Abstract
X-ray crystal structures of human soluble epoxide hydrolase (sEH) complexed with four different dialkylurea inhibitors bearing pendant carboxylate “tails” of varying length have been determined at 2.3–3.0 Å resolution. Similarities among inhibitor binding modes reinforce the proposed roles of Y381 and/or Y465 as general acids that protonate the epoxide ring of the substrate in concert with nucleophilic attack of D333 at the electrophilic epoxide carbon. Additionally, the binding of these inhibitors allows us to model the binding mode of the endogenous substrate 14,15-epoxyeicosatrienoic acid. Contrasts among inhibitor binding modes include opposite orientations of inhibitor binding in the active-site hydrophobic tunnel. Alternative binding orientations observed for this series of inhibitors to human sEH, as well as the binding of certain dialkylurea inhibitors to human sEH and murine sEH, complicate the structure-based design of human sEH inhibitors with potential pharmaceutical applications in the treatment of hypertension. Thus, with regard to the optimization of inhibitor designs targeting human sEH, it is critical that human sEH and not murine sEH be utilized for inhibitor screening, and it is critical that structures of human sEH-inhibitor complexes be determined to verify inhibitor binding orientations that correlate with measured affinities.
Keywords: domain swapped dimer, epoxide hydrolase, inhibition
Microsomal epoxide hydrolase (mEH) and soluble epoxide hydrolase (sEH) catalyze the oxirane ring opening reaction on epoxide-bearing substrates. The catalytic mechanism proceeds via an SN2-type reaction mechanism in which the epoxide is activated by hydrogen bonds with general acids Y381 and/or Y465 with simultaneous nucleophilic attack by D333 to form an alkylenzyme intermediate; a water molecule activated by H523 attacks the alkylenzyme intermediate, forming a tetrahedral intermediate which subsequently collapses to release the product diol (Lacourciere and Armstrong 1993; Hammock et al. 1994; Borhan et al. 1995; Pinot et al. 1995; Arand et al. 1996; Müller et al. 1997; Laughlin et al. 1998; Yamada et al. 2000). One biological function of the epoxide hydrolases is to provide chemical protection against endogenous and environmental epoxides that might otherwise be toxic by alkylation of DNA (Hammock et al. 1997). However, recent studies point to sEH as a key regulator of mammalian blood pressure regulation via the hydrolysis of cis-epoxyeicosatrienoic acids (Zeldin et al. 1993; Fang et al. 2001; Fleming 2001; Imig et al. 2002). Therefore, sEH is a potential target for the treatment of hypertension (Zeldin et al. 1993; Fleming 2001; Fang et al. 2001; Imig et al. 2002). The X-ray crystal structures of murine sEH (Argiriadi et al. 1999) and human sEH (Gomez et al. 2004) each reveal an active site located in the middle of an “L”-shaped hydrophobic tunnel, ~25 Å in length, which appears to be suitable for the binding of extended lipid epoxide substrates and aliphatic dialkylurea inhibitors (Argiriadi et al. 2000).
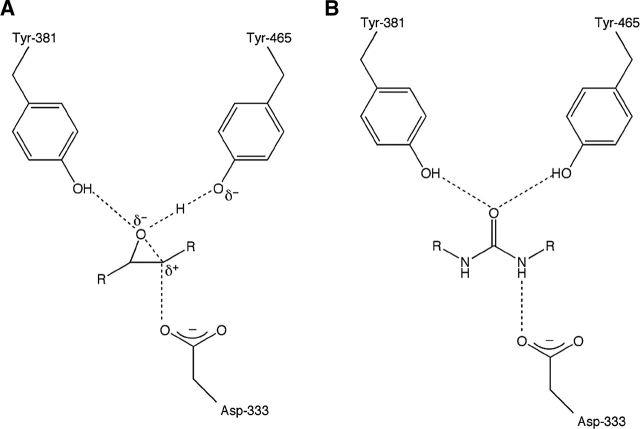
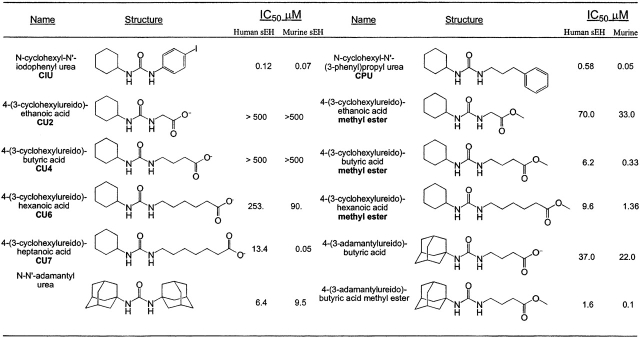
The best class of sEH inhibitors to date are the dialkylureas (Morisseau et al. 1998, 1999; McElroy et al. 2003), which typically bind as analogs of the transition state for epoxide ring opening such that the urea C==O moiety accepts hydrogen bonds from Y381 and Y465 and one urea NH group donates a hydrogen bond to D333 (Fig. 1) (Argiriadi et al. 2000; Gomez et al. 2004). However, there are examples of unexpected binding modes for dialkyurea inhibitors: N-cyclohexyl- N′-(3-phenyl) propylurea (CPU, Table 1) binds to murine sEH with an energetically unfavorable cis-amide linkage (Argiriadi et al. 1999), and N-cyclohexyl- N′-iodophenyl urea (CIU, Table 1), while having similar inhibitory potency toward human sEH and murine sEH (IC50 values of 0.12 μM and 0.07 μM, respectively) (McElroy et al. 2003; Gomez et al. 2004), binds to these enzymes with opposite orientations (Argiriadi et al. 2000; Gomez et al. 2004).
