition to their natural substrate DIMBOAGlc (Fig. 2A). However, Glu2 hydrolyzes certain artificial substrates (e.g., nitrophenyl glucosides) about five to six times less efficiently than Glu1, and it does not hydrolyze 6-bromo-2-naphthyl-β-d-glucoside, which is readily hydrolyzed by Glu1. Similarly, Dhr1 hydrolyzes only its natural substrate dhurrin, whereas Dhr2 hydrolyzes certain artificial substrates in addition to the natural substrate dhurrin (Fig. 2A).
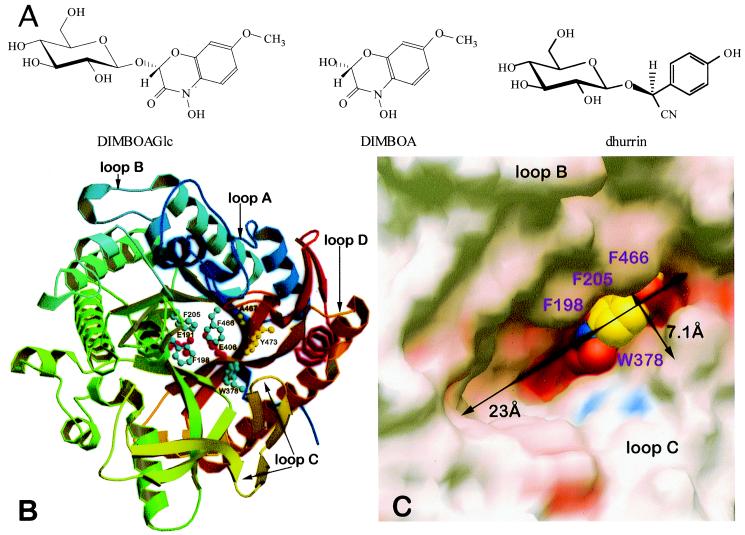
Figure 2.
Structure of the ligands and the active site of Glu1E191D. (A) The natural substrate DIMBOAGlc (Left), the aglycone DIMBOA (Center), and the competitive inhibitor dhurrin (Right). (B) Ribbon diagram of the structure of the maize β-glucosidase Glu1 and its inactive Glu1E191D mutant, showing the catalytic residues E191 (D191 in the mutant) and E406 (red), four residues (F198, F205, W378, and F466) forming the aglycone-binding pocket (blue), and two other residues (A467 and Y473) that are probably important for aglycone recognition (yellow). Different colors and the color transitions in α-helices and β-strands trace the polypeptide backbone in the barrel-shaped three-dimensional structure from the N terminus (dark blue) to the C terminus (dark red) direction. The figure was produced with molscript (35) and raster3d (36). (C) Electrostatic surface representation of the active site region of Glu1E191D showing positively charged regions in blue, negatively charged regions in red, and neutral regions in white. The slot-like active site, measuring 23 Å × 7.1 Å at the entrance, contains the natural substrate DIMBOAGlc in compact representation with standard atom-type colors. In this view, only the aglycone moiety is visible in its binding site as glucose is hidden below aglycone. C was produced with grasp (37).
To address questions about the mechanism and the site of substrate specificity directly, we produced an inactive form (referred as Glu1E191D hereafter) of the maize β-glucosidase isozyme Glu1 by site-directed mutagenesis and used it to obtain crystals of the enzyme-substrate (DIMBOAGlc), enzyme-aglycone [2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one (DIMBOA)], and enzyme-unhydrolyzed competitive inhibitor (dhurrin) complexes for three-dimensional analysis by x-ray crystallography. In this paper, we present data on the identity of the amino acids within the active site of a β-glucosidase that are involved in aglycone recognition and binding (i.e., substrate specificity) and provide insights into the mechanisms of enzyme–substrate complex formation, inhibition, and catalysis.
Materials and Methods
Production and Purification of Glu1E191D Inactive Mutant in Escherichia coli.
