Abstract
The genome of Arabidopsis (Arabidopsis thaliana) encodes over 100 MADS-domain transcription factors, categorized into five phylogenetic subgroups. Most research efforts have focused on just one of these subgroups (MIKCc), whereas the other four remain largely unexplored. Here, we report on five members of the so-called Mδ or Arabidopsis MIKC* (AtMIKC*) subgroup, which are predominantly expressed during the late stages of pollen development. Very few MADS-box genes function in mature pollen, and from this perspective, the AtMIKC* genes are therefore highly exceptional. We found that the AtMIKC* proteins are able to form multiple heterodimeric complexes in planta, and that these protein complexes exhibit a for the MADS-family unusual and high DNA binding specificity in vitro. Compared to their occurrence in promoters genome wide, AtMIKC* binding sites are strongly overrepresented in the proximal region of late pollen-specific promoters. By combining our experimental data with in silico genomics and pollen transcriptomics approaches, we identified a considerable number of putative direct target genes of the AtMIKC* transcription factor complexes in pollen, many of which have known or proposed functions in pollen tube growth. The expression of several of these predicted targets is altered in mutant pollen in which all AtMIKC* complexes are affected, and in vitro germination of this mutant pollen is severely impaired. Our data therefore suggest that the AtMIKC* protein complexes play an essential role in transcriptional regulation during late pollen development.
MADS-domain transcription factors play key roles in the development of higher eukaryotes. Their characteristic feature is the N-terminal MADS-domain, which is responsible for DNA binding and is highly conserved among fungi, animals, and plants. As homo- or heterodimeric complexes, and in some cases as higher-order complexes, MADS-proteins regulate gene expression by binding to CArG-box motifs in promoter regions (Egea-Cortines et al., 1999; Honma and Goto, 2001; Theissen and Saedler, 2001). The so-called serum response element (SRE)-type CArG-box, CC(A/T)6GG, is the DNA motif preferred by most MADS-protein complexes investigated to date (Hayes et al., 1988; Riechmann et al., 1996; de Folter and Angenent, 2006). However, some MADS-protein complexes, such as mammalian myocyte enhancer factor-2A, preferentially bind the related MEF2- or N10-type CArG-box. This motif has as general consensus C(A/T)8G but is usually more strictly defined as CTA(A/T)4TAG (Pollock and Treisman, 1991; Shore and Sharrocks, 1995). The embryo-specific AGAMOUS-LIKE 15 (AGL15) protein from Arabidopsis (Arabidopsis thaliana) and SQUAMOSA from Antirrhinum majus both have a broader spectrum of binding sites, recognizing SRE-, MEF2-, and intermediate motifs (West et al., 1998; Tang and Perry, 2003). In spite of the rather common occurrence of CArG-boxes in promoters, MADS-protein complexes are quite specific in recognizing their target promoters. Although different homodimeric MADS-protein complexes have been crystallized together with their preferred binding motif (e.g. Santelli and Richmond, 2000), the structural basis of these differences in binding specificity is still not fully understood.
In contrast to the situation in animals and fungi, the MADS-family has undergone a spectacular expansion during the evolution of plants. An initial expansion is already apparent in gymnosperms (Theissen et al., 2000; Becker and Theissen, 2003; De Bodt et al., 2003), but in angiosperms it is far more pronounced (see, e.g., Martinez-Castilla and Alvarez-Buylla, 2003; Nam et al., 2004; Irish and Litt, 2005; Shiu et al., 2005). The genomes of Arabidopsis and Oryza sativa encode 107 and 71 putatively functional MADS-proteins, respectively (Pařenicová et al., 2003; Nam et al., 2004), compared to only four in yeast (Saccharomyces cerevisiae), five in Homo sapiens, two in both Drosophila melanogaster and Caenorhabditis elegans, one in the alga Chlamydomonas reinhardtii, 18 in the moss Physcomitrella patens, and at least 19 in Gnetum gnemon (Becker et al., 2000; Nam et al., 2003; Shiu et al., 2005; Tanabe et al., 2005; D. Liebsch and T. Münster, unpublished data). MADS-proteins are therefore expected to play specialized roles in higher plant development, and several members have indeed been associated with seed plant-specific developmental programs (Theissen et al., 2000). The best-studied examples are the classical MIKC MADS-box genes (MIKCc; nomenclature according to Henschel et al., 2002) involved in floral organ patterning, flowering time, and ovule and fruit development (for review, see Theissen et al., 2000; Ng and Yanofsky, 2001).
Two major monophyletic lineages have been defined within the Arabidopsis MADS-family (Alvarez-Buylla et al., 2000b). Type I genes consist of one or two exons, while type II genes always have more than five exons. The genes from both lineages encode proteins that share the MADS-domain as common feature, but in type II proteins, three additional domains are present, namely the I-, K-, and C-domains (Münster et al., 1997). The former two domains mediate protein-protein interactions (dimerization), whereas the C-terminal domain functions in transcriptional activation and in the formation of higher-order protein complexes (Ma et al., 1991; Shore and Sharrocks, 1995). Pařenicová and co-workers (2003) further classified the Arabidopsis MADS-family and identified a total of five subgroups: three belonging to the type I lineage (the Mα, Mβ, and Mγ subgroups) and two within the type II lineage (the MIKCc and Mδ subgroups). The Mδ subgroup consists of only six genes in Arabidopsis, compared to around 40 genes in the MIKCc subgroup. An exceptionally complex intron-exon structure and the lack of a clearly identifiable K-domain are the most striking differences between the Mδ and MIKCc genes (Kofuji et al., 2003; Martinez-Castilla and Alvarez-Buylla, 2003; Nam et al., 2004).
The Arabidopsis Mδ genes are highly homologous to the MIKC* genes from the moss P. patens (Kofuji et al., 2003). The MIKC* genes in moss distinguish themselves from classical type II genes because of their longer I-domain, which is encoded by four or five exons, whereas the shorter I-domain of MIKCc proteins is encoded by a single exon (Henschel et al., 2002; Riese et al., 2005). The Arabidopsis Mδ genes also meet these criteria (Kofuji et al., 2003), and therefore we refer to them as Arabidopsis MIKC* genes (AtMIKC*). They have evolved independently from the MIKCc subgroup for the last 450 million years. Within the AtMIKC* subgroup, the AGL66, AGL104, and AGL67 genes form one monophyletic lineage, and AGL30, AGL65, and AGL94 a second one (Kofuji et al., 2003). These two lineages have previously been named S and P, respectively (Nam et al., 2004).
While the AGL67 gene is expressed in embryos and is a candidate for regulating aspects of late embryo development (de Folter et al., 2004), the five other AtMIKC* genes (AGL30, AGL65, AGL66, AGL94, and AGL104) are almost exclusively expressed in pollen (Kofuji et al., 2003; Honys and Twell, 2004). Pollen development can be divided into four stages (uninuclear microspore, bicellular pollen [BCP], tricellular pollen [TCP], and mature pollen [MPG]; Honys and Twell, 2004; McCormick, 2004), and the AtMIKC* genes are predominantly expressed from the tricellular stage onwards, after the second mitotic division (Honys and Twell, 2004; Supplemental Table S1). Intriguingly, while the MADS-family is altogether underrepresented in the pollen transcriptome (Honys and Twell, 2004), the nonclassical lineages (type I and especially AtMIKC*) are in fact overrepresented in pollen (Pina et al., 2005). The only strongly pollen-expressed MIKCc gene is AGL18 (Kofuji et al., 2003; Alvarez-Buylla et al., 2000a). The coexpression of five of the six AtMIKC* genes during late stages of pollen development suggests that they might constitute a transcription factor network in mature pollen. This assumption is strengthened by the preliminary observation that AGL65 interacts with both AGL66 and AGL104 in yeast (de Folter et al., 2005). Functional characterization of the AtMIKC* genes might therefore provide new insights in the unique transcriptome of mature pollen grains, which several recent studies found to be very different from that of other plant tissues (Becker et al., 2003; Honys and Twell, 2003, 2004; Hennig et al., 2004; Pina et al., 2005).
In a first attempt to functionally characterize the AtMIKC* proteins, we performed protein-interaction and DNA-binding studies for the five pollen-expressed members of this subgroup. We show that between members of the two monophyletic AtMIKC* lineages, five heterodimeric protein complexes can be formed, at least four of which exist in planta. They preferentially bind MEF2-type CArG-boxes in vitro, and these preferred binding motifs are strongly overrepresented in the proximal region of promoters that are activated during the last stages of pollen development, at the time of AtMIKC* expression. Our results suggest that the AtMIKC* protein complexes play an important role in the regulation of transcription during late pollen development, and this is confirmed by a preliminary functional analysis of mutant pollen in which all five AtMIKC* complexes are either absent or strongly reduced in abundance.
RESULTS
Cloning of AtMIKC* cDNAs from Pollen
We cloned the full-length cDNAs of five AtMIKC* genes (AGL30, AGL65, AGL66, AGL94, and AGL104) from a cDNA pool obtained from mature Arabidopsis pollen grains, using gene-specific primers. AGL30 had previously been regarded a pseudogene based on the cloning of a truncated splice variant with a stop codon in the fifth exon (Kofuji et al., 2003). However, we amplified and cloned a splice form of AGL30 that consists of 10 exons, encoding a protein with much higher homology to AGL65 and AGL94. Our AGL30 sequence is identical to the recently created GenBank accession DQ446459. Protein alignments are shown in Supplemental Figure S1.