Figure 1.
(A) Transition state for oxirane ring opening as catalyzed by soluble epoxide hydrolase. (B) General binding mode of a dialkylurea inhibitor, which mimics the transition state.
Table 1.
Epoxide hydrolase inhibitors
Data from Kim et al. 2004.
Quantitative structure–activity relationships (QSAR) point to further ambiguities in the binding of inhibitors to human sEH and murine sEH. For instance, QSAR analysis of dialkylurea inhibitors yields equations for murine sEH and human sEH with r2 values of 0.955 and 0.616, respectively (Nakagawa et al. 2000; McElroy et al. 2003), suggesting that the binding of dialkylurea inhibitors to the human enzyme is not as readily predictable as the binding of inhibitors to the murine enzyme. To further probe the complexities of dialkylurea inhibition we now report the X-ray crystal structures of human sEH complexed with four dialkylurea inhibitors bearing pendant carboxylate “tails” of varying lengths: 4-(3-cyclohexylureido)- ethanoic acid (CU2), 4-(3-cyclohexylureido) -butyric acid (CU4), 4-(3-cyclohexylureido)-hexanoic acid (CU6), and 4-(3-cyclohexylureido)-heptanoic acid (CU7) (Table 1). Although the addition of the carboxylate functionality to each compound enhances solubility (and may make them better mimics of fatty acid epoxides, the suspected endogenous substrates), this series of inhibitors shows different affinity trends toward human sEH and murine sEH. The structures of these enzyme– inhibitor complexes allow us to rationalize the observed affinity trends with human sEH and reveal the structural basis for the complexity of QSAR analysis.
Results and Discussion
Although the pendant carboxylate groups of CU2, CU4, CU6, and CU7 differ by only five methylene units in length, the crystal structures of enzyme–inhibitor complexes exhibit some remarkable differences. Two distinct new inhibitor binding modes are observed that, taken together with the previously reported structure of the human sEH–CIU complex (Gomez et al. 2004), and the structures of murine sEH complexed with dialkylurea inhibitors (Argiriadi et al. 1999, 2000), illuminate important structure–affinity relationships in the sEH active site tunnel. The ensemble of structures also reveals the molecular basis for the variable inhibition exhibited by 4-(3- cyclohexylureido)-carboxylic acids against sEH (Table 1).
Crystal structures of human sEH–inhibitor complexes
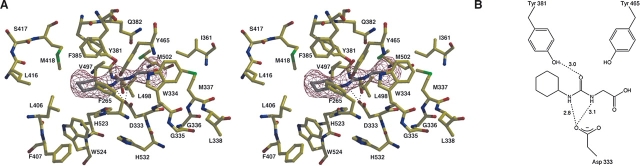
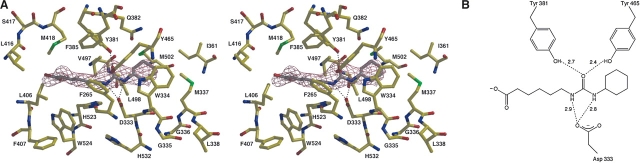
The binding of 4-(3-cyclohexylureido)-ethanoic acid (CU2) does not induce any major structural changes in human sEH, as indicated by the root mean square (RMS) deviation of 0.4 Å for the 320 Cα atoms of the C-terminal domains between the native enzyme and the enzyme–inhibitor complex. Despite the modest 3.0 Å resolution and the poor inhibition exhibited by CU2 (IC50>500 μM), clear electron density defines the location and conformation of the inhibitor cyclohexyl group; a slight bulge in the electron density of the carboxylate may reflect some modest disorder (Fig. 2A). The cyclohexyl group of CU2 makes van der Waals interactions in a hydrophobic region defined by F265, Y381, L406, M418, V497, and W524 (henceforth designated the “F265 pocket”), and the carboxylic acid group lies adjacent to a hydrophobic surface defined by W334, M337, and L498 (henceforth designated the “W334 niche”). This inhibitor-binding orientation is identical to that observed for the binding of N-cyclohexylurea inhibitors such as CIU (Table 1) to murine sEH (Argiriadi et al. 2000), which bind with their cyclohexyl groups in the corresponding F265 pocket. It is possible that the carboxylic acid of CU2 is protonated as it binds to human sEH, since the binding of a negatively charged carboxylate in a hydrophobic environment would be destabilizing. Inhibitor binding is weakly stabilized by poorly oriented hydrogen bonds with Y381 and D333 (Fig. 2B). Poor hydrogen-bond geometries and the placement of a carboxylate deep within a hydrophobic tunnel likely contribute to the poor inhibitory potency of CU2.