The region of the glu1 (GenBank no.U25157) cDNA coding for the mature Glu1 was cloned and expressed in E. coli as described (21). To obtain a catalytically inactive form of Glu1, the codon for the acid-base catalyst E191 in the glu1 cDNA was changed to that for aspartic acid by site-directed mutagenesis, and the resulting mutant cDNA was cloned and expressed as described (21). The Glu1E191D protein was purified to homogeneity according to the procedure described (22). It was then dissolved (15 mg/ml) in 20 mM Hepes, pH 7, and used in the crystallization trials. The natural substrate DIMBOAGlc, its free aglycone DIMBOA, and the unhydrolyzed substrate analogue (inhibitor) dhurrin were purified from methanol extracts of etiolated, frozen seedlings as described (9, 22). Purified DIMBOAGlc, DIMBOA, and dhurrin were dissolved in 20 mM Hepes buffer (pH 7.09) for use in the crystallization experiments.
Crystallization and Structure Determination.
All crystallization trials were performed by the vapor diffusion method (hanging drops). Crystals of the Glu1E191D mutant were obtained in 0.1 M Hepes (pH 7.5) containing 22% polyethylene glycol 4000 and 5% isopropanol. The crystals belong to space group P212121 and have unit cell parameters a = 92.1 Å, b = 94.9 Å, and c = 117.5 Å. Cocrystallization trials involving all three ligands (i.e., DIMBOAGlc, DIMBOA, and dhurrin) were set up at different ligand concentrations, which ranged from 1 to 3 molar equivalents of the ligand with respect to the protein. These conditions yielded cocrystals only for the Glu1E191D–dhurrin complex. The Glu1E191D–DIMBOAGlc and Glu1E191D–DIMBOA complexes were obtained in soaking experiments, in which 92 μl of the crystallization solution was mixed with 5 μl 5% glycerol (the cryoprotectant) and 3 μl ligand (10 mM) solution. Subsequently, 45.5 μl of this mixture was supplemented with 2.5 μl 5% glycerol and 2 μl ligand solution. The crystals were soaked in these two solutions for 15 min and then frozen in a stream of N2 at 100 K. All data sets were collected at the European Synchrotron Radiation Facility (Grenoble, France) on beam-line ID14-EH2. The data sets were treated with the program denzo (23) and scaled with scala (24). The statistics on the data collections are given (see Table 1, which is published as supplemental data on the PNAS web site, www.pnas.org).
The molecules of the Glu1E191D mutant were positioned in the new unit cell with respect to the native β-glucosidase by molecular replacement by using the program amore (25) and the native maize β-glucosidase (Protein Data Bank ID code 1e1e) as the model. Two solutions were obtained, which correspond to the two molecules in the asymmetric unit related by a noncrystallographic 2-fold symmetry, with a correlation coefficient of 60% and an R factor of 38.7% in the range of resolution 10.0–2.6 Å. The complexes have been analyzed with sigmaa (24) weighted Fobs − Fcalc maps calculated with model phases. The model is based on the native structure from which water and glycerol molecules in the active site have been removed. A direct interpretation of a difference–Fourier synthesis was hampered by the presence of a glycerol molecule and well-ordered water molecules in the active site. All four structures were refined with the program cns (26), restraining the noncrystallographic symmetry (supplemental data). The final R and Rfree factors for the uncomplexed Glu1E191D are 20.7% and 24.4%, and for Glu1E191D in complex with DIMBOAGlc, DIMBOA, and dhurrin, the values are 22.8 and 26.7, 21.8 and 26.6, and 19.6 and 23.5%, respectively.
Results
The Overall Structure of Glu1E191D Inactive Mutant.
The Glu1E191D mutant had no detectable catalytic activity on any of the substrates hydrolyzed by wild-type Glu1. The three-dimensional structure of Glu1E191D and its complexes, resolved at 2.2–2.0 Å, are identical to that of the wild-type enzyme whose structure was solved recently (M.C. et al., unpublished data). Both forms have the classical (β/α)8 barrel fold, where β strands and α helices within each β/α repeat are connected by loops at the top of the barrel (Fig. 2B). The quaternary structure of Glu1E191D is a 120-kDa homodimer, as is its parental wild-type enzyme.
Active-Site Architecture.
Four extended loops (A, B, C, and D in Fig. 2B) form a cleft-like gate to the active site of Glu1 (M.C. et al., unpublished data) and its Glu1E191D mutant, as in other family 1 enzymes. These loops are the sites of highest variability in β-glucosidase sequences. The catalytic machinery, formed by the conserved acid/base (E191) and nucleophilic (E406) glutamates, is invariably located at the bottom of the active-site pocket.
