Abstract
Megakaryocytes lacking transcription factor GATA-1 fail to complete maturation in vivo and hyperproliferate. To define how GATA-1 regulates megakaryocyte cell growth we searched for mRNA transcripts expressed in primary wild-type, but not GATA-1−, megakaryocytes. One differentially expressed transcript encodes inositol polyphosphate 4-phosphatase type I (4-Ptase I). This enzyme hydrolyses phosphatidylinositol 3,4-bisphosphate and also has lesser activity against soluble analogues of this lipid, inositol 3,4-bisphosphate and inositol 1,3,4-triphosphate. Reintroduction of 4-Ptase I into both primary GATA-1− and wild-type megakaryocytes significantly retards cell growth, suggesting that absence of 4-Ptase I may contribute to the hyperproliferative phenotype of GATA-1− megakaryocytes. Overexpression of 4-Ptase I also markedly reduces growth of NIH 3T3 fibroblasts. Taken together, these data indicate that 4-Ptase I is a regulator of cell proliferation.
Controlling the number of terminally differentiated cells produced from stem/progenitor cells is vital for homeostasis in all tissues. In the hematopoietic system where ≈3–5 × 1010 differentiated cells are produced daily in the adult human this is especially important. Many control mechanisms govern hematopoiesis as safeguards for regulating cell numbers in the stem/progenitor compartment, for selection of lineage fates by stem/progenitor cells, and lastly for expansion of differentiated progeny from a progenitor cell. Failure of these control mechanisms can lead to blood disorders including anemias and leukemias (1).
In hematopoietic cells the C2C2 zinc finger containing transcription factor GATA-1 plays a nonredundant role in differentiation of selected lineages. In erythroid cells analysis of GATA-1− mice, complemented by in vivo and in vitro analysis of GATA-1− embryonic stem cells, demonstrates that GATA-1 is required for differentiation and survival of erythroid cells beyond the proerythroblast stage (2–5). Moreover, a threshold level of GATA-1 is required for this process (6). Although many erythroid-specific GATA-1 target genes are known, the pivotal GATA-1 effector genes regulating growth differentiation and apoptosis remain poorly defined. Recently, GATA-1 has been shown to augment expression of the antiapoptotic gene Bcl-XL in erythroid cells, indicating one potential mechanism to forestall apoptosis (7).
GATA-1 expression is also essential for full megakaryocyte differentiation and platelet release (8, 9). Numerous megakaryocyte-specific genes harbor functionally important GATA sites in their cis elements (10–12). However, in contrast to red cells, loss of megakaryocyte-specific GATA-1 expression does not lead to apoptosis but to marked accumulation of immature megakaryocytes (8, 9). In vivo, mutant mice exhibit a 10- to 100-fold increase in megakaryocyte numbers in bone marrow and spleen. Although the mechanism responsible for this megakaryocytosis is uncertain, evidence suggests it is consequent to abnormal expansion of megakaryocytes from a normal number of committed progenitors. Colony assays reveal normal numbers of GATA-1− megakaryocyte progenitors. However, a proportion of these progenitors produce abnormally large colonies composed of thousands of immature megakaryocytes. Such colonies are never observed from wild-type progenitors. Excessive growth of GATA-1− megakaryocytes expanded from progenitors also is seen in liquid culture; maximal megakaryocyte numbers are 16-fold higher from GATA-1− cultures compared with wild-type, and mutant cultures persist for much longer than controls (9). Taken together, prior studies indicate that growth and survival of developing erythroid and megakaryocytic precursors are regulated in part through GATA-1. Moreover, its role in these processes is lineage-context-dependent.
To search for genes controlled by GATA-1 in megakaryocytes, we performed subtractive hybridization between wild-type and GATA-1− primary megakaryocyte cDNAs. Among the transcripts identified, one encoded inositol polyphosphate 4-phosphatase type I (4-Ptase I), which normally was expressed in megakaryocytes but was absent in GATA-1- megakaryocytes. In platelets, 4-Ptase I has been previously implicated in signaling by regulating phosphatidylinositol 3-kinase (PI 3-kinase; refs. 13, 14). The cDNA of the two isoforms of 4-Ptases (types I and II) that encode proteins with 37% amino acid identity have been cloned and characterized (15, 16). Both are widely expressed, although their patterns of expression vary among tissues.