Figure 2.
(A) Simulated annealing omit map of the human sEH–CU2 complex (contoured at 4.0 σ). Poorly oriented hydrogen bonds are indicated by dotted lines. (B) Schematic representation of hydrogen bond interactions in the CU2 complex.
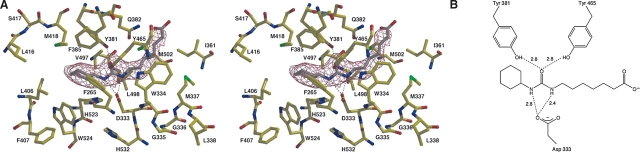
Neither CU4 nor CU6 induce any major structural differences upon binding to sEH, as indicated by RMS deviations of 0.4 Å for the 320 Cα atoms of the C-terminal domains between the native enzyme and each enzyme– inhibitor complex. The position and orientation of the inhibitor CU4 is established by unambiguous electron density at 2.3 Å resolution (Fig. 3A). The moderate 2.7 Å resolution of the sEH–CU6 complex similarly allows for the unambiguous assignment of the position and orientation of inhibitor CU6, although the electron density for the hexanoic acid moiety weakens somewhat toward the terminal carboxylate group (Fig. 4A). Surprisingly, these inhibitors each bind with opposite orientation relative to that of CU2, such that the cyclohexyl group of each binds in the W334 niche instead of the F265 pocket. In the human sEH–CIU complex, the cyclohexyl group of CIU similarly binds in the W334 niche instead of the F265 pocket (Gomez et al. 2004). Thus, both sides of the active site tunnel can accommodate the bulky cyclohexyl substituent in human sEH.
Figure 3.
(A) Simulated annealing omit map of the human sEH–CU4 complex (contoured at 4.0 σ). Hydrogen bonds are indicated by dotted lines. (B) Schematic representation of hydrogen bond interactions in the CU4 complex.
Figure 4.
(A) Simulated annealing omit map of the human sEH–CU6 complex (contoured at 4.0 σ). Hydrogen bonds are indicated by dotted lines. (B) Schematic representation of hydrogen bond interactions in the CU6 complex.
Both CU4 and CU6 share a similar array of hydrogen bond interactions: the urea C==O moiety of each inhibitor accepts hydrogen bonds Y381 and Y465, and a urea NH group donates a hydrogen bond to D333 (Figs. 3B, 4B). Both CU4 and CU6 are relatively poor inhibitors of human sEH and murine sEH, presumably due in part to the interactions of their pendant carboxylate groups. The carboxylate tail of CU4 is relatively constrained by H523, W524, and M418, but the carboxylate moiety appears to be poorly oriented for electrostatic interactions with H523 and W524 that could otherwise stabilize the negatively charged carboxylate in a hydrophobic environment. The additional methylene units in the carboxylate tail of CU6 inhibitor extend the carboxylate toward the opening of the active site tunnel, but the carboxylate makes no direct interactions with the protein or ordered solvent molecules. The enhanced affinity of CU6 relative to CU4 may arise in part from weaker destabilizing interactions resulting from the placement of the inhibitor carboxylate group closer to the mouth of the hydrophobic tunnel.