The active site of Glu1 and its inactive Glu1E191D mutant (Fig. 2C) resemble a flattened crater or slot in which one can recognize two distinct regions starting from its surface entrance and proceeding down to the bottom. The first region, about half the depth of the active site, is identified as the aglycone-binding pocket and is like a slot 23 Å long and 7.1 to 7.6 wide, where one finds F198, F205, and F466 on one side and W378 on the other. The aglycone occupies only the narrowest part of the aglycone-binding pocket (Fig. 2C). The second region, the bottom half of the active site slot, is the glycone-binding site. This buried region, 11.6 Å × 9.6 Å, has a larger width than the aglycone-binding site and contains the residues that are involved in glycone binding and catalysis. When looking down the active site from above, the aglycone and glycone planes of the natural substrate DIMBOAGlc are positioned end to end, such that the glycone is completely hidden by the aglycone (Fig. 2C).
The Mechanism of Substrate (Aglycone) Specificity.
The catalytically inactive Glu1E191D mutant–DIMBOAGlc, –DIMBOA, and –dhurrin complexes are the first glycoside hydrolase–substrate, aglycone, or substrate analogue complexes in which the aglycone moiety has been visualized in the active site of a family 1 enzyme. The electron density of the difference–Fourier maps (see Fig. 5, supplemental data), observed for the different complex structures and contoured at 1.8σ level, showed that the active sites are occupied by DIMBOAGlc, DIMBOA and dhurrin molecules, respectively. The atomic occupation parameters have been refined as one grouped variable for each complexed molecule and led to the values of 0.7, 0.8, and 0.6 for DIMBOAGlc, DIMBOA and dhurrin, respectively. However, not all of the atoms of the molecules could be defined by electron density. In all three complexes, the aglycone moiety is better defined than the glycone moiety. In the complex structure Glu1E191D-DIMBOA (Fig. 5B, supplemental data), the aglycone moiety lacks definition for the 2-hydroxyl group, some atoms of the aromatic ring, and the 7-methyl group. This lack of definition is either because of the lowered occupation or some positional disorder of this molecule in the aglycone-binding pocket or a combination of both.
The electron density in the glycone-binding pocket of this complex was modeled as water molecules and could also be caused, to some extent, by the presence of glycerol, which was used as the cryoprotectant. However, the different appearance of the electron density in the active site of Glu1E191D in complex with DIMBOAGlc (Fig. 5A, supplemental data), in comparison to the complex with DIMBOA alone, indicates that the unhydrolyzed substrate is present. The density is sufficiently continuous to account for most of the atoms of the O-glycosidic linkage and the glucopyranoside ring. However, no clearcut conformation can be modeled to bring all atoms into the existing electron density. Mainly, O5, C6, O6, C5, O4, and O3 are defined by the electron density, whereas C4, C3, and C2 lack electron density. The atoms O2, C1, and O1 are slightly displaced with respect to the electron density present in the glycone-binding pocket of the active site. The models of two conformations for the glucopyranoside ring, 4C1 chair- and the 1S3 skew-boat, led to identical R and Rfree values. The electron density is probably the trace of the mean value of these two conformations and possibly all intermediate conformations. Because the occupation of the molecules in the active site is not 100%, the superimposition with “empty” active sites that contain water molecules and/or glycerol is also a plausible explanation for the lack of definition. Furthermore, the soaking experiments lead to slightly mosaic crystals, reflected by low quality Rsym values at high resolution. This also explains less well-defined electron density.
In the active site of the structure of Glu1E191D complexed with dhurrin (Fig. 5C, supplemental data), the electron density for the glucopyranoside ring is also lacking several atoms. Here, O1, C1, O5, C5, C6, O6, C4, and O2 are defined by density, but the connection C1–O1 is missing, as well as O4, C3, and C2. The continuous electron density is not located in exactly the same position as in the active site of the complex structure of Glu1E191D with DIMBOAGlc. A separated electron density, in the position where O4 normally stands probably corresponds to a water molecule. The aglycone moiety of dhurrin is rather well defined, except for the para-hydroxyl group, indicating that the aromatic ring has limited rotational freedom.