AtMIKC* Proteins Form Specific Heterodimeric Complexes
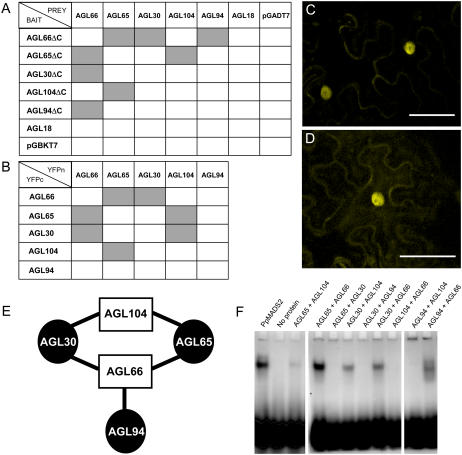
Given that MADS-proteins generally function in dimeric complexes, we tested the interactions between AGL30, AGL65, AGL66, AGL94, and AGL104 in the yeast two-hybrid (Y2H) system. All bait constructs showed autoactivation in yeast, which was abolished after deletion of a C-terminal fragment (ΔC), including suspected activation domains. Using these truncated bait constructs and full-length prey constructs, we found four reciprocally interacting pairs: AGL66 dimerizes with AGL30, AGL65, and AGL94, whereas AGL104 interacts only with AGL65. The interactions of AGL65 with AGL66 and with AGL104 in yeast was previously reported (de Folter et al., 2005). None of the AtMIKC* proteins is able to form a homodimer or to interact with AGL18, which is the only highly expressed MIKCc protein in pollen (Fig. 1A). This indicates that in spite of their cooccurrence in mature pollen, AGL18 and the AtMIKC* proteins are unlikely to interact physically, whereas specific heterodimeric complexes can be formed between the AtMIKC* proteins.
Figure 1.
Identification of AtMIKC* protein complexes. A and B, Overview of AtMIKC* protein interactions, as tested by Y2H (A) and BiFC (B). Successful interactions are represented by gray boxes. C and D, Visualization of the interaction of AGL30 with AGL66 (C) and AGL30 with AGL104 (D) in the nucleus of transiently transformed tobacco leaf cells using BiFC. Scale bars = 50 μm. E, Schematic summary of the interactions between all pollen-expressed AtMIKC* proteins. Members of the two monophyletic lineages within the AtMIKC* subgroup can be distinguished by shape and color. F, EMSA with different AtMIKC* protein combinations and a randomized mixture of SRE and MEF2 CArG-boxes as DNA probe. In the first lane, the PpMADS2 homodimer from P. patens (R. Hallinger, W. Verelst, W. Faigl, H. Saedler, and T. Münster, unpublished data) is included as a positive control. All protein combinations that can interact are also able to bind DNA, suggesting that they form functional transcription factor complexes.
To verify AtMIKC* protein interactions in planta, bimolecular fluorescence complementation (BiFC) was performed using the full-length open reading frame (ORF) sequences (Fig. 1B). The AGL30/66, AGL65/66, and AGL65/104 complexes, which we demonstrated in yeast, could be detected in the nuclei of transiently transformed tobacco (Nicotiana benthamiana) leaf cells (Fig. 1C), indicating that no pollen-specific factors are required for their targeting to the nucleus. The fluorescence signal colocalized with the 4′,6-diamidino-2-phenylindole nuclear stain and had the exact spectral properties of the yellow fluorescent protein (YFP; data not shown). We were unable to confirm interaction between AGL66 and AGL94 using this technique, even though these proteins strongly associated in yeast. On the other hand, interaction between AGL30 and AGL104, which was not evident in yeast, could be reliably detected in planta (Fig. 1D). In agreement with our Y2H results, no interaction could be observed between other AtMIKC* protein combinations. All interactions between AtMIKC* proteins are summarized in Figure 1E.
AtMIKC* Protein Complexes Preferentially Bind MEF2-Type CArG-Boxes in Vitro
To test their ability to bind DNA, the five pollen-expressed AtMIKC* proteins were synthesized in vitro in a cell-free system and used in electrophoretic mobility shift assay (EMSA) experiments. The five protein combinations capable of physical interaction (namely AGL30/66, AGL65/66, AGL94/66, AGL30/104, and AGL65/104; Fig. 1E) showed appreciable binding to a randomized mixture of SRE- and MEF2-type CArG-boxes (Fig. 1F). No DNA interaction could be observed for other AtMIKC* protein combinations (Fig. 1F) nor for any of the proteins alone (data not shown).
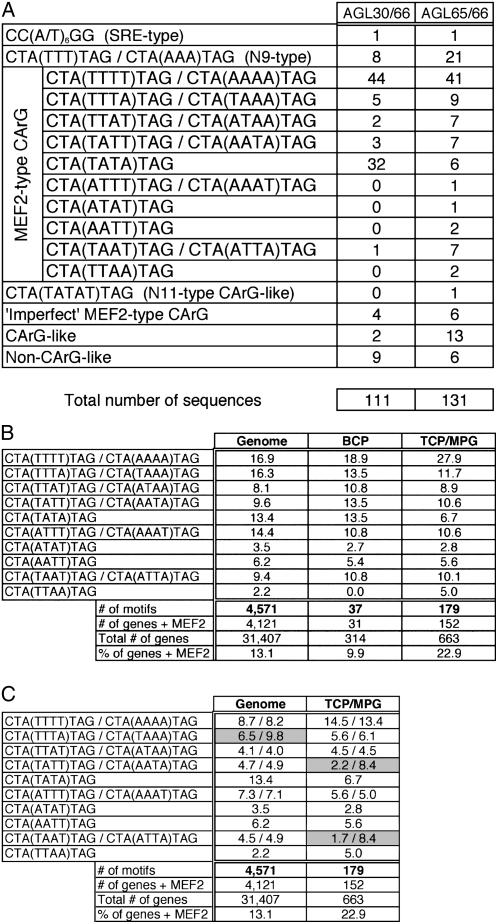
The AGL30/66 and AGL65/66 protein complexes, which exhibited the strongest DNA binding in this experiment, were chosen for a random binding site selection (RBSS) experiment to determine their preferred binding motifs. After five iterative rounds of RBSS, both complexes showed a very pronounced preference for the MEF2-type CArG-box with consensus CTA(A/T)4TAG (Fig. 2A). This motif accounted for 78% and 63% of all sequences enriched by the AGL30/66 and AGL65/66 complexes, respectively (87 out of 111 and 83 out of 131 sequences). One particular motif, CTA(TTTT)TAG/CTA(AAAA)TAG, was preferred most strongly by both complexes. The second most preferred motif was CTA(TATA)TAG, in the case of AGL30/66, and the N9-type CArG-like motif CTA(TTT)TAG/CTA(AAA)TAG for AGL65/66. Only 1% of the sequences was of the SRE type [CC(A/T)6GG], indicating that this motif is not preferred by AtMIKC* complexes. The complete RBSS datasets are listed in Supplemental Figure S2.
Figure 2.
DNA-binding preference of AtMIKC* complexes. A, RBSS results for the AGL30/66 and AGL65/66 complexes. Sequences selected by the AtMIKC* complexes are listed and subdivided into relevant categories. All original sequences are listed in Supplemental Figure S2. B, Relative occurrence (in percent) of the 16 different MEF2 CArG-boxes in Arabidopsis 3-kb promoters genome wide and in BCP and TCP/MPG-specific promoters. The total number of MEF2 motifs is mentioned, as well as the percentage of genes containing MEF2 motifs in their 3-kb upstream region. C, Relative occurrence of the two complements of each MEF2 motif in Arabidopsis 3-kb promoters (in percent of the total number of MEF2 motifs, which is indicated below the table). Motifs for which there is a clear inequality in the occurrence of both complements are shaded in gray. This analysis was irrelevant for BCP-specific promoters, due to the small number of MEF2 motifs present there (n = 37). All original values used to create this table are presented in Supplemental Table S2.
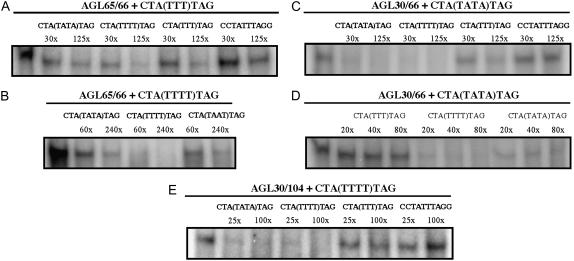
We corroborated our RBSS results with competitive EMSA experiments, confirming the strong preference of the AGL30/66 and AGL65/66 complexes for MEF2 motifs and in particular for the CTA(TTTT)TAG/CTA(AAAA)TAG motif (Fig. 3). We also used the competitive EMSA approach to investigate the DNA-binding preference of the three other AtMIKC* protein complexes. The AGL30/104 complex showed a similar behavior as AGL30/66, clearly preferring the MEF2-type CArG-box motifs CTA(TTTT)TAG and CTA(TATA)TAG to the N9-type motif CTA(TTT)TAG and the SRE-type motif CCTATTTAGG (Fig. 3). For the AGL94/66 complex, accurate quantification of DNA binding was impossible, because it produced very diffuse shifted bands in our hands and also for AGL65/104, which in general bound very weakly to DNA in vitro (Fig. 1F).