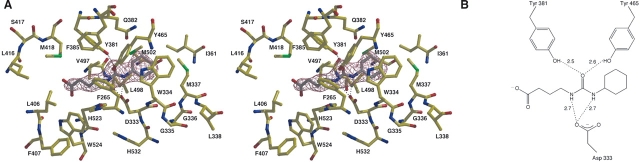
Although the structure of the sEH–CU7 complex reveals the expected hydrogen bond interactions between the urea C==O moiety and Y381 and Y465, and the urea NH groups and D333, the inhibitor binds in the same orientation as CU2 with its cyclohexyl group in the F265 pocket (Fig. 5). The heptanoic acid moiety of CU7 extends around M337 through the active site tunnel. Elevated thermal B-factors for the carboxylate group suggest some degree of disorder for the aliphatic carboxylate group.
Figure 5.
(A) Simulated annealing omit map of the human sEH–CU7 complex (contoured at 4.0 σ). Hydrogen bonds are indicated by dotted lines. (B) Schematic representation of hydrogen bond interactions in the CU7 complex.
The opposite inhibitor binding orientations represented by CU4/CU6 and CU2/CU7, as well as the binding of CIU with opposite orientations in the active sites of human sEH (Gomez et al. 2004) and murine sEH (Argiriadi et al. 2000), underscore the caution with which QSAR must be applied in understanding and predicting structure–affinity relationships for human sEH inhibitors. Comparison of the human sEH–CIU complex (Gomez et al. 2004) with the CU2, CU4, CU6, and CU7 complexes suggests that the slight “pinch” in the active-site tunnel in the vicinity of M337 may guide the orientation of dialkylurea inhibitors in the active site of human sEH. Inhibitors with a carboxylate tail too short (CU2) or too long (CU7) to interact with M337 bind with their cyclohexyl groups in the F265 pocket. However, inhibitors CU4 and CU6 each bear a carboxylate tail of sufficient length to interact unfavorably with M337 and therefore bind with opposite orientation to avoid an unfavorable interaction with M337. The side chain of M337 in human sEH also causes CIU to bind with the opposite orientation relative to that observed in its complex with murine sEH (which has the less bulky Val 337 side chain) in order to avoid a steric clash between Met 337 and the bulky iodophenyl substituent of the inhibitor (Gomez et al. 2004).
One key finding of the current work is that inhibitor potency is not predictive of inhibitor-binding orientation in the active site of human sEH. However, the crystal structures of human sEH complexed with dialkylurea inhibitors bearing pendant carboxylate groups allow us to explain some of the observed affinity trends (Kim et al. 2004). For example, although addition of the pendant carboxylate group blunts inhibitory potency due to the binding of the carboxylate in the hydrophobic active-site tunnel, this destabilizing interaction is readily neutralized by esterification of the carboxylate: The methyl esters of CU2, CU4, and CU6 bind with much higher affinities than the carboxylic acids (Table 1), and this is the case regardless of whether inhibitors bind with the aliphatic carboxylate in the F 265 pocket or the W334 niche. Important structural inferences can also be made regarding structure–affinity relationships for a second family of inhibitors characterized by a bulky N-adamantyl substituent (Table 1). The N,N′-diadamantylurea inhibitor exhibits high potency against human sEH, with IC50=6.4 μM (Morisseau et al. 2002). This suggests that the bulky adamantyl group can occupy either the F265 pocket or the W334 niche. Although the F265 pocket is larger, the free-energy cost of structural changes in the W334 niche triggered by the binding of N,N′-diadamantylurea must be partially offset by multiple van der Waals interactions with the bulky hydrocarbon moiety. Notably, 4-(3-adamantylureido)- butyric acid binds much more tightly than 4-(3- cyclohexylureido)-butyric acid (Table 1), so the additional van derWaals interactions afforded by an adamantyl group compared with a cyclohexyl group significantly contribute to enzyme–inhibitor affinity.
Model of human sEH-14,15-epoxyeicosatrienoic acid complex
Epoxyeicosatrienoic acids (EETs) are arachidonic acid metabolites of the cytochrome P-450 epoxygenases that produce vasodilatation by hyperpolarization of smooth muscle cells via activation of Ca2+-activated K+ channels (Hu and Kim 1993; Harder et al. 1995; Campbell et al. 1996; Fisslthaler et al. 1999; Capdevila et al. 2000; Quilley and McGiff 2000). As such, 14,15-EET has antihypertensive properties that are not attributed to their sEH hydrolyzed diol products. The targeted disruption of sEH is therefore a potential therapeutic strategy for the treatment of hypertension (Sinal et al. 2000).