In vitro, 4-Ptases catalyze the hydrolysis of the D-4 position phosphate of the PI 3-kinase second messenger, phosphatidylinositol 3,4-bisphosphate [PtdIns(3,4)P2]. In addition, these enzymes catalyze the hydrolysis of the soluble analogues of this lipid, inositol 3,4-bisphosphate [Ins(3,4)P2] and inositol 1,3,4-triphosphate [Ins(1,3,4)P3] (15–18). The PI 3-kinase signal pathway has been shown to regulate cellular growth, differentiation, and apoptosis in many cell types (19). PtdIns(3,4)P2, together with phosphatidylinositol 3,4,5-triphosphate [PtdIns(3,4,5)P3], are rapidly produced after activation of PI 3-kinase. Several intracellular targets of these two lipids have been identified including isozymes of protein kinase C, protein kinase B (PKB/Akt), and protein kinase PDK1. These protein kinases have multiple potential substrates involved in regulation of cell proliferation, differentiation, and apoptosis. Genetic studies have indicated that phosphatases of these lipid second messengers are essential for the regulation of PI 3-kinase signaling. The human tumor suppressor gene PTEN is a phosphatase that catalyzes the hydrolysis of the D-3 position phosphate of PtdIns(3,4)P2 and PtdIns(3,4,5)P3 (for a review see ref. 19). Moreover, mutant mice lacking SHIP, a phosphatase that dephosphorylates the D-5 position of PtdIns(3,4,5)P3 (20) develop a lethal myeloproliferative syndrome, strongly implicating PI 3-kinase signaling in the regulation of hematopoietic cell numbers (21). The work described here suggests that 4-Ptase I expression in mouse megakaryocytes is GATA-1-dependent and that this phosphatase is involved in the regulation of megakaryocyte and fibroblast proliferation.
Methods
Generation of GATA-1− Mice.
Megakaryocytes from GATA-1− (neoΔHS) mice lack GATA-1 and have been described (8).
Generation and Analysis of the Subtracted Library.
Single-cell fetal liver suspensions from five neoΔHS fetuses and 15 normal littermate controls at embryonic day 12.5 were cultured as described (9). Cells were harvested at day 3 when the percentage of megakaryocytes is maximal (≈70% of the culture). A total of 1,248 library clones from a subtracted cDNA library (PCR-Select protocol, CLONTECH) were gridded onto nylon membranes. Duplicate membranes were hybridized with 32P dCTP-labeled aliquots of the original normal and mutant megakaryocyte cDNA used for subtraction. Clones hybridizing to wild-type, but not GATA-1−, cDNA were selected for sequencing.
Semiquantitative Reverse Transcriptase–PCR (RT-PCR) Analysis of 4-Ptase I Expression.
Semiquantitative RT-PCR was performed as described (9) with one modification. As the amount of starting hypoxanthine phosphoribosyl transferase (HPRT) cDNA is greater than 4-Ptase I cDNA, 10-fold less 32P tracer was added to reactions with HPRT primers. cDNA amounts in PCRs were normalized for HPRT expression. No reaction products were detected in mRNA samples without RT (data not shown). The oligonucleotide primer pair used to amplify HPRT has been described (22). The positions of oligonucleotides F and G used to amplify murine 4-Ptase I are shown. Their sequences are: F, 5′-GGCATGCTGCTGCGGGTG-3′ and G, 5′-TCGGACCCCGTTAAGGCGGCG-3′.
Western Blot Analysis of 4-Ptase I Expression.
Western blots were performed as described (9, 15). One filter was sequentially used to detect expression of 4-Ptase I and c-src.
Cloning and Sequencing of Murine 4-Ptase I.