In each of these complexes, the aglycone moiety is unequivocally found sandwiched in a slot formed by W378 on one side and F198, F205, and F466 on the other side. The natural aglycone DIMBOA fits best into the aglycone-binding site, because its molecular curvature follows the hydrophobic surface formed by the edges of the three phenyl rings, and its volume neatly fills the space of the slot. Although the interactions of both free DIMBOA and DIMBOAGlc within the aglycone pocket are similar, they are not identical, because the molecular plane of free DIMBOA differs from that of DIMBOAGlc by about 1 Å, and it has a 13° maximum angle of rotation in the plane (Fig. 3 B and C).
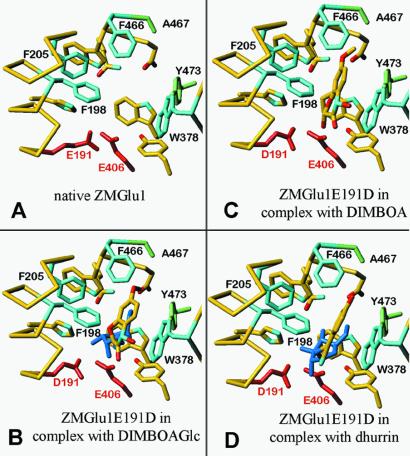
Figure 3.
Aglycone recognition and binding in β-glycosidases as revealed by DIMBOAGlc–, DIMBOA–, and dhurrin–Glu1E191D inactive mutant complexes. (A) Closeup view of the active site of Glu1, showing the catalytic glutamates E191 and E406 (red), the four residues (F198, F205, W378, and F466) forming the aglycone-binding pocket (light blue), and the additional residues (A467 and Y473) that are probably important for aglycone recognition (light green). (B) Glu1E191D with bound DIMBOAGlc. The glycone moiety is in blue, whereas the aglycone is in atom-type colors. The bulky aryl group is sandwiched between W378 on one side and F198, F205, and F466 on the other. (C) Same as B but with bound DIMBOA, showing a slightly different orientation than DIMBOA in DIMBOAGlc, which is constrained by the glycosidic linkage. (D) Same as B but with bound dhurrin. The aglycone moiety of the inhibitor dhurrin is in the same position as the aglycone of the natural substrate. Figs. 3. and 4 were produced with turbo-frodo (38).
The major mechanism of aglycone recognition and specificity appears to be aromatic stacking and π-interactions between DIMBOA or other aromatic aglycones and W378, F198, F205, and F466 (Fig. 3 C and D). In addition, the OH-group of Y473 forms a hydrogen bond with W378 NE1 (3.74 Å), and in this way, Y473 may indirectly affect substrate specificity and catalytic efficiency because it orients the plane of W378 for optimum interaction with the aglycone moiety of DIMBOAGlc. Finally, A467 is in close contact with the 7-methoxy group of DIMBOA (A467CB-methyl 3.97 Å), which might also provide a stabilizing interaction. Surprisingly, none of the polar groups of any of the aglycone moieties forms hydrogen bonds with the amino acid side chains lining the aglycone-binding site. It must be emphasized that the disulphide bridge between C210 and C216 stabilizes the loop containing F198 and F205, and this loop shields the cluster of the three phenylalanines (F198, F205, and F466) from the solvent. Another source of stabilization for the three phenylalanines that interact directly with DIMBOA is a large hydrophobic cluster formed by F56, W53, W143, F195, and W465. These five residues are within ≈5 Å around F205 and, interestingly, they are invariant in maize and sorghum enzymes.
Our data from the Glu1E191D–dhurrin complex show clearly that the dhurrin aglycone, parahydroxy-(S)-mandelonitrile, binds at the same site as does DIMBOA in the aglycone-binding pocket (Figs. 3D and 4). It appears that the aromatic group of the aglycone moiety is responsible for the positioning of the entire dhurrin molecule. Consequently, the glucose moiety of dhurrin is not correctly positioned, forming longer or shorter hydrogen bonding distances with the glycone binding residues (see below) than are seen with DIMBOAGlc (Table 3, supplemental data). Therefore, the glycosidic bond is not at the same position with respect to the catalytic residues (i.e., E191 and E406) as in the complex with the substrate DIMBOAGlc (Fig. 4 A and B).