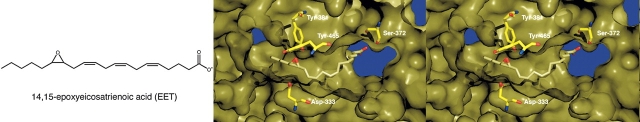
The binding modes of EET substrates in the active site of sEH are potentially exemplified by the binding of cyclohexylureido carboxylate inhibitors bearing longer aliphatic tails, e.g., CU7. Assuming that the transition state analogy of Figure 1 holds for the EET, then this substrate threads through the active site tunnel until the epoxide moiety hydrogen bonds with Y381 and Y465. It appears that either orientation of the EET could be accommodated in the active-site tunnel of human sEH. However, given that the inhibitor N-cyclohexyl-N-decylurea binds with its long aliphatic chain exclusively on the Met 337 side of the active-site tunnel (Argiriadi et al. 2000), and given that the aliphatic carboxylate chain of the inhibitor CU7 binds with a similar orientation (Fig. 5), we hypothesize that the long unsaturated chain of the proposed endogenous sEH substrate, 14,15-EET, similarly binds on the Met 337 side of the active-site tunnel. A model of this putative enzyme–substrate complex is presented in Figure 6.
Figure 6.
Model of the proposed endogenous substrate 14,15-EET bound to the active site of human sEH.
Conclusions
X-ray crystal structures of sEH complexes with dialkylurea inhibitors bearing pendant carboxylate “tails” illustrate the difficulties in accurately predicting inhibitor- binding orientations in the human sEH active site based solely on crystal structures of murine sEH–inhibitor complexes. Although human and murine sEH enzymes are related by 73% amino acid sequence identity, sufficient structural differences in their respective active-site tunnels cause certain dialkylurea inhibitors to bind to human sEH with opposite binding orientations. For the inhibitor CIU, opposite binding orientations in the active site of human sEH (Gomez et al. 2004) and murine sEH (Argiriadi et al. 2000) make relatively no impact on affinity; for the dialkylurea carboxylate inhibitors CU6 and CU7 (Table 1), opposite binding orientations in complexes with human sEH complicate the analysis of quantitative structure–affinity relationships. These results demonstrate that murine sEH is limited in its use as a model system for studying inhibitor binding to human sEH. Moreover, these results highlight the importance of X-ray crystal structures in determining conclusive structure–affinity relationships for human sEH as this enzyme is explored as a potential target for hypertension therapy.
Materials and methods
Recombinant human sEH was produced in a baculovirus expression system and purified as described (Beetham et al. 1993; Wixtrom et al. 1988), with the addition of a final step using a Superdex 200 size exclusion column (Amersham Pharmacia) in 0.1 M sodium phosphate (pH 7.4) and 3 mM dithiothreitol (DTT). For crystallization by vapor diffusion, a sitting drop containing 5.0 μL of protein solution (12–16 mg/mL sEH, 3 mM DTT, 0.1 M sodium phosphate at pH 7.4), 5.0 μL of precipitant buffer (0.1 M Tris at pH 8.4, 34% [wt/vol] PEG 3350), and 0.5 μL n-hexadecyl-β-D-maltoside (Hampton) was equilibrated against a 1-mL reservoir of precipitant buffer at 4 °C. Hexagonal rods appeared in ~7 d with dimensions of 0.4 mm × 0.1 mm × 0.1 mm. Crystals belonged to space group P6522 with unit cell parameters a=b ≈ 93 Å and c ≈ 244 Å . Crystals were then soaked in precipitation buffer containing 30 mM inhibitor for 3 d. Following transfer to a 20% sucrose cryoprotectant and flash-cooling, crystals yielded diffraction data to 3.0 Å resolution for 4-(3-cyclohexylureido)- ethanoic acid (CU2), 2.3 Å resolution for 4-(3- cyclohexylureido)-butyric acid (CU4), and 2.6 Å resolution for 4-(3-cyclohexylureido)-heptanoic acid (CU7) at the Advanced Light Source, Berkeley (beamline 5.0.3 with an ADSC Quantum 4R detector at 100 K). Crystals of human sEH complexed with 4-(3-cyclohexylureido)-hexanoic acid (CU6) yielded diffraction to 2.7 Å resolution on our home X-ray source (Rigaku RU-200HB rotating X-ray generator operating at 100 mA, 50 kV, equipped with an R-Axis IV++ image plate detector).