The initial murine 4-Ptase I clone was used to screen a primary murine megakaryocyte-enriched (≈70% megakaryocytes) cDNA library constructed in the vector λ Ziplox (GIBCO/BRL). A 1.7-kb clone corresponding to one of the 3′ ends of 4-Ptase I was isolated. The rest of the murine cDNA was obtained by RT-PCR. The position of the oligonucleotide primers is shown in Fig. 1A. The 5′ end of the coding region was cloned by seminested PCR. Primer sequences were: A, 5′-ATGACAGCAAGAGAGCACAGC-3′; B, 5′-TCCTGTGGTATGTAGATTATGGACTG-3′; C, 5′-TTGATCTGAGCAATGATCTCC-3′; D, 5′-CAGTCCATAATCTACATACCACAGGA-3′; E, 5′-TCATGTCTCAACTTTTCCGTAAGTCCC-3′; H, 5′-CCCAAGCATTACAGGCCTCCA-3′; and I, 5′-CTCACAGCTGGCTTTACATCT-3′.
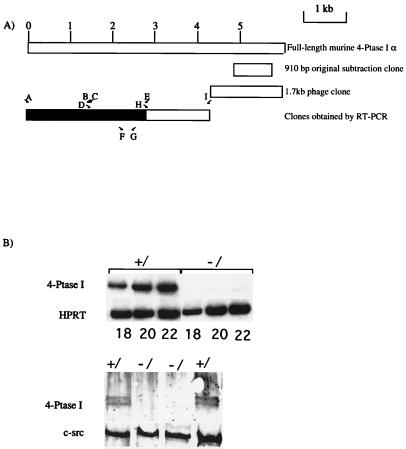
Figure 1.
(A) Schematic representation of murine 4-Ptase I cDNA clones. (Upper) The assembled 4-Ptase I α cDNA is depicted by the full open box. (Lower) The location of the initial clone obtained from subtraction analysis; the phage clone that contained a poly(A) tail and lastly a composite cDNA of clones obtained by RT-PCR (shaded box corresponds to the coding region). The locations of the oligonucleotide primers (A–E, H, and I) used to clone the cDNA and oligonucleotide primers F and G used in the RT-PCR to document expression of 4-Ptase I in normal and GATA-1- megakaryocytes are shown. (B, Upper) mRNA expression of 4-Ptase I in normal (+/) and GATA-1− (−/) megakaryocytes. cDNA from these two megakaryocyte populations was used in a PCR. Oligonucleotide primers F and G (see A for positions) in the 3′ end of the coding region of mouse 4-Ptase I were used. The amount of input cDNA was normalized with respect to HPRT expression as shown. Numbers of PCR cycles performed to obtain the cDNA product are shown below each lane. (Lower) Western blot analysis of 4-Ptase I protein expression from normal (+/) and GATA-1− (−/) primary fetal liver cells containing ≈ 70% megakaryocytes. Above, 4-Ptase I protein was detected, as a doublet, by antibody to the C terminal of 4-Ptase I. This antibody reacts with the C termini of both 4-Ptase I and II. Below, the same blot was stripped and reprobed with an antibody to c-src to document protein loading in all of the lanes.
cDNA from primary megakaryocytes was used as template in RT-PCRs. NotI and XhoI restriction enzyme sites were incorporated into all oligonucleotide primers (except C). NotI- and XhoI-digested PCR products were cloned into NotI and XhoI sites of pcDNA 3.1 (Invitrogen). To reduce PCR sequence errors, two independent PCRs were performed with Pfu DNA polymerase. Multiple clones from each of the two PCRs were sequenced completely on both strands.
Northern Blot Analysis.
A mouse multitissue Northern blot (CLONTECH) was hybridized with a mixture of fragments corresponding to all of cloned mouse 4-Ptase I cDNA. To demonstrate equivalent loading and integrity of mRNAs, the blot was stripped and rehybridized to a mouse β actin probe.
Cloning of Retroviral Vectors.
The vector PGDpuro (gift from M. Weiss, Children's Hospital, Philadelphia), containing a puromycin selection cassette, was linearized by digestion with NotI and blunt-ended. Full-length XhoI–EcoRI human 4-Ptase I fragment (15) was blunt-ended and cloned into this vector. The cloning site was sequenced to ensure the orientation and junctions of the cloned fragment were correct. The same XhoI–EcoRI human 4-Ptase I fragment was cloned into the XhoI-EcoRI sites of the vector MSIG (gift from Merav Socolovsky, Massachusetts Institute of Technology, Cambridge), which has a MSCV backbone with an IRES-green fluorescent protein (GFP) cassette.
Production of Retroviral Supernatants.