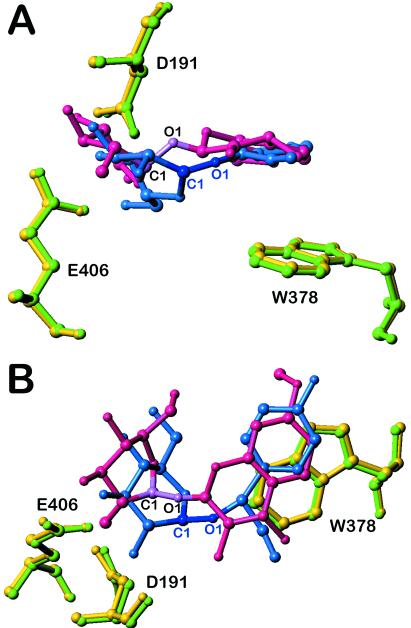
Figure 4.
Superimposition of the natural substrate DIMBOAGlc and the competitive inhibitor dhurrin in the active site of Glu1E191D. The catalytic residues E191 (D) and E406 and the conserved aglycone-binding residue W378 are in yellow (for Glu1E191D–DIMBOAGlc complex) and in green (for Glu1E191D–dhurrin complex), DIMBOAGlc in pink and dhurrin in blue. Two views (A, side view; B, top view, perpendicular to A) show that the three amino acids perfectly superimpose in both complexes, and the aromatic aglycone ring of dhurrin is almost at the same position as that of DIMBOAGlc. However, the anomeric carbon C1 and the O-glycosidic linkage at O1 do not superimpose; they are pulled away from the catalytic residues E406 and E191 (D) in dhurrin, precluding hydrolysis.
The binding of the glycone (glucose) moiety involves predominantly hydrogen bonding interactions with a set of universally conserved amino acids such as Q38 (H-bonded to O4), H142 (H-bonded to O3), N190 (H-bonded to O3), D191 (H-bonded to O2), and E464 (H-bonded to O6). Moreover, W465 occupies a position right above O4 and separates the glycone-binding pocket at that end from the aglycone-binding pocket like a halfway valve. The ring-plane of W465 is perpendicular to that of glucose (no hydrophobic interaction), and the NE1 forms a hydrogen bond with O4.
Discussion
The importance of the aglycone moiety for substrate specificity in β-glycosidases in general and plant β-glucosidases in particular has long been documented (27, 28). It is also intuitively apparent that the glycone moiety (i.e., glucose) of the substrate is invariant and therefore cannot be the basis for substrate specificity. The question of how β-glucosidases recognize and bind their specific substrates has defied answer in the past. This is because crystals of a β-glucosidase in complex with a substrate or substrate analogue have not been available to address the question until the present study. Crystals of the Glu1E191D mutant enzyme–DIMBOAGlc, DIMBOA–, and –dhurrin complexes allowed us to identify not only the aglycone-binding pocket of the active site but also the specific amino acids that form such a pocket, as well as those that directly or indirectly interact with the aglycone moiety of the natural substrate. Most importantly, the identification of the aglycone-binding pocket now provides a basis for rational explanation of aglycone (substrate) specificity differences not only between two maize isozymes but also those between sorghum and maize β-glucosidases.
The Active-Site Structure.
The active site of maize β-glucosidase is a highly organized cleft starting on the surface with a wide entrance gate followed by specialized aglycone- and glycone-binding pockets, respectively. The aglycone-binding pocket is well suited to accommodate aryl groups varying in bulkiness by sandwiching them between two hydrophobic “walls” formed by W378 on one side and F198, F205, and F466 on the other (Fig. 2C). The glycone-binding pocket contains the highly conserved residues Q38, H142, E191(D), E406, E464, and W465, as in all family 1 enzymes, which form hydrogen bonds to the hydroxyl groups of the glucopyranoside ring. The orientation of the glucose ring in the DIMBOAGlc complex and its hydrogen bonding network are very similar to those observed in the complexes of myrosinase, trapped as the glycosyl-enzyme intermediate, and the β-glucosidase from Bacillus polymyxa with d-glucono-1,5-lactone (13, 14).
The Aglycone-Binding Pocket.