X-ray intensity data reduction was achieved with Denzo/ Scalepack and Crystal Clear (Otwinowski and Minor 1997; Pflugrath 1999). Molecular replacement calculations utilized AMoRe (Navaza 1994) with the native human sEH monomer (1S8O) (Gomez et al. 2004) as the search probe. Iterative cycles of refinement and model building using CNS (Brünger et al. 1998) and O (Jones et al. 1991), respectively, improved each protein structure as monitored by Rfree. Group B-factors were utilized for the refinement of human sEH complexed with CU2 and CU6, and individual B-factors were utilized for the refinement of the human sEH–CU4 and sEH–CU7 complexes. Buffer molecules, ions, and water molecules were included in later cycles of refinement. Inhibitor molecules were added in the final stages of refinement. Data reduction and refinement statistics are recorded in Table 2.
Table 2.
Data collection and refinement statistics
| Structure | Human sEH–CU2 complex | Human sEH–CU4 complex | Human sEH–CU6 complex | Human sEH–CU7 complex |
| Limiting resolution (Å ) | 3.0 | 2.3 | 2.7 | 2.6 |
| Rmerge/last shella | 0.110/0.461 | 0.068/0.370 | 0.133/0.354 | 0.091/0.358 |
| Completeness/last shell | 99.9/100 | 50.8/86.1 | 87.0/98.2 | 76.9/93.3 |
| I/σI/last shell | 20.8/5.25 | 23.6/3.0 | 15.3/6.8 | 21.6/3.25 |
| No. of reflections, work/test | 11,485/892 | 23,477/1365 | 16,828/853 | 17,175/935 |
| R/Rfreeb | 0.232/0.283 | 0.212/0.254 | 0.233/0.283 | 0.231/0.274 |
| Protein atomsc | 4332 | 4332 | 4332 | 4332 |
| Water moleculesc | 17 | 131 | 41 | 31 |
| Metal ions/ligand atomsb | 1/19 | 1/29 | 1/23 | 1/24 |
| RMS deviations | ||||
| Bond lengths (Å ) | 0.008 | 0.007 | 0.009 | 0.008 |
| Bond angles (°) | 1.4 | 1.4 | 1.4 | 1.5 |
| Dihedral angles (°) | 22.6 | 22.5 | 22.9 | 22.4 |
| Improper dihedral angles (°) | 1.0 | 0.9 | 1.0 | 1.1 |
aRmerge= ∑|I – 〈I〉|/∑I, where I is the observed intensity and 〈I〉 is the average intensity calculated for replicate data.
b Crystallographic R factor, R=∑|Fo| – |Fc|/∑|Fo|, for reflections contained in the working set. The free R factor, Rfree=∑|Fo| − |Fc|/∑|Fo|, is calculated in the same manner for reflections contained in the test set excluded from refinement (8% of total). |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively.
c Per asymmetric unit.
Disordered segments at the N and C termini (M1 and P548-M554) were absent in electron density maps and were excluded from each final model. Figures were prepared with MOLSCRIPT2 and BOBSCRIPT with Raster3D and Pymol (Kraulis 1991; Merritt and Murphy 1994; Esnouf 1997; DeLano 2002).
Acknowledgments
We thank the Advanced Light Source, Berkeley, for synchrotron beamline access. This research was supported by NIH research grant GM63106 (D.W.C.), NIEHS grant R37 ES 02710, NIEHS grant P30 ES05707, and NIEHS Superfund Basic Research Program grant P42 ES04699 (B.D.H.). Coordinates of the human sEH–CU2, human sEH–CU4, human sEH– CU6, and the human sEH–CU7 complexes have been deposited in the Protein Data Bank (http://www.rcsb.org/pdb) with accession codes 1ZD2, 1ZD3, 1ZD4, and 1ZD5, respectively.
Article published online ahead of print. Article and publication date are at http://www.proteinscience.org/cgi/doi/10.1110/ps.051720206.
References
- Arand, M., Wagner, H., and Oesch, F. 1996. Asp333, Asp495, and His523 form the catalytic triad of rat soluble epoxide hydrolase. J. Biol. Chem. 271: 4223–4229. [DOI] [PubMed] [Google Scholar]
- Argiriadi, M.A., Morisseau, C., Hammock, B.D., and Christianson, D.W. 1999. Detoxification of environmental mutagens and carcinogens: Structure, mechanism, and evolution of liver epoxide hydrolase. Proc. Natl. Acad. Sci. 99: 10637–10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argiriadi, M.A., Morisseau, C., Goodrow, M.H., Dowdy, D.L., Hammock, B.D., and Christianson, D.W. 2000. Binding of alkylurea inhibitors to epoxide hydrolase implicates active site tyrosines in substrate activation. J. Biol. Chem. 275: 15265–15270. [DOI] [PubMed] [Google Scholar]
- Beetham, J.K., Tian, T., and Hammock, B.D. 1993. cDNA cloning and expression of a soluble epoxide hydrolase from human liver. Arch. Biochem. Biophys. 305: 197–201. [DOI] [PubMed] [Google Scholar]
- Borhan, B., Jones, A.D., Pinot, F., Grant, D.F., Kurth, M.J., and Hammock, B.D. 1995. Mechanism of soluble epoxide hydrolase. J. Biol. Chem. 270: 26923–26930. [DOI] [PubMed] [Google Scholar]
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.-S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography and NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Campbell, W.B., Gebremedhin, D., Pratt, P.F., and Harder, D.R. 1996. Identification of epoxyeicosatrienoic acids as endothelium-derived hyperpolarizing factors. Circ. Res. 78: 415–423. [DOI] [PubMed] [Google Scholar]
- Capdevila, J.H., Falck, J.R., and Harris, R.C. 2000. Cytochrome P450 and arachidonic acid bioactivation: Molecular and functional properties of the arachidonate monooxygenase. J. Lipid Res. 41: 163–181. [PubMed] [Google Scholar]
- DeLano, W.L. 2002. The PyMOL user’s manual. DeLano Scientific, San Carlos, CA.