Figure 3.
Competitive EMSA for AtMIKC* complexes. A radioactively labeled DNA probe competed with an excess of unlabeled probe for binding to a protein complex. For each segment, the protein complex and labeled probe are mentioned above the horizontal line, and the different cold competitor probes and the excess at which they were used in each lane are shown under that line. A, An unlabeled CTA(TTTT)TAG probe successfully competes with a labeled CTA(TTT)TAG probe for binding to the AGL65/66 protein complex (complete competition at 125-fold excess). Unlabeled CTA(TATA)TAG and CTA(TTT)TAG probes compete to a lesser extent with CTA(TTT)TAG, while CCTATTTAGG hardly competes at all. B, CTA(TTTT)TAG can compete with itself for binding to AGL65/66, and is a stronger competitor than the other MEF2-type CArG-boxes CTA(TATA)TAG and CTA(TAAT)TAG. C, The MEF2-type probes CTA(TTTT)TAG and CTA(TATA)TAG both compete much stronger with the labeled CTA(TATA)TAG probe for binding to the AGL30/66 complex than CTA(TTT)TAG and CCTATTTAGG do. D, The AGL30/66 complex has a slightly higher affinity for CTA(TTTT)TAG than for CTA(TATA)TAG, as suggested by RBSS (Fig. 2A). E, The AGL30/104 protein complex shows a similar behavior as AGL30/66, clearly preferring the MEF2-type CArG-box motifs CTA(TTTT)TAG and CTA(TATA)TAG to the N9-type motif CTA(TTT)TAG and the SRE-type motif CCTATTTAGG.
MEF2 Motifs Are Specifically Overrepresented in Late Pollen-Specific Promoters
The AtMIKC* genes are highest expressed during the tricellular and mature stages of pollen development (Honys and Twell, 2004; Supplemental Table S1). As a consequence, the AtMIKC* protein complexes are expected to regulate transcription during that period of pollen development. Within the MADS family, they are quite exceptional in this respect; apart from AGL18, AGL29, and a small number of low expressed type-I proteins (Kofuji et al., 2003; Pina et al., 2005), they are the only MADS-proteins that are expected to function in mature pollen. In combination with our knowledge of the DNA-binding behavior of the AtMIKC* complexes in vitro, this low background of other MADS-box genes in pollen enabled us to identify genes that are potentially under direct transcriptional regulation by the AtMIKC* complexes. Such genes are expected to contain a MEF2 motif in their promoter and to be either down- or up-regulated at the time of AtMIKC* gene expression, i.e. in the tricellular and/or mature stages of pollen development.
To address this matter, we reanalyzed the pollen transcriptome dataset from Honys and Twell (2004) containing the expression levels of the 22,464 Arabidopsis genes represented on the ATH1 microarray chip (Affymetrix) in the four stages of pollen development and in seven other (nonpollen) tissues. We first selected all genes with a strongly pollen-specific expression (at least 5 times higher in one or more stages of pollen development than in any of the seven other tissues). Subsequently, we arbitrarily defined BCP-specific genes, whose expression is down-regulated at least 2-fold during the TCP stage, and TCP/MPG-specific genes, whose expression is up-regulated at least 2-fold during the TCP and/or MPG stages, relative to the BCP stage. Using these stringent criteria, we identified 314 BCP-specific and 663 TCP/MPG-specific genes (Supplemental Table S1). Because these selections were based on the ATH1 microarray chip, which covers only around 70% of the Arabidopsis genome, there are certainly more pollen-specific genes in Arabidopsis. Nevertheless, our selected gene lists can be regarded as highly representative for the entire pollen transcriptome. We verified the pollen specificity of these selected genes using Genevestigator (Zimmermann et al., 2004), which includes the pollen transcriptome dataset (mature pollen only) from the AtGenExpress developmental series (Schmid et al., 2005), which is completely independent from the dataset of Honys and Twell (2004). Most BCP-specific genes were confirmed as stamen specific, and nearly all TCP/MPG-specific genes were mature pollen specific (data not shown).
Subsequently, we screened the 3-kb upstream regions of these genes for the presence of MEF2-type CArG-boxes, and found them in 31 BCP-specific and in 152 TCP/MPG-specific promoters (Fig. 2B). Remarkably, 22.9% of the TCP/MPG-specific promoters contain a MEF2 motif, while this is only the case for 13.1% of all Arabidopsis promoters genome wide and 9.9% of the BCP-specific promoters (Fig. 2B). In contrast, SRE-type CArG-boxes, which are not bound in vitro by the AtMIKC* complexes (Fig. 2A), are overall not more abundant in BCP- and TCP/MPG-specific promoters (17.2% and 19.0%, respectively) than in all promoters genome wide, where they occur in 18.8% of all promoters (Supplemental Table S2). This percentage is in agreement with the report from de Folter and Angenent (2006). Similarly, no deviations from the genome-wide situation were observed for N9-type CArG-like motifs.
Some MEF2 Motifs Occur More Frequently in Promoters
Because AtMIKC* protein complexes are able to discriminate between the 16 different MEF2 motifs in vitro (Fig. 2A), we investigated whether these motifs occur with comparable frequencies in Arabidopsis promoters, or whether some of them occur more frequently than the rest. We first screened the 3-kb upstream regions of all Arabidopsis genes for the presence of MEF2-type CArG-boxes using the Patmatch tool on The Arabidopsis Information Resource (TAIR) Web site (www.arabidopsis.org) and identified 4,571 such motifs in 4,121 different promoters (Fig. 2B). For our study, we defined a promoter as the entire region upstream of the ATG start codon, including the 5′ untranslated region (5′UTR). We found that the CTA(TTTT)TAG/CTA(AAAA)TAG and CTA(TTTA)TAG/CTA(TAAA)TAG motifs each account for over 16% of all MEF2 motifs in 3-kb promoters, whereas the palindromic CTA(TTAA)TAG motif only represents 2% of the cases (Fig. 2B). Hence, some of the MEF2 motifs occur very frequently in Arabidopsis promoters, while others are clearly underrepresented.
Next, we repeated this analysis with all BCP- and TCP/MPG-specific promoters and found that the relative occurrence of the 16 different MEF2 motifs in these promoters differs from that in all promoters genome wide. In particular, the CTA(TTTT)TAG/CTA(AAAA)TAG motif, which is the most preferred binding motif of the AtMIKC* complexes in vitro (Fig. 2A), is strongly overrepresented in TCP/MPG-specific promoters (27.9% compared to 16.9% in all Arabidopsis promoters; Fig. 2B). None of the other MEF2 motifs shows this tendency. The palindromic CTA(TATA)TAG motif, bound quite well in vitro by the AGL30/66 complex (Fig. 2A), is underrepresented; it accounts for just 6.7% of the motifs in TCP/MPG-specific promoters, compared to 13.4% in all 3-kb promoter regions genome wide and 13.5% in BCP-specific promoters. The limited occurrence of this motif in TCP/MPG-specific (but not in BCP-specific) promoters might be related to the function of AGL18 in mature pollen. Because AGL18 is closely homologous to AGL15 (Alvarez-Buylla et al., 2000a), which shows preference for the CTA(TATA)TAG motif (Tang and Perry, 2003), it is possible that AGL18 also binds to this motif in pollen-specific promoters.
To allow comparison, we also screened all Arabidopsis 3-kb promoters for N9- and SRE-type CArG-boxes (Supplemental Table S2). The relative occurrence of the 64 possible SRE motifs is also nonrandom, with the CCTTTTTTGG/CCAAAAAAGG and CCATTTTTGG/CCAAAAATGG motifs accounting for over 8% of all SRE motifs in Arabidopsis promoter regions, while several other SRE motifs represent less than 1% of the total number. In BCP-specific, but not in TCP/MPG-specific, promoters, one particular motif (CCTTTTTTGG/CCAAAAAAGG) is strongly overrepresented (accounting for 17.5% of all SRE motifs in BCP-specific promoters and for only 8.1% in TCP/MPG-specific promoters and 8.9% in all Arabidopsis promoters; Supplemental Table S2), suggesting that it could be the preferred binding site for a non-AtMIKC* MADS-protein complex functioning during the BCP and/or TCP stage of pollen development. AGL18 would, again, be a good candidate for being part of such a complex, based on its expression profile throughout pollen development and the fact that the related AGL15 protein binds well to SRE motifs (Tang and Perry, 2003).