The alignment of 61 family 1 plant β-glucosidase sequences available in public databases (not shown) reveals that the sites homologous to three of the aromatic residues (F198, F205, and F466) involved in binding to the aglycone of DIMBOAGlc are not conserved, whereas the fourth aromatic residue (W378) is highly conserved. These four hydrophobic residues form a well-tailored pocket for interaction with the bulky aryl aglycone moiety of the natural substrate DIMBOAGlc. Four additional residues (T334, M374, Y473, and A467) may also affect aglycone binding. Of these, T334 and M374 are not in the pocket, but they are involved in the positioning of loop C, which contains W378, needed for substrate binding. In contrast, Y473 and A467 are in the aglycone-binding site; they probably maintain the hydrophobic integrity of the aglycone-binding pocket for binding to the natural substrate. A467 is likely to be critical to DIMBOAGlc specificity, because only the maize isozymes Glu1 and Glu2 and the noncyanogenic β-glucosidase of white clover have alanine in this position among 61 plant β-glucosidases examined. That the wild-type Glu1 hydrolyzes p-nitrophenyl glucoside (pNPGlc) and oNPGlc with high catalytic efficiency and that the aglycones of these NPGlcs are strong competitive inhibitors suggest that the aglycone-binding pocket of Glu1 has considerable flexibility for binding aryl groups less bulky than DIMBOA.
It should be noted that the maize isozyme Glu2 has a Y at position F466 of Glu1, and the sorghum isozymes Dhr1 and Dhr2 (not shown) each have V/L, L, and S, respectively, at sites homologous to F198, F205, and F466 of Glu1 (Fig. 1). As stated before, all three enzymes (Glu2, Dhr1, and Dhr2) differ from Glu1 with respect to substrate specificity. Thus, the alignment and crystallographic data together establish the importance of W378, F198, F205, and F466 and their homologues in determining substrate (aglycone) specificity and explain the basis of the subtle aglycone (substrate)-specificity differences among β-glucosidases.
Our data from the Glu1E191D–dhurrin complex now provide a plausible explanation for the basis of dhurrin binding to the enzyme (Ki = 76 μM) as well as its lack of hydrolysis (22). The crystallographic data show that the interaction of the parahydroxy-S-mandelonitrile moiety of dhurrin with the aglycone-binding pocket is such that it forces the glucose ring to take a position slightly different from that observed with the natural substrate DIMBOAGlc (Figs. 3D and 4). This change in orientation is because of the aglycone moiety, which pulls the glucopyranoside ring out of the glycone-binding pocket. The major consequence is that the two atoms (the anomeric C1 and the O1 of the sugar) that are crucial for hydrolysis are positioned too far from the catalytic residues, and this is most likely why dhurrin acts as a competitive inhibitor and is not hydrolyzed.
The Mechanism of Substrate Specificity, Binding, and Catalysis.
Our interpretation of the electron density in the active site of the structures of the three complexes leads us to postulate that it is primarily the aglycone binding that is responsible for substrate specificity. That we have not been able to define a specific conformation, distorted or not, of the glycone ring in any of the enzyme–substrate or substrate analogue complexes suggests a conformational flexibility of the glucose ring. This is likely because the active site must be able not only to recognize the ground state (in 4C1, chair conformation), but also a distorted skew-boat conformation, such as that found in unhydrolyzed enzyme–substrate and enzyme–inhibitor complexes of a chitobiase and of retaining cellulases (29, 30). Thus, the density we observe would be the average of several conformations that are at different points on the path from the ground state to the transition state, where such atoms like the O6, O4, and O3 and most of the aglycone atoms undergo less displacement and are therefore more clearly defined by density. In fact, previous studies (31) showed that glucose alone was a poor competitive inhibitor of Glu1 (Ki = 170 mM), suggesting that glucose cannot bind tightly without an aglycone attached to it. That d-glucono-1,5-lactone, which inherently has a half-chair conformation and is a transition state analogue, is a better competitive inhibitor than glucose for Glu1 by about 3,700 times (31), suggests that the ground state of the glycone-binding pocket has a conformation favorable to binding a glycone in the half-chair conformation either directly or through induced fit. Thus, the lack of definition in the glucose-ring based on the electron density for the complexes of Glu1E191D with dhurrin and DIMBOAGlc suggests that the glucose moiety is deformed during catalysis (32, 33), and a weakly bound glucose is necessary for stabilizing several reaction intermediates.