Retroviral constructs were transiently transfected into the BOSC 23 producer cells by using Fugene (Roche Diagnostics). Supernatants were prepared as described (23).
Infection of Primary Megakaryocyte Progenitors.
Single cell suspension of ≈5 × 107 fetal liver cells from GATA-1− and wild-type mice were resuspended in PBS/1% FCS. To partially deplete lineage committed cells, cells were incubated at 4°C for 30 min with the following biotinylated antibodies: 30 μl of TER-119 (PharMingen), 10 μl of Mac-1 (PharMingen), and 10 μl of Gr-1 (PharMingen). Cells were washed once in PBS/1% FCS. Two hundred microliters of avidin-coated magnetic beads (PerSeptive Biosystems, Framingham, MA) was added, and cells binding to beads were removed by a magnetic separator. A total of ≈1–5 × 105 lineage-depleted cells were cultured for 48 h in DMEM, with 1% recombinant human thrombopoietin tissue culture supernatant (24) and stem cell factor (R&D Technologies, Minneapolis, MN). Cells then were incubated for 24 or 48 h with a mixture of 50% concentrated retroviral supernatant, 50% DMEM with 2% recombinant human thrombopoietin tissue culture supernatant supplemented with 4 μg/ml of polybrene.
Colony Assays of Infected Megakaryocyte Progenitors.
Equal numbers of lineage-depleted GATA-1− fetal liver cells (between 1 and 5 × 105) were infected for 48 h with three sets of PGD retroviral supernatants, containing either human full-length 4-Ptase I and a puromycin selectable marker, or the selectable marker alone, or empty virus. Colony assays were preformed as described (22), but in addition, puromycin (1 μg/ml) was added. After 5–6 days, the numbers of macroscopic and normal-size CFU-Mk colonies were counted. Normal microscopic colonies were defined as those less than 0.3 mm in diameter, and abnormal macroscopic colonies were defined as those greater than 1.5 mm in diameter.
Growth Curves of Infected Megakaryocyte Progenitors.
Wild-type or GATA-1− fetal liver cells were infected for 24 h with one of three sets of retroviral MSCV supernatants; supernatant containing human full-length 4-phosphatase and a GFP cassette, or just the GFP cassette or the vector alone. Infected cells were selected by sorting for cells expressing GFP on a high-speed Flomation FACS machine and gated to exclude 99.9% of cells that had been infected with vector alone. Equal numbers of infected cells were plated in 2 ml of DMEM supplemented with 1% recombinant human thrombopoietin tissue culture supernatant. Growth curves were determined as described (9).
Human 4-Ptase I expression was measured in infected GATA-1− megakaryocytes expanded in liquid culture. Twenty microliters of the culture was removed every 2 days and added to the same tube containing 200 μl of RNAZOL B (Tel-Test, Friendwood, TX). RNA was extracted according to the manufacturer's protocol, and cDNA was made using oligo(dT) and random hexamer primers (Roche Diagnostics). Semiquantitative RT-PCR was performed as described above. Sequences of the oligonucleotide primers used to amplify human 4-Ptase I in infected progenitors are: 5′-CATTGCGAAAGGACACTTTG-3′ and 5′-GCTCCTCCAGCTCACACACT-3′.
Infection of 3T3 Cells.
Viral supernatants containing the virus PGDpuro-4-Ptase I (see above) or the PDGpuro control were used to infect 5 × 105 NIH 3T3 cells (23). Populations of infected cells were selected in DMEM containing 2 μg/ml puromycin, and overexpression of 4Ptase Iα was confirmed by immunofluorescence and Western blot analysis using rabbit antiserum against the C-terminal peptide of 4-Ptase I (15).
Growth Curves of Infected 3T3 Cells.
Stable populations of NIH 3T3 cells infected with control retrovirus or retrovirus expressing 4-Ptase I were established by puromycin selection. Cells were plated in triplicate at a density of 5,000/well in 12-well plates and grown in DMEM containing 10% heat-inactivated calf serum. At the time points indicated, cells were washed twice in PBS and cell numbers were determined by using the Quantos Cell Proliferation Assay (Stratagene), using the manufacturer's protocol.
Measurement of Apoptosis of Infected 3T3 Cells.