Whether the AtMIKC* complexes preferentially bind one of the two complements of a nonpalindromic MEF2 motif or whether they bind both complements to the same extent is a question that remained unanswered after our RBSS experiments, which are essentially nondirectional in nature. Our in silico analyses showed that the two complements of nearly all MEF2 motifs occur equally often in all promoters genome wide (Fig. 2C). In TCP/MPG-specific promoters, the CTA(TTTT)TAG and CTA(AAAA)TAG complements also occur with similar frequencies (26 and 24 times, respectively), indicating that the directionality of this motif in a promoter plays no role in its recognition by the AtMIKC* protein complexes in vivo. However, the complements of two other motifs, CTA(TATT)TAG/CTA(AATA)TAG and CTA(TAAT)TAG/CTA(ATTA)TAG, are found in quite unequal proportions in TCP/MPG-specific promoters (2.2% versus 8.4% and 1.7% versus 8.4%, respectively; Fig. 2C), suggesting that the AtMIKC* complexes might have a different affinity for binding to the two complements of these motifs. Both motifs are moderately represented in the RBSS dataset for the AGL65/66 complex (Fig. 2A).
MEF2 Motifs Are Enriched in the Proximal Region of TCP/MPG-Specific Promoters
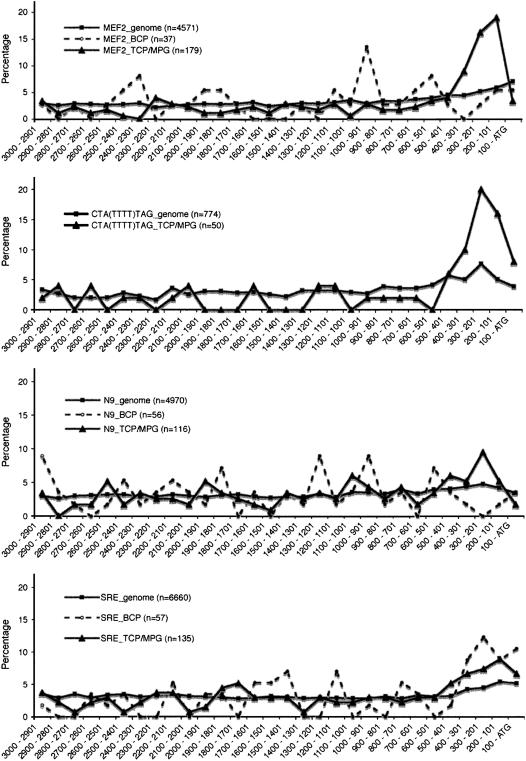
Since spatial positioning of cis-acting elements in a promoter is often an important factor in the regulation of gene expression, we investigated the spatial distribution of CArG-boxes in Arabidopsis 3-kb upstream regions. We found that MEF2, SRE, and N9-type motifs are distributed quite homogeneously across the promoters genome wide (Fig. 4). For example, 50% of all MEF2 motifs are found within 1,200 bp upstream of the ATG and 27% are positioned in the most proximal 500 bp of a promoter (Supplemental Table S2). In BCP-specific promoters, the spatial distribution of MEF2 motifs is comparable to that in the promoters genome wide. In contrast, 51% of the MEF2 motifs in TCP/MPG-specific promoters are located within 500 bp upstream of the ATG, and for the CTA(TTTT)TAG/CTA(AAAA)TAG motif, this is even 60% (Fig. 4; Supplemental Table S2). SRE motifs, in contrast, tend to have a proximal positioning in BCP-specific, but not in TCP/MPG-specific, promoters.
Figure 4.
Spatial distribution of CArG-boxes in Arabidopsis 3-kb promoters genome wide and in the promoters of all BCP- and TCP/MPG-specific genes represented on the ATH1 microarray chip. The x axis shows the distance (in base pairs) from the ATG, and the y axis the percentage of the motifs that occur in a given distance interval. The total number of motifs of each type is mentioned in the legend. All original values are presented in Supplemental Table S2.
Loss of AtMIKC* Complexes Affects Pollen Germination in Vitro
We identified T-DNA insertion lines for four of the AtMIKC* genes (as described in “Materials and Methods”) and obtained homozygous plants for each line. We then combined the different mutant alleles by crossing, ultimately resulting in the following single and double mutants: agl65, agl66, agl94, agl104, agl65/66, agl65/104, and agl66/104. Only for the agl104 mutant residual expression of the full-length transcript could be observed (Fig. 6; consistently less than 30% of the wild-type level), while the transcripts of the other AtMIKC* genes were expressed as truncated forms and/or were completely absent in their respective mutants (see “Materials and Methods”). Consistent with the male gametophyte-specific expression of the AtMIKC* genes, no sporophytic phenotype could be observed for any of the mutant lines. All mutant plants also exhibited a normal fertility.
Figure 6.
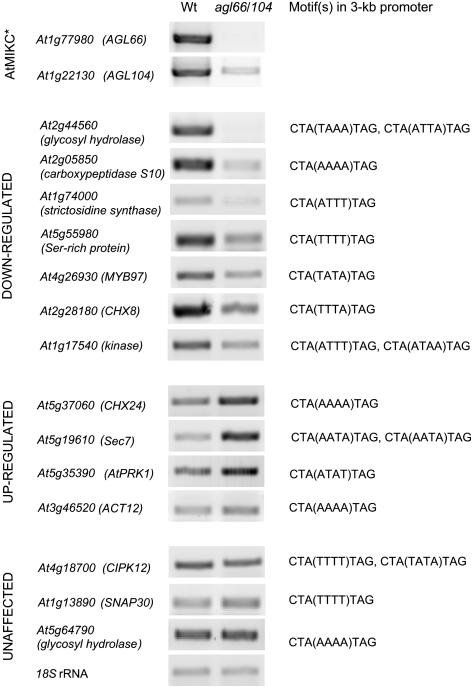
Predicted target genes are downstream of AtMIKC* complexes in vivo. RT-PCR for a selection of 14 in silico predicted direct targets of the AtMIKC* complexes, in wild-type (Wt) and agl66/104 double mutant pollen. As control, 18S ribosomal RNA is amplified, and the strongly reduced expression of the AGL66 and AGL104 transcripts in the mutant is confirmed. For each of the putative target genes, the MEF2 motif(s) present in the 3-kb promoter are listed. PCR conditions are described in “Materials and Methods.”
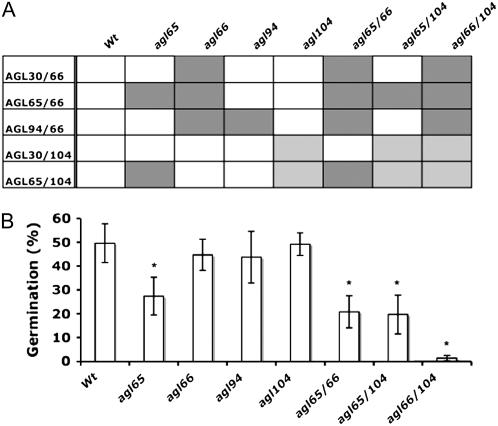
Assuming that the late pollen-expressed AtMIKC* complexes might be important for pollen germination, we performed in vitro germination assays with pollen grains from our different single and double mutant Arabidopsis lines lacking one or more functional AtMIKC* complexes. In Figure 5A, the different genotypes are listed, together with the AtMIKC* complexes still present in each mutant. Pollen viability, as scored with fluorescein-3′,6′-diacetate, was unaffected in all mutants and always higher than 90% (data not shown). Each of the AtMIKC* complexes contains either the AGL66 or AGL104 protein (Fig. 1E), and the loss of functional AGL66 (in the agl66 mutant) or the strong reduction in AGL104 abundance (in the agl104 mutant) does not affect pollen germination in vitro (Fig. 5B). This indicates that the complexes containing AGL66 are completely functionally redundant with those containing AGL104. On the other hand, loss of functional AGL65 protein (in the agl65 mutant) results in a significant reduction of pollen germination efficiency (Fig. 5B). This demonstrates that the remaining three complexes (AGL30/66, AGL94/66, and AGL30/104) are unable to completely compensate for the loss of AGL65/66 and AGL65/104 in this mutant. The AGL65/66 and AGL65/104 complexes are therefore most likely functionally redundant with each other but not with the other three AtMIKC* complexes.
Figure 5.
AtMIKC* complexes are essential for pollen germination in vitro. A, Overview of the different Arabidopsis genotypes used for in vitro pollen germination assays, and the AtMIKC* complexes present (white), strongly reduced (light gray), and absent (dark gray) in each of these mutants. B, In vitro pollen germination efficiency for various Arabidopsis genotypes lacking different combinations of AtMIKC* complexes. Asterisks indicate a statistically significant decrease in germination efficiency relative to wild-type (Wt) pollen, as found in a t test, for α = 0.001.
The additional loss of two other complexes (AGL30/66 and AGL94/66) in the agl65/66 double mutant only slightly reduces the germination efficiency further compared to the agl65 mutant, as does the additional reduction of AGL30/104 complex abundance in the agl65/104 double mutant (Fig. 5B). This observation indicates that the AGL30/66 and AGL94/66 complexes are largely functionally redundant with the AGL30/104 complex and that they are able to sustain around 40% of the wild-type pollen germination efficiency in vitro. When all five AtMIKC* complexes are affected (either functionally lost or strongly reduced in abundance, in the agl66/104 double mutant), pollen grains are virtually unable to germinate in vitro (Fig. 5B). This confirms the essential role of the AtMIKC* complexes in late pollen development and pollen tube growth.