In conclusion, we believe that it is the specific conformations of the four key amino acids (W378, F198, F205, and F466) and the shape of the aglycone-binding pocket they form that determines aglycone recognition and substrate specificity in maize Glu1. Besides these four residues, only A467 interacts directly with DIMBOA. That these sites, with the exception of W378, are highly variable among β-glucosidases supports the conclusion that they are involved in aglycone recognition and binding in all family 1 enzymes and should be the target of future studies aimed at altering substrate specificity through genetic engineering. Although W378 is highly conserved among family 1 β-glucosidases, it may still be important for specific recognition of the aglycone moiety, because its specific conformation may vary depending on the nature of neighboring amino acids with which it has contact. Apparently W378 interacts with the aglycone moieties (e.g., p- or o-nitrophenyl, 6-bromonaphthyl, indoxyl, methylumbelliferyl) of all substrates hydrolyzed by Glu1 as well as with those of unhydrolyzed competitive inhibitors (e.g., dhurrin). However, stacking may not be as perfect with other aglycones as with the aglycone (DIMBOA) of the natural substrate DIMBOAGlc. In fact, the stabilization of the loop B by the disulfide bond between C210 and C216 and the kink produced by P377 that is unique to maize β-glucosidases and precedes W378 may position W378 for “perfect” stacking with DIMBOA. Future studies will introduce amino acid substitutions by site-directed mutagenesis in each of the substrate-specificity determining positions identified in our study. These studies should clarify further the relative importance and contributions of each of the residues that interact with the aglycone moiety and are implicated in substrate specificity.
Supplementary Material
Acknowledgments
We thank S. Arzt and E. Mitchell at the European Synchrotron Radiation Facility for assistance on beam-line ID14-EH2. This research is supported by National Science Foundation Grants MCB-9906698 and INT-991088 and Centre National de la Recherche Scientifique (INT-9147) Cooperative Research Program.
Abbreviations
- DIMBOA
2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one
- Glc
glucose
- Glu1
maize β-glucosidase-1
- Glu1E191D
inactive Glu1 mutant
- Glu2
maize β-glucosidase-2
- Dhr
sorghum β-glucosidase dhurrinase
Footnotes
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes GluE191D 1e4n; GluE191D + DIMBOA 1e4l; GluE191D + DIMBOAGlc 1e56; and GluE191D + dhurrin 1e55).
References
- 1.Henrissat B. Biochem J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henrissat B, Davies G J. Curr Opin Struct Biol. 1997;7:637–644. doi: 10.1016/s0959-440x(97)80072-3. [DOI] [PubMed] [Google Scholar]
- 3.Conn E E. In: Biochemistry of Plants. Stumpf P K, Conn E E, editors. Vol. 7. New York: Academic; 1981. pp. 479–500. [Google Scholar]
- 4.Niemeyer H M. Phytochemistry. 1988;27:3349–3358. [Google Scholar]
- 5.Poulton J E. Plant Physiol. 1990;94:401–405. doi: 10.1104/pp.94.2.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dharmawardhana D P, Ellis B E, Carlson J E. Plant Physiol. 1995;107:331–339. doi: 10.1104/pp.107.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leah R, Kigel J, Svendsen I, Mundy J. J Biol Chem. 1995;270:15789–15797. doi: 10.1074/jbc.270.26.15789. [DOI] [PubMed] [Google Scholar]
- 8.Bandaranayake H, Esen A. Plant Physiol. 1996;110:1048. [Google Scholar]
- 9.Cicek M, Esen A. Plant Physiol. 1998;116:1469–1478. doi: 10.1104/pp.116.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hösel W, Tober I, Eklund S H, Conn E E. Arch Biochem Biophys. 1987;252:152–162. doi: 10.1016/0003-9861(87)90019-1. [DOI] [PubMed] [Google Scholar]