Apoptotic death in NIH 3T3 cells was determined by measuring the percentage of cells with DNA content less than that of G1 cells by FACS analysis using fixed cells stained with propidium iodide as described (25).
Results
Absence of 4-Ptase I in GATA-1− Megakaryocytes.
To identify novel GATA-1-regulated genes in megakaryocytes, we used subtractive cDNA hybridization and searched for mRNA transcripts expressed more abundantly in primary normal megakaryocytes compared with those lacking GATA-1. One clone from this screen contained a 910-bp fragment that corresponded to the 3′ noncoding region of murine 4-Ptase I (Fig. 1A). Semiquantitative RT-PCR using cDNAs from normal and GATA-1− megakaryocyte colonies (CFU-Mk) as templates with oligonucleotide primers, F and G (Fig. 1A), confirmed differential expression of 4-Ptase I (Fig. 1B). 4-Ptase I is also differentially expressed at the protein level. Whole-cell protein extracts of purified wild-type and mutant megakaryocytes were subjected to Western blot analysis. Antibody to 4-Ptase I C-terminal (15) detected a doublet in wild-type, but not mutant, samples (Fig. 1B). As neither 4-Ptase I mRNA nor protein is expressed in megakaryocytes lacking GATA-1, 4-Ptase I is likely to be either a direct or indirect downstream target of GATA-1. We also confirmed that 4-Ptase-1 expression was not reduced in cells that normally do not express GATA-1 (brain, not shown).
Conservation of 4-Ptase I Sequence.
The initial clone isolated from the subtracted library mapped to sequences in the 3′ untranslated region (UTR) of mouse 4-Ptase I (Fig. 1A). Further sequence of mouse 4-Ptase I was obtained by two approaches. First, a 1.7-kb clone containing a poly(A) tail was obtained (Fig. 1A) by screening a primary murine megakaryocyte cDNA library. This clone extended the 3′ UTR sequence and suggested that at least one bona fide 3′ end of the gene had been cloned. Both 4-Ptases I and II possess alternative 3′ ends (α and β). The murine 3′ end we have cloned corresponds to 4-Ptase Iα. Second, as previously cloned human and rat 4-Ptases I (15) show extensive nucleotide homology, the remainder of the murine gene was cloned by RT-PCR. Oligonucleotide primers (A-F, H, and I, Fig. 1A) were designed at intervals along the cDNA, either in areas of high nucleotide sequence conservation or in known sequence in the 3′ UTR (Fig. 1A) and used in RT-PCRs with primary murine megakaryocyte cDNA as template.
Alignment of the deduced mouse, rat, and human 4-Ptase I protein sequences reveals remarkable sequence conservation not just within the phosphatase domain, but also throughout the entire length of the protein (data not shown). Mouse and human proteins are 96% identical in amino acid sequence. Sixty percent of the amino acid substitutions are conservative.
Pattern of Expression of 4-Ptase I.
cDNA fragments encompassing mouse 4-Ptase I were hybridized to a multitissue Northern blot (data not shown). 4-Ptase I is widely expressed as a 6.3-kb transcript, save for testes where a 4.8-kb transcript is predominant. The tissue-specific pattern of murine 4-Ptase I mRNA expression closely mirrors that of rat (15) with the exception of kidney, where 4-Ptase I expression was not seen in the mouse.
Effect of 4-Ptase I Expression on Megakaryocyte Growth.
GATA-1− megakaryocytes obtained from fetal liver progenitors accumulate in vivo and in vitro and exhibit enhanced proliferation and prolonged culture lifespan in the presence of the megakaryocyte cytokine thrombopoietin. Given the potential role of 4-Ptase I in cell growth control (see Introduction), we asked whether failure to express 4-Ptase I might account, at least in part, for hyperproliferation of GATA-1− megakaryocytes. To address this, we introduced full-length human 4-Ptase I cDNA into primary GATA-1− megakaryocyte progenitors by retroviral-mediated gene transfer and assessed megakaryocyte growth both in liquid culture and colony assays.