Predicted AtMIKC* Target Genes Are Downstream of AtMIKC* Complexes in Vivo
The 152 TCP/MPG-specific genes with a MEF2 motif in their 3-kb promoter (listed in Supplemental Table S2), whose expression is up-regulated following the appearance of the AtMIKC* complexes during the TCP stage, are potential direct targets of the AtMIKC* transcription factor complexes. Among them are various genes with a function related to vesicle transport and cytoskeleton, cell wall, and signal transduction (Supplemental Fig. S3). These classes are essential for pollen germination and are generally overrepresented in the transcriptome of mature pollen (Honys and Twell, 2003). We tested the expression of 14 of these in silico predicted target genes by reverse transcription (RT)-PCR, in wild-type and agl66/104 double mutant pollen. For 11 of these 14 genes, we observed differences in expression level between mutant and wild-type pollen (Fig. 6), indicating that several of the in silico predicted target genes are indeed downstream of the AtMIKC* complexes in vivo.
DISCUSSION
MADS-box genes are key regulators of a range of higher plant-specific developmental programs. However, our knowledge of the MADS family in Arabidopsis is mainly limited to the MIKCc subgroup, as only two non-MIKCc genes have been functionally characterized to date (namely AGL37 and AGL80, both from the Mγ subgroup; Köhler et al., 2003; Portereiko et al., 2006). In this study, we initially characterized five additional non-MIKCc MADS-box genes, all belonging to the Mδ or AtMIKC* subgroup and specifically expressed during the tricellular and mature stages of pollen development (Honys and Twell, 2004). We found that five heterodimeric complexes were formed between these AtMIKC* proteins in yeast, at least four of which also exist in planta (Fig. 1, A–D). The AGL30/104 complex could be observed by EMSA and BiFC, but not in yeast. This apparent discrepancy might be caused by the weakness of their interaction, because complementation of YFP (in BiFC) has been reported to stabilize transient and weak protein interactions (Walter et al., 2004), and interaction with DNA (in EMSA) can have a comparable effect. Conversely, we could demonstrate an interaction between AGL94 and AGL66 in yeast but not in planta. In EMSA experiments, the AGL94-AGL66 protein combination consistently produced diffuse shifted bands (Fig. 1F), indicating suboptimal DNA binding. Taken together, these observations indicate that the AGL94/66 complex might not exist in vivo, but because it does bind DNA in vitro, we are currently unable to fully exclude a role for AGL94/66 in Arabidopsis pollen.
Interestingly, interactions exclusively occur between members of the two monophyletic lineages within the AtMIKC* subgroup (AGL30, AGL65, and AGL94 on one hand, and AGL66 and AGL104 on the other hand) and never between members of the same lineage (Fig. 1E). This indicates an ancestral scenario in which only one heterodimeric AtMIKC* complex existed, formed between an AGL65-like and an AGL66-like protein. Gene duplication events have subsequently led to the expansion of both MIKC* lineages and resulted in the five AtMIKC* complexes present in extant Arabidopsis pollen.
Our experiments suggest a high degree of functional redundancy between the five AtMIKC* complexes. The AGL30/66 and AGL65/66 complexes, which can be considered representative for all five AtMIKC* complexes, most avidly bind the same MEF2 motif CTA(TTTT)TAG/CTA(AAAA)TAG and show hardly any affinity for SRE motifs (Fig. 2A), which are preferred by most reported MADS-proteins (de Folter and Angenent, 2006). Nevertheless, the DNA-binding specificity of AGL65/66 is not completely identical to that of AGL30/66 and AGL30/104 (Figs. 2A and 3), and these subtle differences likely result from the various amino acid changes in the MADS and I domains of AGL30 and AGL65 that occurred since their duplication and divergence (Supplemental Fig. S1).
Our in vitro pollen germination experiments (Fig. 5) further clarified the redundancy between the different complexes. They revealed that the AGL65/66 and AGL65/104 complexes are functionally redundant with each other, but not with the other three complexes, and that AGL30/66 and AGL94/66 are redundant with AGL30/104. Our in vitro observations based on DNA-binding preferences (suggesting redundancy between AGL30/66 and AGL30/104, but not between AGL30/66 and AGL65/66) are therefore in good agreement with the in vivo situation. The functional interchangeability of the AGL66 and AGL104 proteins is actually not that surprising, considering the high homology they share at the protein level. Their MADS and I domains are nearly identical in sequence (Supplemental Fig. S1), indicating that the AGL66 and AGL104 genes originated from a relatively recent gene duplication event.
MADS complexes have always been found to bind a relatively broad spectrum of DNA motifs. An in-depth analysis of the occurrence of their binding motifs in the genome therefore seemed irrelevant. Only the AGL15 homodimer has been reported to exhibit preference for one particular MEF2 motif, CTA(TATA)TAG, but it also binds SRE- and N9-type motifs with comparable affinity (Tang and Perry, 2003). The DNA-binding specificities of AGL30/66 and AGL65/66 are higher than those reported for other MADS-protein complexes, as they hardly show any affinity for SRE-type CArG-boxes, and they can even distinguish between different MEF2 motifs to some extent (Figs. 2A and 3). This observation prompted us to examine the relative occurrence of the different CArG-boxes in Arabidopsis promoters. In general, 16 and 64 distinct motifs correspond to the MEF2- and SRE-type consensus sequence, respectively (Fig. 2B; Supplemental Table S2). Surprisingly, we found that some of the motifs occur far more frequently than others (Fig. 2B), and in some cases, even the directionality of a motif might play a role (Fig. 2C). This nonrandom occurrence strongly suggests that there are biologically meaningful differences between the individual CArG-box motifs.
We then carried out a large-scale in silico analysis in which we examined the overall and relative abundance of the individual CArG-boxes, as well as their spatial positioning in Arabidopsis promoters genome wide and in pollen-specific promoters. Thereby, we took advantage of a unique single-cell transcriptome dataset covering the different stages of pollen development (Honys and Twell, 2004). Two additional factors increased the reliability of this analysis: the high DNA-binding specificities of the AtMIKC* complexes and the fact that pollen grains express only a small number of MADS-box genes (Honys and Twell, 2004; Pina et al., 2005). The latter implies that, apart from the AtMIKC* complexes, only few other proteins are expected to bind CArG-boxes in pollen-specific promoters. We observed that MEF2-type CArG-boxes are overall overrepresented in TCP/MPG-specific promoters (Fig. 2B) and tend to be positioned in the proximal region of these promoters (Fig. 4). Both tendencies are even more pronounced for the CTA(TTTT)TAG/CTA(AAAA)TAG motif, which is most preferred by AGL30/66 and AGL65/66 in vitro. It accounts for nearly 28% of all MEF2 motifs in TCP/MPG-specific promoters, and 60% of these motifs are located within 500-bp distance of the ATG start codon (Figs. 2 and 4). Similarly, we found that a particular SRE-type CArG-box (CCTTTTTTGG/CCAAAAAAGG) is exceptionally frequent in BCP-specific promoters (Supplemental Table S2) and tends to be positioned in proximity of the start codon (data not shown), as is, in general, the case for SRE motifs in BCP-specific promoters (Fig. 4). In conclusion, our in silico approach indicates that the MEF2-binding AtMIKC* complexes play an important role in transcriptional regulation during the TCP and MPG stages of pollen development, whereas a non-MIKC* MADS complex, preferentially binding the CCTTTTTTGG/CCAAAAAAGG motif, might play a role in regulating gene expression during the BCP and/or TCP stages.
Our example illustrates that specific binding sites can be overrepresented in promoters that share a similar, narrow expression pattern, and this reflects the binding specificity of transcription factors functioning in that specific cell type or during a predefined developmental stage. We think this in silico approach is in general a powerful, indirect tool to help uncover new transcriptional regulatory networks in specific cell types or developmental stages. The concept of using experimentally well-defined transcription factor binding sites as a starting point for a large-scale genomics and single cell-type transcriptomics in silico analysis also seems feasible for other cell types than pollen, and for any transcription factor with a clearly defined DNA-binding preference. The main difficulty lies in obtaining transcriptome data that allow such in silico analyses, but recent technical advances have enabled the purification of trichomes (Zhang and Oppenheimer, 2004), various cell types of the root (Birnbaum et al., 2005), and different stages in xylem development (Kubo et al., 2005), and single cell-type transcriptomics is feasible for all these examples (Birnbaum et al., 2005; Kubo et al., 2005). Laser capture microdissection (e.g. see Wu et al., 2006) will most likely enable the isolation of additional single cell types for transcriptome studies in the near future.