- 11.Esen A, Gungor G. In: β-Glucosidases: Biochemistry and Molecular Biology. Esen A, editor. Washington, DC: Am. Chem. Soc.; 1993. pp. 214–239. [Google Scholar]
- 12.Barrett T, Suresh C G, Tolley S P, Dodson E J, Hughes M A. Structure (London) 1995;3:951–960. doi: 10.1016/s0969-2126(01)00229-5. [DOI] [PubMed] [Google Scholar]
- 13.Burmeister W P, Cottaz S, Driquez H, Iori R, Palmieri S, Henrissat B. Structure (London) 1997;5:663–675. doi: 10.1016/s0969-2126(97)00221-9. [DOI] [PubMed] [Google Scholar]
- 14.Sanz-Aparicio J, Hormoso J A, Martinez-Ripoll M, Lequerica J L, Polaina J. J Mol Biol. 1998;275:491–502. doi: 10.1006/jmbi.1997.1467. [DOI] [PubMed] [Google Scholar]
- 15.Davies G, Henrissat B. Structure (London) 1995;3:853–859. doi: 10.1016/S0969-2126(01)00220-9. [DOI] [PubMed] [Google Scholar]
- 16.Aguilar C F, Sanderson I, Moracci M, Ciaramella M, Nucci R, Rossi M, Pearl L H. J Mol Biol. 1997;271:789–802. doi: 10.1006/jmbi.1997.1215. [DOI] [PubMed] [Google Scholar]
- 17.Withers S G, Warren R A J, Street I P, Rupitz K, Kempton J B, Aebersold R. J Am Chem Soc. 1990;112:5887–5889. [Google Scholar]
- 18.Wang Q, Trimbur D, Graham R, Warren R A J, Withers S G. Biochemistry. 1995;34:14554–14562. doi: 10.1021/bi00044a034. [DOI] [PubMed] [Google Scholar]
- 19.Moracci M, Capalbo L, Ciaramella M, Rossi M. Protein Eng. 1996;9:1191–1195. doi: 10.1093/protein/9.12.1191. [DOI] [PubMed] [Google Scholar]
- 20.Cottaz S, Henrissat B, Driguez H. Biochemistry. 1996;35:15256–15259. doi: 10.1021/bi9622480. [DOI] [PubMed] [Google Scholar]
- 21.Cicek M, Esen A. Biotechnol Bioeng. 1999;63:392–400. doi: 10.1002/(sici)1097-0290(19990520)63:4<392::aid-bit2>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 22.Cicek M, Blanchard D J, Bevan D R, Esen A. J Biol Chem. 2000;275:20002–20011. doi: 10.1074/jbc.M001609200. [DOI] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter C W, Sweet R M, editors. 276, Part A. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 24.CCP4 (Collaborative Computational Project Number 4) Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 25.Navazza J. Acta Crystallogr A. 1994;50:157–163. [Google Scholar]
- 26.Brünger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M, Pannu N S, et al. Acta Crystallogr D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 27.Hösel W, Conn E E. Trends Biochem Sci. 1982;6:219–221. [Google Scholar]
- 28.Conn E E. In: β-Glucosidases: Biochemistry and Molecular Biology. Esen A, editor. Washington, DC: Am. Chem. Soc.; 1993. pp. 15–26. [Google Scholar]
- 29.Tews I, Perrakis A, Oppenheim A, Dauter Z, Wilson K S, Vorgias C E. Nat Struct Biol. 1996;3:638–648. doi: 10.1038/nsb0796-638. [DOI] [PubMed] [Google Scholar]
- 30.Sulzenbacher G, Schülein M, Davies G J. Biochemistry. 1997;36:5902–5911. doi: 10.1021/bi962963+. [DOI] [PubMed] [Google Scholar]
- 31.Babcock G D, Esen A. Plant Sci. 1994;101:31–39. [Google Scholar]
- 32.Davies G J, Dauter M, Brzozowski A M, Bjornvad M E, Anderson K V, Schülein M. Biochemistry. 1998;37:1926–1932. doi: 10.1021/bi972162m. [DOI] [PubMed] [Google Scholar]
- 33.Davies G J, Mackenzie L, Varrot A, Dauter M, Brzozowski A M, Schülein M, Withers S G. Biochemistry. 1998;37:11707–11713. doi: 10.1021/bi981315i. [DOI] [PubMed] [Google Scholar]
- 34.Barton G J. Protein Eng. 1993;6:37–40. doi: 10.1093/protein/6.1.37. [DOI] [PubMed] [Google Scholar]
- 35.Kraulis P J. J Appl Crystallogr. 1991;24:946–947. [Google Scholar]
- 36.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 37.Nicholls A, Sharp K A, Honig B. Proteins. 1991;11:281–296. doi: 10.1002/prot.340110407. [DOI] [PubMed] [Google Scholar]
- 38.Roussel A, Cambillau C. TURBO-FRODO, Silicon Graphics Geometry Partners Directory. Mountain View, CA: Silicon Graphics; 1991. pp. 86–87. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.