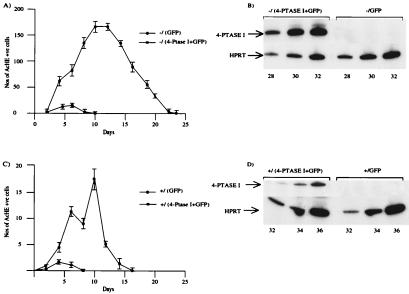
As shown in Fig. 2A, primary GATA-1− megakaryocytes transduced with control retrovirus (expressing GFP) proliferate abundantly for >20 days in liquid culture. Forced expression of human 4-Ptase I markedly reduces total megakaryocyte growth and culture lifespan (Fig. 2A). We asked whether expression of human 4-Ptase I also might affect growth of normal megakaryocytes. As shown in Fig. 2C, forced expression of exogenous 4-Ptase I also suppressed growth of normal megakaryocytes. However, the effect is less marked, most probably because the number of megakaryocytes generated from wild-type progenitors is far smaller (9). Note the different ordinates in Fig. 2 A versus C. Human 4-Ptase I mRNA expression in cells infected with virus containing human 4-Ptase I was confirmed by RT-PCR (Fig. 2 B and D). Thus, reexpression of 4-Ptase I in GATA-1− megakaryocyte precursors markedly inhibits growth of megakaryocytes.
Figure 2.
(A) Growth curves of primary megakaryocytes from GATA-1− progenitors. Progenitors infected with one of two different retroviruses, one containing human 4-Ptase I and GFP and the other just GFP on its own. Selected progenitors then were expanded in liquid culture with 1% recombinant thrombopoietin-conditioned tissue culture medium. The number of cells with acetylcholinesterase activity in 20-μl aliquots of the culture is shown as a function of the duration of culture. The values shown are an average ± 1 SD from three experiments. (B) Human 4-Ptase I expression was assayed by RT-PCR in aliquots of the two retrovirally infected GATA-1− cultures in A. (Upper) Human 4-Ptase I expression was assayed, and the input amount of cDNA was normalized with respect to HPRT expression (Lower). The number of PCR cycles is shown at the bottom. (C) Growth curves of primary megakaryocytes from retrovirally infected progenitors of normal littermate embryos of GATA-1− embryos. The growth curves were performed in the same manner as A. Note the different ordinates in A versus C. (D) Human 4-Ptase I expression was assayed by RT-PCR in aliquots of the two retrovirally infected cultures of normal progenitors in C. The RT-PCR analysis is illustrated as in B.
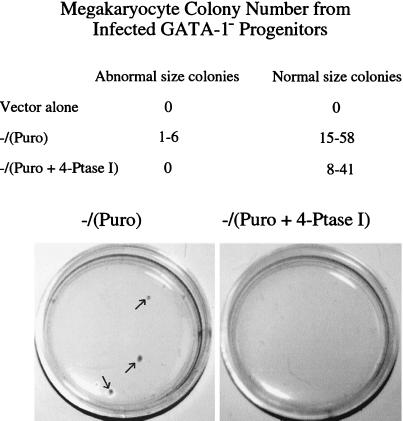
Effects of 4-Ptase I expression on megakaryocyte colony formation also were assessed. Three sets of retroviral infections were performed. Retroviral supernatants containing either human 4-Ptase I and a puromycin selection marker, or the selection marker, or the retroviral vector alone, were used to infect GATA-1− fetal liver hematopoietic progenitors. A day thereafter, progenitors were plated in methylcellulose containing puromycin and recombinant thrombopoietin. Colonies were enumerated 5–6 days later (Fig. 3). Progenitors infected with virus containing puromycin-resistance gene alone produced two types of colonies: normal microscopic and abnormal, macroscopic colonies, as anticipated from prior studies (8). No puromycin-resistant colonies were seen with infection of cells with vector alone. Forced 4-Ptase I expression led to a complete loss of macroscopic colonies, and a slight reduction of normal microscopic CFU-Mk was also slightly reduced (≈30% reduction in the average colony number). This difference was significant, χ2 = 12.3, P < 0.01. Taken together, these studies demonstrate that reintroduction of 4-Ptase I into GATA-1− megakaryocytes markedly retards megakaryocyte growth both at the level of the whole population of cells and megakaryocyte colony growth from progenitors.
Figure 3.