- Esnouf, R.M. 1997. An extensively modified version of Mol Script that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model. 15: 132–134. [DOI] [PubMed] [Google Scholar]
- Fang, X., Kaduce, T.L., Weintraub, N.L., Harmon, S., Teesch, L.M., Morisseau, C., Thompson, D.A., Hammock, B.D., and Spector, A.A. 2001. Pathways of epoxyeicosatrienoic acid metabolism in endothelial cells. J. Biol. Chem. 276: 14867–14874. [DOI] [PubMed] [Google Scholar]
- Fisslthaler, B., Popp, R., Kiss, L., Potente, M., Harder, D.R., Fleming, I., and Busse, R. 1999. Cytochrome P450 2C is an EDHF synthase in coronary arteries. Nature 401: 493–497. [DOI] [PubMed] [Google Scholar]
- Fleming, I. 2001. Cytochrome P450 enzymes in vascular homeostasis. Circ. Res. 89: 753–762. [DOI] [PubMed] [Google Scholar]
- Gomez, G.A., Morisseau, C., Hammock, B.D., and Christianson, D.W. 2004. Structure of human epoxide hydrolase reveals mechanistic inferences on bifunctional catalysis in epoxide and phosphate ester hydrolysis. Biochemistry 43: 4716–4723. [DOI] [PubMed] [Google Scholar]
- Hammock, B.D., Pinot, F., Beetham, J.K., Grant, D.F., Arand, M.E., and Oesch, F. 1994. Isolation of a putative hydroxyacyl enzyme intermediate of an epoxide hydrolase. Biochem. Biophys. Res. Commun. 198: 850–856. [DOI] [PubMed] [Google Scholar]
- Hammock, B.D., Grant, D.F., and Storms D.H. 1997. Epoxide hydrolases. In Comprehensive toxicology: Biotransformation (eds. I. Sipes et al.), pp. 283–305. Elsevier, New York.
- Harder, D.R., Campbell, W.B., and Roman, R.J. 1995. Role of cytochrome P-450 enzymes and metabolites of arachidonic acid in the control of vascular tone. J. Vasc. Res. 32: 79–92. [DOI] [PubMed] [Google Scholar]
- Hu, S. and Kim, H.S. 1993. Activation of K+ channel in vascular smooth muscles by cytochrome P450 metabolites of arachidonic acid. Eur. J. Pharmacol. 230: 215–221. [DOI] [PubMed] [Google Scholar]
- Imig, J.D., Zhao, X., Capdevila, J.H., Morisseau, C., and Hammock, B.D. 2002. Soluble epoxide hydrolase inhibition lowers arterial blood pressure in angiotensin II hypertension. Hypertension 39: 690–694. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.-Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Kim, I.-H., Morisseau, C., Watanabe, T., and Hammock, B.D. 2004. Design, synthesis, and biological activity of 1,3-disubstituted ureas as potent inhibitors of the soluble epoxide hydrolase of increased water solubility. J. Med. Chem. 47: 2110–2122. [DOI] [PubMed] [Google Scholar]
- Kraulis, P.J. 1991. Molscript: A program to produce both detailed and schematic plots of protein structures. J. Appl. Cryst. 24: 946–950. [Google Scholar]
- Lacourciere, G.M. and Armstrong, R.N. 1993. The catalytic mechanism of microsomal epoxide hydrolase involves an ester intermediate. J. Am. Chem. Soc. 115: 10466–10467. [Google Scholar]
- Laughlin, L.T., Tzeng, H.-F., Lin, S., and Armstrong, R.N. 1998. Mechanism of microsomal epoxide hydrolase. Semifunctional site-specific mutants affecting the alkylation half-reaction. Biochemistry 37: 2897–2904. [DOI] [PubMed] [Google Scholar]
- McElroy, N.R., Jurs, P.C., Morisseau, C., and Hammock, B.D. 2003. QSAR and classification of murine and human soluble epoxide hydrolase inhibition by urea-like compounds. J. Med. Chem. 46: 1066–1080. [DOI] [PubMed] [Google Scholar]