With our in silico approach, we identified 152 putative direct target genes of the AtMIKC* complexes in Arabidopsis pollen. These genes are specifically expressed during the last two stages of pollen development and contain at least one MEF2 motif in their 3-kb promoter (Supplemental Table S2). We tested the expression of a random selection of these genes by RT-PCR and found that the majority of them are affected in agl66/104 double mutant pollen (Fig. 6), implying that these genes are truly downstream of the AtMIKC* complexes, and that our in silico approach is reliable for predicting target genes of transcription factors. Two of the most strongly affected genes (At2g44560 and At5g19610) contain the CTA(TAAA)TAG and CTA(AATA)TAG motifs in their proximal promoter, respectively. In vitro, these motifs are both bound rather well by the AGL30/66 and/or AGL65/66 complexes (Fig. 2A), but they are not overrepresented in TCP/MPG-specific promoters (Fig. 2B). This observation raises the possibility that MEF2 motifs other than the most preferred and overrepresented CTA(TTTT)TAG/CTA(AAAA)TAG motif can be bound by the different AtMIKC* complexes in vivo, or, alternatively, that these affected genes are actually indirect targets rather than direct targets. We are currently unable to exclude either possibility, but chromatin immunoprecipitation experiments could help to further clarify this matter. In addition, the expression of some of the in silico predicted direct target genes with proximal CTA(TTTT)TAG/CTA(AAAA)TAG motif in their promoter is unaffected in agl66/104 double mutant pollen (Fig. 6), suggesting that the presence of AtMIKC* binding sites in the proximal promoter is not always sufficient to make a late pollen-specific gene a direct target of the AtMIKC* complexes. How MADS-protein complexes achieve their high degree of target specificity and how they distinguish the CArG-boxes in their target promoters from all other CArG-boxes in the genome are important questions in plant development. Current hypotheses assume the involvement of accessory DNA-binding factors associating with MADS-protein complexes or a role for the nucleotides flanking CArG-boxes in promoters (de Folter and Angenent, 2006). Alternatively, these unaffected genes might be direct targets of the AtMIKC* complexes, requiring only low amounts of these complexes for their regulation. In the agl66/104 double mutant, used for our RT-PCR experiment, the AGL104 transcript is strongly reduced in abundance but not completely absent (Fig. 6). This is the result of a T-DNA insertion in the fifth intron of the AGL104 gene, allowing the occasional transcription of the full-length mRNA. Therefore, low amounts of the AGL30/104 and AGL65/104 protein complexes are still expected to be present in agl66/104 double mutant pollen, and these might be sufficient to regulate the expression of some, but not all, of the AtMIKC* direct target genes.
One of the genes demonstrated to be down-regulated in agl66/104 double mutant pollen is MYB97, one of the seven transcription factor-encoding genes among the predicted direct targets (Fig. 6; Supplemental Fig. S3). The fact that other transcription factors are downstream of the AtMIKC* complexes highlights their importance in transcriptional regulation during late pollen development. It suggests that the AtMIKC* complexes rank high in the hierarchy of a pollen-specific transcriptional network and that many more BCP- and TCP/MPG-specific genes are likely under indirect control by the AtMIKC* complexes. The profound effect of the virtual loss of all AtMIKC* complexes on pollen germination efficiency in vitro (Fig. 5) clearly illustrates the pivotal role these complexes play in pollen.
The underlying cause of the pollen germination defect remains to be elucidated, but it is likely the result of multiple factors, because quite a few of the putative AtMIKC* target genes have reported or proposed functions in pollen germination. For example, seven cation/proton exchanger-encoding genes are among the putative direct targets (Supplemental Fig. S3), and at least two of them (CHX8 and CHX24) are indeed differentially expressed in agl66/104 double mutant pollen (Fig. 6). These proteins are postulated to allow osmotic adjustment and ion homeostasis during pollen desiccation, rehydration, and germination (Sze et al., 2004). Expression of the At5g19610 gene, encoding a Sec7 domain-containing protein, is strongly elevated in agl66/104 mutant pollen. Sec7 domain proteins function as guanidine exchange factors, which generally play an important role in Golgi structure and function (Peyroche et al., 2001). Proper regulation of the Golgi vesicle trafficking process is essential for pollen tube growth (Cheung et al., 2002).
Based on the severity of the in vitro pollen germination phenotype of the agl66/104 double mutant (Fig. 5), one might expect this mutant to be male sterile. Intriguingly, however, it shows no impaired fertility, making it unlikely that the AtMIKC* complexes play a role in determining pollen fertility itself. The mutant phenotype might therefore be more subtle in vivo, perhaps affecting the speed and efficiency of pollen germination and tube growth. A more detailed molecular and functional characterization of Arabidopsis mutants lacking the different AtMIKC* complexes should further clarify the exact biological role of these atypical MADS-box genes in pollen development.
MATERIALS AND METHODS
Isolation and Cloning of MIKC* cDNAs
Mature pollen grains were isolated from open flowers of the Arabidopsis (Arabidopsis thaliana) Columbia accession (grown in a growth chamber at 22°C, with 16 h of light at around 140 μmol m−2 s−1), using the protocol of Honys and Twell (2003). Subsequently, total RNA was isolated with the RNeasy Plant Mini kit (Qiagen), and 5 μg of RNA was used for cDNA synthesis with the Superscript kit (Invitrogen) with oligo(dT) primers. RT-PCR was performed with gene-specific primers to obtain the full-length cDNAs of AGL30 (At2g03060), AGL65 (At1g18750), AGL66 (At1g77980), AGL94 (At1g69540), AGL104 (At1g22130), and AGL18 (At3g57390). All fragments were cloned into the pCR2.1-TOPO vector (Invitrogen), verified by sequencing on a PE Biosystems ABI Prism 377 sequencer by the Max Planck Institute DNA core facility (ADIS), and found to be identical to the sequences reported in the most recent version of the TAIR database. The AGL30 transcript we cloned is identical to the recently created GenBank accession DQ446459 and different from the truncated splice form reported by Kofuji et al. (2003).
Y2H
The full-length ORF sequences of the five pollen-expressed AtMIKC* genes were cloned into the pGADT7 prey vector and the pGBKT7 bait vector (CLONTECH), which carry the LEU2 and TRP1 selection markers, respectively (AGL30 with NcoI and BamHI, AGL65 with NdeI and BamHI, AGL66 with NcoI, AGL94 with EcoRI and BamHI, and AGL104 with XmaI). The ORF of the pollen-expressed MIKCc gene AGL18 was also cloned into both vectors (with NcoI). Yeast (Saccharomyces cerevisiae) strain AH109 (CLONTECH) was sequentially transformed with all available combinations of bait and prey constructs. The transformation mixtures were plated out onto synthetic dropout medium lacking Leu and Trp to test for transformation efficiency and onto synthetic dropout medium lacking Leu, Trp, and Ade to test for protein-protein interactions. For all AtMIKC* proteins, a strong autoactivation of the bait constructs was observed, and to circumvent this problem, a C-terminal part was removed (by restriction digestion with PstI in case of AGL94 and by PCR with nested primers and recloning in case of the other genes), resulting in a series of ΔC constructs with abolished autoactivation (118 amino acids deleted for AGL30ΔC, 133 for AGL65ΔC, 141 for AGL66ΔC, 133 for AGL94ΔC, and 145 for AGL104ΔC).
BiFC
We used the BiFC system described by Bracha-Drori et al. (2004) and Walter et al. (2004) to investigate AtMIKC* protein complex formation in planta. The full-length AtMIKC* ORFs were first cloned into the pCR8/GW/TOPO entry vector (Invitrogen) and introduced into the pBaTL-YFPc and pBaTL-YFPn vectors (Hackbusch et al., 2005), which were kindly donated by Drs. J. Uhrig and K. Richter (Max Planck Institute), by Gateway technology (Invitrogen). The constructs were subsequently electroporated into Agrobacterium tumefaciens strain ABI (Koncz and Schell, 1986). Leaves from 3- to 4-week-old Nicotiana benthamiana plants were coinfiltrated with all possible combinations of the available constructs (Fig. 1B), using a 1-mL syringe. To avoid cosuppression, an A. tumefaciens strain carrying the p19 viral silencing suppressor gene was always included in the A. tumefaciens infiltration mixture, according to Voinnet et al. (2003). The infiltrated leaves remained attached to the plants, which were returned to greenhouse conditions. After 2, 3, and 5 d, the leaves were examined for YFP fluorescence, using a Leica TCS SP2 AOBS confocal laser scanning microscope. Fluorescence was usually strongest at day three, and therefore, this time point was used to score for interactions and to make the pictures in Figure 1.
EMSA and RBSS
The AtMIKC* ORFs were cloned into the pSPUTK vector (Promega) using NcoI (AGL30 and AGL66), SalI and EcoRI (AGL94), HindIII and BamHI (AGL65), and SmaI (AGL104). These constructs (500 ng of plasmid) were used for coupled in vitro transcription and translation with the TnT SP6-Coupled Reticulocyte Lysate system (Promega). Radioactively labeled Met ([35S]Met) was included in this reaction to allow detection of the synthesized proteins on SDS-PAGE gel. In samples meant for testing the DNA-binding properties of heterodimeric complexes, two proteins were synthesized together in one reaction. As a control PpMADS2, a homodimerizing MIKC* protein from the moss Physcomitrella patens was included (R. Hallinger, W. Verelst, W. Faigl, H. Saedler, and T. Münster, unpublished data). In an EMSA reaction, 2 μL of protein sample was mixed with 1 μL of DNA probe in a reaction mixture containing 2.5% (w/v) CHAPS, 9 mm HEPES, pH 7.3, 1.4 mm EDTA, pH 8.0, 8% (w/v) glycerol, 1.33 mm spermidine, 0.9 mm dithiothreitol, 75 ng/μL bovine serum albumin (New England Biolabs), and 11.5 μg/μL autoclaved calf thymus DNA (Serva) and incubated on ice for 30 min. Subsequently, protein-DNA complexes were separated from unbound probes by electrophoresis on a 4% nondenaturing polyacrylamide gel. Bands were visualized using a phosphor screen and a Typhoon 8600 phosphor imager (Molecular Dynamics). For the EMSA experiment illustrated in Figure 1F, an equimolar mixture of two randomized probes was used: GATCCTGTCGNNNCC-(A/T)6-GGNNNGAGGCGAAT (SRE-type CArG-box) and GATCCTGTCGNNNC-(A/T)8-GNNNGAGGCGAAT (relaxed MEF2-type CArG-box). DNA probes were labeled with [α-32P]dCTP in a single PCR cycle (using 500 ng of a primer complementary to the 3′ linker fragment and 400 ng of template probe), purified by ethanol precipitation, and dissolved in 10 mm Tris, pH 8.0, to a specific activity of 100,000 cpm/μL.