Equal numbers of GATA-1− hematopoietic progenitors were infected with retroviruses containing either empty retrovirus (vector alone), the puromycin selection marker [−/(puro)], or human 4-Ptase I plus the puromycin-resistance gene [−/(4-Ptase + Puro)]. In each experiment equal numbers of lineage-depleted GATA-1− fetal liver cells (1–5 × 105) were infected for 48 h with the retroviral supernatants indicated. The cells then were plated in methylcellulose containing 1% recombinant thrombopoietin conditioned tissue culture medium with 1 μg/ml puromycin. The number and size of the colonies was assessed 5–6 days after plating. The numbers of abnormal and normal size colony from three independent experiments are shown. (Lower) Plates of GATA-1− mutant progenitors infected with [−/(4-Ptase + Puro)] or without [−/(puro)] 4-Ptase I are shown. At this magnification (×2), only the abnormal-size megakaryocyte colonies can be seen and are present only in the −/(puro) plate (indicated by the arrows). Infection with 4-Ptase I abolishes growth of these colonies in the −/(4-Ptase + Puro) plate.
Growth Suppressive Effect of 4-Ptase I in Fibroblasts.
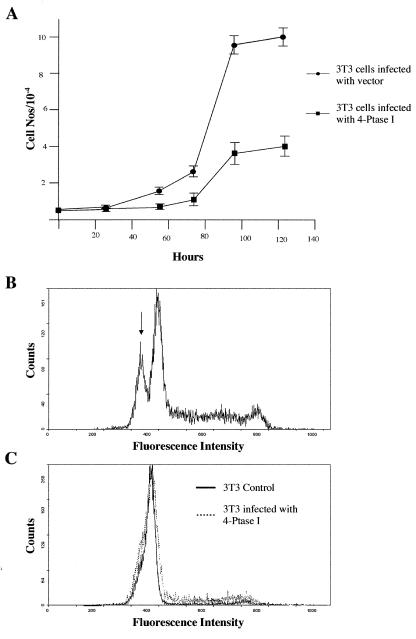
As 4-Ptase I is widely expressed, we asked whether 4-Ptase I might influence cell growth in other contexts. NIH 3T3 fibroblasts were infected with retroviruses containing either human 4-Ptase I and a puromycin selection marker or the selection marker alone. After selection in puromycin, equal numbers of cells were plated and growth curves were determined by measuring DNA content of the cell population at different time points (Fig. 4A). Maximal cell numbers produced were reduced 2.5-fold upon forced expression of 4-Ptase I. To determine whether this resulted from decreased proliferation or increased cell death, numbers of apoptotic cells in cultures infected with 4-Ptase I to those with vector alone were determined (Fig. 4 B and C). No difference was observed providing evidence that stable exogenous expression of 4-Ptase I in NIH 3T3 cells primarily suppresses proliferation.
Figure 4.
(A) Effect of 4-Ptase I overexpression on the proliferation of NIH 3T3 cells. Time course for the growth of NIH 3T3 cells infected with control retrovirus (●) or retrovirus overexpressing 4-Ptase I (■). Points represent the average of three values. Error bars indicate the range of values measured for each time point. 4-Ptase I overexpression does not enhance apoptosis of NIH 3T3 cells. (B) FACS analysis of the DNA content of NIH 3T3 cells infected with control retrovirus 24 h after apoptosis induction by trypsin treatment. The arrow indicates the population of apoptotic cells with DNA content less than G1 cells. (C) FACS analysis of the DNA content of NIH 3T3 cells infected with control retrovirus (solid line) or retrovirus overexpressing 4-Ptase I (dashed line).
Discussion
As a critical transcriptional regulator GATA-1 coordinates differentiation and proliferation in two related, but distinct, hematopoietic lineages (erythroid and megakaryocytic). By defining its downstream molecular targets, we hoped to gain insight into this aspect of GATA-1 function and, more generally, into biochemical pathways activated in terminal maturation. To identify potential target genes we searched for mRNA transcripts expressed in normal, but not in GATA-1−, primary megakaryocytes. Among sequences isolated, those encoding 4-Ptase I met these criteria. Three important conclusions derive from our findings. First, 4-Ptase I is likely to be either a direct or indirect target gene of GATA-1 in megakaryocytes. Second, overexpression of 4-Ptase I can substantially modulate cell growth. Third, these results suggest that GATA-1 influences megakaryocyte proliferation, in part, by interfacing with inositol phosphate signaling.