- Merritt, E.A. and Murphy, M.E.P. 1994. Raster 3D Version 2.0. A program for photorealistic molecular graphics. Acta. Crystallogr. D Biol. Crystallogr. 50: 869–873. [DOI] [PubMed] [Google Scholar]
- Morisseau, C., Du, G., Newman, J.W., and Hammock, B.D. 1998. Mechanism of mammalian soluble epoxide hydrolase inhibition by chalcone oxide derivatives. Arch. Biochem. Biophys. 356: 214–228. [DOI] [PubMed] [Google Scholar]
- Morisseau, C., Goodrow, M.H., Dowdy, D., Zheng, J., Greene, J.F., Sanborn, J.R., and Hammock, B.D. 1999. Potent urea and carbamate inhibitors of soluble epoxide hydrolases. Proc. Natl. Acad. Sci. 96: 8849–8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morisseau, C., Goodrow, M.H., Newman, J.W., Wheelock, C.E., Dowdy, D.L., and Hammock, B.D. 2002. Structural refinement of inhibitors of urea-based soluble epoxide hydrolases. Biochem. Pharmacol. 63: 1599– 1608. [DOI] [PubMed] [Google Scholar]
- Müller, F., Arand, M., Frank, H., Seidel, A., Hinz, W., Winkler, L., Hänel, K., Blée, E., Beetham, J.K., Hammock, B.D., et al. 1997. Visualization of a covalent intermediate between microsomal epoxide hydrolase, but not cholesterol epoxide hydrolase, and their substrates. Eur. J. Biochem. 245: 490–496. [DOI] [PubMed] [Google Scholar]
- Nakagawa, Y., Wheelock, C.E., Morisseau, C., Goodrow, M.H., Hammock, B.G., and Hammock, B.D. 2000. 3-D QSAR analysis of inhibition of murine soluble epoxide hydrolase (MsEH) by benzoylureas, arylureas, and their analogues. Bioorg. Med. Chem. 8: 2663–2673. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Pflugrath, J.W. 1999. The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55: 1718–1725. [DOI] [PubMed] [Google Scholar]
- Pinot, F., Grant, D.F., Beetham, J.K., Parker, A.G., Borhan, B., Landt, S., Jones, A.D., and Hammock, B.D. 1995. Molecular and biochemical evidence for the involvement of the Asp-333–His-523 pair in the catalytic mechanism of soluble epoxide hydrolase. J. Biol. Chem. 270: 7968–7974. [DOI] [PubMed] [Google Scholar]
- Quilley, J. and McGiff, J.C. 2000. Is EDHF an epoxyeicosatrienoic acid? Trends Pharmocol. Sci. 21: 121–124. [DOI] [PubMed] [Google Scholar]
- Sinal, C.J., Miyata, M., Tohkin, M., Nagata, K., Bend, J.R., and Gonzalez, F.J. 2000. Targeted disruption of soluble epoxide hydrolase reveals a role in blood pressure regulation. J. Biol. Chem. 275: 40504–40510. [DOI] [PubMed] [Google Scholar]
- Wixtrom, R.N., Silva, M.H., and Hammock, B.D. 1988. Affinity purification of cytosolic epoxide hydrolase using derivatized epoxy-activated sepharose gels. Anal. Biochem. 169: 71–80. [DOI] [PubMed] [Google Scholar]
- Yamada, T., Morisseau, C., Maxwell, J.E., Argiriadi, M.A., Christianson, D.W. and Hammock, B.D. 2000. Biochemical evidence for the involvement of tyrosine in epoxide activation during the catalytic cycle of epoxide hydrolase. J. Biol. Chem. 275: 23082–23088. [DOI] [PubMed] [Google Scholar]
- Zeldin, D.C., Kobayashi, J., Falck, J.R., Winder, B.S., Hammock, B.D., Snapper, J.R., and Capdevila, J.H. 1993. Regio- and enantiofacial selectivity of epoxyeicosatrienoic acid hydration by cytosolic epoxide hydrolase. J. Biol. Chem. 268: 6402–6407. [PubMed] [Google Scholar]