For RBSS (Pollock and Treisman, 1990), a 62-nucleotide long single-stranded DNA molecule (GGTCAGTTCAGCGGATCCTGTCG-N16-GAGGCGAATTCAGTGCAACTGCG; Birkenbihl et al., 2005), consisting of a completely random core of 16 nucleotides flanked by linkers with EcoRI and BamHI restriction sites, respectively, was radioactively labeled and used in an EMSA experiment as described above. Shifted bands were excised from the polyacrylamide gel, and DNA was eluted by overnight incubation of the gel slices in 1× Tris-EDTA buffer, pH 8.0, 10 mm MgCl2, 0.1% (w/v) SDS, 500 mm Na acetate, pH 5.6, at room temperature. DNA was recovered by phenol chloroform extraction and ethanol precipitation and resuspended in 20 μL 10 mm Tris-HCl, pH 8.0. Of this enriched DNA fraction, 8 μL was relabeled with [α-32P]dCTP in 16 PCR cycles using primers annealing to both linker sequences of the randomized probe and purified by ethanol precipitation. The EMSA procedure was repeated four additional times until the shifted band was intense and no additional background bands were visible anymore. Then the shifted DNA pool was subjected to eight PCR cycles and subsequently cloned into pCR2.1-TOPO (Invitrogen) and transformed into Escherichia coli. Colonies were screened for the presence of inserts by colony PCR, and the inserts of positive clones were sequenced with a vector-specific primer.
In competitive EMSA experiments, the only difference with the regular EMSA approach was that the proteins were incubated on ice with unlabeled double-stranded 62-mer fragments for 20 min, prior to the addition of a radioactively labeled probe to the EMSA mixture, followed by another 20 min of incubation. Both the labeled and unlabeled probes were identical to the randomized probe used for RBSS but with a more defined core sequence [NNNCTA(TTTT)TAGNNN, NNNCTA(TATA)TAGNNN, NNNCTA(TTT)TAGNNNN, NNNCTA(TAAT)TAGNNN, and NNNCCTATTTAGGNNN]. These sequences were chosen based on the results of the RBSS experiments (Fig. 2A; Supplemental Fig. S2). The double-stranded competitor probes were produced by boiling a mixture of two complementary single-stranded oligonucleotides, followed by gradual cooling to room temperature and subsequent purification over columns designed for double-stranded DNA isolation (Roche). The resulting double-stranded probes were diluted to a concentration of 100 ng/μL and their concentration was verified both spectrophotometrically and on agarose gel. These cold competitor probes were generally used at an excess of 30- and 125-fold relative to the labeled probe, unless stated otherwise.
RT-PCR for Putative Target Genes
Total RNA was isolated from mature pollen (as described above), which had been harvested from wild-type and agl66/104 double mutant plants grown in a greenhouse during summer, with temperature controlled at 22°C and 16 h of light at around 120 μmol m−2 s−1. Primers for RT-PCR on predicted target genes were designed in the 3′ region of the ORF, they usually coincided with probe sets for these genes used on the ATH1 microarray chip (Affymetrix), and wherever possible spanned an intron. RT-PCR was performed for 14 predicted target genes using 150 ng cDNA and gene-specific primers. As a control, 18S ribosomal RNA was amplified using the QuantumRNA primer-competimer approach from Ambion, with a 2:8 ratio of primer-competimer and 35 PCR cycles. The number of PCR cycles was optimized for each gene: 20 cycles for At1g74000, At1g17540, At4g18700, At5g35390, At3g46520, At1g13890, and At5g64790; 25 cycles for At4g26930, At5g55980, At2g28180, At5g19610, and At2g05850; 30 cycles for At2g44560, AGL66, and AGL104; and 35 cycles for At5g37060.
In Vitro Pollen Germination
For in vitro germination of pollen the protocol of Li et al. (1999) was used. The experiment was performed three times and averages are presented in Figure 5B. Each time, at least 500 pollen grains of each genotype were scored for germination. Arabidopsis lines with disrupted AtMIKC* loci were obtained from the Nottingham Arabidopsis Stock Centre (Alonso et al., 2003) and genotyped using the standard T-DNA left border primer and a gene-specific primer pair. The T-DNA insertion lines we used are agl65 (SALK_009651, located in the ninth exon of the At1g18750 gene, resulting in the reduced expression of a truncated transcript), agl66 (SALK_072108, located in the ninth exon of the At1g77980 gene, resulting in a complete loss of AGL66 expression, see Fig. 6), agl94 (SALK_016078, located in the last exon of the At1g69540 gene, resulting in the expression of a truncated transcript), and agl104 (SALK_098698, located in the fifth intron of the At1g22130 gene, resulting in a strongly reduced expression of AGL104; see Fig. 6).
Viability staining of pollen was performed with fluorescein-3′,6′-diacetate according to Eady et al. (1995), and at least 300 pollen grains were observed.
In Silico Analyses
The protein sequence alignment in Supplemental Figure S1 was created with ClustalW (Chenna et al., 2003). Arabidopsis promoter and 5′UTR sequences genome wide were screened for CArG-boxes using the Patmatch tool on the TAIR Web site (www.arabidopsis.org), and for BCP- and TCP/MPG-specific genes they were downloaded using the Bulk downloads option. For genes with available 5′UTR information, the length of the 5′UTR was added to the distance of CArG-boxes from the end of the 3-kb promoter sequence to obtain their correct distance to the ATG (see Supplemental Table S2). The dataset from Honys and Twell (2004) was downloaded from the Genome Biology Web site (http://genomebiology.com) and reanalyzed using Access and Excel software (Microsoft). The pollen-specific expression of genes identified as BCP- or TCP/MPG-specific in this dataset was verified against all publicly available microarray datasets, using the Genevestigator tool (Zimmermann et al., 2004).
The sequence data from this article can be found in the GenBank data library, under accession numbers DQ446459 (AGL30), NM_101733 (AGL65), NM_106447 (AGL66), NM_105623 (AGL94), NM_102063 (AGL104), and NM_115599 (AGL18).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. Sequence alignment of the members of both monophyletic lineages within the AtMIKC* subgroup.
Supplemental Figure S2. Overview of all sequences enriched by two of the AtMIKC* complexes in RBSS, subdivided into relevant categories.
Supplemental Table S1. List of the AtMIKC* genes, and of all BCP-specific and TCP/MPG-specific genes represented on the ATH1 microarray chip, with their expression levels in all stages of pollen development and in other plant tissues.
Supplemental Table S2. Additional information and original data concerning the occurrence and distribution of CArG-boxes in Arabidopsis promoters.
Supplemental Table S3. Selection of in silico predicted direct target genes of the AtMIKC* complexes in pollen. The complete list is presented in Supplemental Table S2.
Supplementary Material
Acknowledgments
We thank Drs. Joachim Uhrig and Klaus Richter for vectors and practical help with BiFC, Dr. Suzanne Kuijt for assistance with confocal microscopy, and Drs. Zsuzsanna Schwarz-Sommer, Rainer Birkenbihl, Malgorzata Domagalska, and two anonymous reviewers for helpful comments and critically reading the manuscript.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Wim Verelst (verelst@mpiz-koeln.mpg.de).
The online version of this article contains Web-only data.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657 [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Liljegren SJ, Pelaz S, Gold SE, Burgeff C, Ditta GS, Vergara-Silva F, Yanofsky MF (2000. a) MADS-box gene evolution beyond flowers: expression in pollen, endosperm, guard cells, roots and trichomes. Plant J 24 1–11 [DOI] [PubMed] [Google Scholar]
- Alvarez-Buylla ER, Pelaz S, Liljegren SJ, Gold SE, Burgeff C, Ditta GS, de Pouplana LR, Martinez-Castilla L, Yanofsky MF (2000. b) An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc Natl Acad Sci USA 97 5328–5333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker A, Theissen G (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29 464–489 [DOI] [PubMed] [Google Scholar]
- Becker A, Winter K-U, Meyer B, Saedler H, Theißen G (2000) MADS-box gene diversity in seed plants 300 million years ago. Mol Biol Evol 17 1425–1434 [DOI] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijo JA (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of the pollen transcriptome. Plant Physiol 133 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birkenbihl RP, Jach G, Saedler H, Huijser P (2005) Functional dissection of the plant-specific SBP-domain: overlap of the DNA-binding and nuclear localization domains. J Mol Biol 352 585–596 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN (2005) Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods 2 615–619 [DOI] [PubMed] [Google Scholar]
- Bracha-Drori K, Shichrur K, Katz A, Oliva M, Angelovici R, Yalovsky S, Ohad N (2004) Detection of protein-protein interactions in plants using bimolecular fluorescence complementation. Plant J 40 419–427 [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Chen CY, Glaven RH, de Graaf BHJ, Vidali L, Hepler PK, Wu H (2002) Rab2 GTPase regulates vesicle trafficking between the endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell 14 945–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Raes J, Van de Peer Y, Theißen G (2003) And then there were many: MADS goes genomic. Trends Plant Sci 8 475–481 [DOI] [PubMed] [Google Scholar]
- de Folter S, Angenent GC (2006) Trans meets cis in MADS science. Trends Plant Sci 11 224–231 [DOI] [PubMed] [Google Scholar]
- de Folter S, Busscher J, Colombo L, Losa A, Angenent GC (2004) Transcript profiling of transcription factor genes during silique development in Arabidopsis. Plant Mol Biol 56 351–366 [DOI] [PubMed] [Google Scholar]
- de Folter S, Immink RGH, Kieffer M, Pařenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al (2005) Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady C, Lindsey K, Twell D (1995) The significance of microspore division and division symmetry for vegetative cell-specific transcription and generative cell differentiation. Plant Cell 7 65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egea-Cortines M, Saedler H, Sommer H (1999) Ternary complex formation between the MADS-box proteins SQUAMOSA, DEFICIENS and GLOBOSA is involved in the control of floral architecture in Antirrhinum majus. EMBO J 18 5370–5379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackbusch J, Richter K, Muller J, Salamini F, Uhrig JF (2005) A central role of Arabidopsis thaliana ovate family proteins in networking and subcellular localization of 3-aa loop extension homeodomain proteins. Proc Natl Acad Sci USA 102 4908–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes TE, Sengupta P, Cochran BH (1988) The human c-FOS serum response factor and the yeast factors GRM/pRTF have related DNA-binding specificities. Genes Dev 2 1713–1722 [DOI] [PubMed] [Google Scholar]
- Hennig L, Gruissem W, Grossniklaus U, Köhler C (2004) Transcriptional programs of early reproductive stages in Arabidopsis. Plant Physiol 135 1765–1775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henschel K, Kofuji R, Hasebe M, Saedler H, Münster T, Theißen G (2002) Two ancient classes of MIKC-type MADS-box genes are present in the moss Physcomitrella patens. Mol Biol Evol 19 801–814 [DOI] [PubMed] [Google Scholar]
- Honma T, Goto K (2001) Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 525–529 [DOI] [PubMed] [Google Scholar]
- Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D (2004) Transcriptome analysis of haploid male gametophyte development in Arabidopsis. Genome Biol 5 R85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish V, Litt A (2005) Flower development and evolution: gene duplication, diversification and redeployment. Curr Opin Genet Dev 15 454–460 [DOI] [PubMed] [Google Scholar]
- Kofuji R, Sumikawa N, Yamasaki M, Kondo K, Ueda K, Ito M, Hasebe M (2003) Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses. Mol Biol Evol 20 1963–1977 [DOI] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U (2003) The polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Schell J (1986) The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet 204 383–396 [Google Scholar]
- Kubo M, Udagawa M, Nishikubo N, Horiguchi G, Yamaguchi M, Ito J, Mimura T, Fukuda H, Demura T (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev 19 1855–1860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Lin Y, Heath RM, Zhu MX, Yang Z (1999) Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11 1731–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Yanofsky MF, Meyerowitz EM (1991) AGL1-AGL6, an Arabidopsis gene family with similarity to floral homeotic and transcription factor genes. Genes Dev 5 484–495 [DOI] [PubMed] [Google Scholar]
- Martinez-Castilla LP, Alvarez-Buylla ER (2003) Adaptive evolution in the Arabidopsis MADS-box gene family inferred from its complete resolved phylogeny. Proc Natl Acad Sci USA 100 13407–13412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S (2004) Control of male gametophyte development. Plant Cell 16 S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Münster T, Pahnke J, Di Rosa A, Kim JT, Martin W, Saedler H, Theissen G (1997) Floral homeotic genes were recruited from homologous MADS-box genes preexisting in the common ancestor of ferns and seed plants. Proc Natl Acad Sci USA 94 2415–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J, de Pamphilis CW, Ma H, Nei M (2003) Antiquity and evolution of the MADS-box gene family controlling flower development in plants. Mol Biol Evol 20 1435–1447 [DOI] [PubMed] [Google Scholar]
- Nam J, Kim J, Lee S, An G, Ma H, Nei M (2004) Type I MADS-box genes have experienced faster birth-and-death evolution than type II MADS-box genes in angiosperms. Proc Natl Acad Sci USA 101 1910–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng M, Yanofsky MF (2001) Function and evolution of the plant MADS-box gene family. Nat Rev Genet 2 186–195 [DOI] [PubMed] [Google Scholar]
- Pařenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyroche A, Courbeyrette R, Rambourg A, Jackson CL (2001) The ARF exchange factors Gea1p and Gea2p regulate Golgi structure and function in yeast. J Cell Sci 114 2241–2253 [DOI] [PubMed] [Google Scholar]
- Pina C, Pinto F, Feijó JA, Becker JD (2005) Gene family analysis of the Arabidopsis pollen transcriptome reveals biological implications for cell growth, division control, and gene expression regulation. Plant Physiol 138 744–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R, Treisman R (1990) A sensitive method for the determination of protein-DNA binding specificities. Nucleic Acids Res 21 4769–4776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock R, Treisman R (1991) Human SRF-related proteins: DNA-binding properties and potential regulatory targets. Genes Dev 5 2327–2341 [DOI] [PubMed] [Google Scholar]
- Portereiko MF, Lloyd A, Steffen JG, Punwani JA, Otsuga D, Drews GN (2006) AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18 1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Wang MQ, Meyerowitz EM (1996) DNA-binding properties of Arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA and AGAMOUS. Nucleic Acids Res 24 3134–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riese M, Faigl W, Quodt V, Verelst W, Matthes A, Saedler H, Münster T (2005) Isolation and characterization of new MIKC*-type MADS-box genes from the moss Physcomitrella patens. Plant Biol 7 307–314 [DOI] [PubMed] [Google Scholar]
- Santelli E, Richmond TJ (2000) Crystal structure of MEF2A core bound to DNA at 1.5Å resolution. J Mol Biol 297 437–449 [DOI] [PubMed] [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Schölkopf B, Weigel D, Lohmann J (2005) A gene expression map of Arabidopsis development. Nat Genet 37 501–506 [DOI] [PubMed] [Google Scholar]
- Shiu S-H, Shih M-C, Li W-H (2005) Transcription factor families have much higher expansion rates in plants than in animals. Plant Physiol 139 18–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P, Sharrocks AD (1995) The MADS-box family of transcription factors. Eur J Biochem 229 1–13 [DOI] [PubMed] [Google Scholar]
- Sze H, Padmanaban S, Cellier F, Honys D, Cheng NH, Bock KW, Conejero G, Li X, Twell D, Ward JM, et al (2004) Expression profiles of a novel AtCHX gene family highlight potential roles in osmotic adjustment and K+ homeostasis in pollen development. Plant Physiol 136 2532–2547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanabe Y, Hasebe M, Sekimoto H, Nishiyama T, Kitani M, Henschel K, Münster T, Theissen G, Nozaki H, Ito M (2005) Characterization of MADS-box genes in charophycean green algae and its implication for the evolution of MADS-box genes. Proc Natl Acad Sci USA 102 2436–2441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Perry SE (2003) Binding site selection for the plant MADS-domain protein AGL15: an in vitro and in vivo study. J Biol Chem 278 28154–28159 [DOI] [PubMed] [Google Scholar]
- Theissen G, Becker A, Di Rosa A, Kanno A, Kim JT, Münster T, Winter KU, Saedler H (2000) A short history of MADS-box genes in plants. Plant Mol Biol 42 115–149 [PubMed] [Google Scholar]
- Theissen G, Saedler H (2001) Plant biology: floral quartets. Nature 409 469–471 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas R, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33 949–956 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40 428–438 [DOI] [PubMed] [Google Scholar]
- West AG, Causier BE, Davies B, Sharrocks AD (1998) DNA binding and dimerization determinants of Antirrhinum majus MADS-box transcription factors. Nucleic Acids Res 26 5277–5287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu YR, Machado AC, White RG, Llewellyn DJ, Dennis ES (2006) Expression profiling identifies genes expressed early during lint fibre initiation in cotton. Plant Cell Physiol 47 107–127 [DOI] [PubMed] [Google Scholar]
- Zhang X, Oppenheimer DG (2004) A simple and efficient method for isolating trichomes for downstream analyses. Plant Cell Physiol 45 221–224 [DOI] [PubMed] [Google Scholar]
- Zimmermann P, Hirsch-Hoffmann M, Hennig L, Gruissem W (2004) GENEVESTIGATOR: Arabidopsis microarray database and analysis toolbox. Plant Physiol 136 2621–2632 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.