At least three reasons may explain how GATA-1 influences 4-Ptase I expression. First, failure to express 4-Ptase I might simply reflect the more immature status of megakaryocytes from a GATA-1− background compared with wild type. We think this possibility is unlikely, however, as 4-Ptase I expression does not markedly change during megakaryocytic differentiation of the megakaryoblastic cell lines L8057 and Meg-01 (P.V., F.A.N., P.W.M., and S.H.O., unpublished data). Conversely, expression of 4-Ptase I could directly depend on GATA-1 and thus represent a bona fide GATA-1 transcriptional target. Lastly, expression might be regulated indirectly further downstream of GATA-1.
Our results demonstrate that overexpression of 4-Ptase I inhibits cell growth. In fibroblasts in particular, growth suppression appears principally because of impaired cell proliferation rather than increased apoptosis. Although biochemical evidence implicates 4-Ptases as the major enzymes regulating metabolism of the lipid second messenger, phosphatidylinositol 3,4-bisphosphate [PtdIns(3,4)P2] and, with less efficiency, the soluble analogues of this lipid (15, 18), their in vivo function remains ill defined. To address this issue, future studies will determine the effect of 4-Ptase overexpression on the intracellular levels of PtdIns(3,4)P2 and the soluble substrates of this enzyme. Given the extensive conservation of 4-Ptase I proteins in regions outside the phosphatase domain, it is possible that this enzyme influences cell growth by a mechanism(s) other than that of metabolism of inositol phosphates. A related question still to be addressed is whether 4-Ptase I's growth suppressive action is coupled with enhanced differentiation. To begin addressing these issues it will be necessary to test the effects of wild type 4-Ptase I and a phosphatase-inactive mutant on primary megakaryocyte proliferation and differentiation.
Acknowledgments
We thank Joy Walker for technical assistance with the NIH 3T3 cell proliferation assays. This work was supported in part by fellowships and grants from the Wellcome Trust (P.V.), National Institutes of Health (F.A.N., P.W.M., and S.H.O.), and the American Heart Association (F.A.N.). S.H.O. is an Investigator of the Howard Hughes Medical Institute.
Abbreviations
- 4-Ptase I
inositol polyphosphate 4-phosphatase type I
- PI 3-kinase
phosphatidylinositol 3-kinase
- RT-PCR
reverse transcriptase–PCR
- HPRT
hypoxanthine phosphoribosyl transferase
- GFP
green fluorescent protein
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF317838).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250476397.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250476397
References
- 1.Enver T, Greaves M. Cell. 1998;94:9–12. doi: 10.1016/s0092-8674(00)81215-5. [DOI] [PubMed] [Google Scholar]
- 2.Pevny L, Simon M C, Robertson E, Klein W H, Tsai S-F, D'Agati V, Orkin S H, Costantini F. Nature (London) 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 3.Weiss M J, Orkin S H. Proc Natl Acad Sci USA. 1995;92:9623–9627. doi: 10.1073/pnas.92.21.9623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weiss M J, Keller G, Orkin S H. Genes Dev. 1994;8:1184–1197. doi: 10.1101/gad.8.10.1184. [DOI] [PubMed] [Google Scholar]
- 5.Fujiwara Y, Browne C P, Cunniff K, Goff S C, Orkin S H. Proc Natl Acad Sci USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDevitt M A, Shivdasani R A, Fujiwara Y, Yang H, Orkin S H. Proc Natl Acad Sci USA. 1997;94:6781–6785. doi: 10.1073/pnas.94.13.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregory T, Yu C, Ma A, Orkin S H, Blobel G A, Weiss M J. Blood. 1999;94:87–96. [PubMed] [Google Scholar]
- 8.Shivdasani R A, Fujiwara Y, McDevitt M A, Orkin S H. EMBO J. 1997;16:3965–3973. doi: 10.1093/emboj/16.13.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vyas P, Ault K, Jackson C W, Orkin S H, Shivdasani R A. Blood. 1999;93:2867–2875. [PubMed] [Google Scholar]
- 10.Lemarchandel V, Ghysdael J, Mignotte V, Rahuel C, Romeo P-H. Mol Cell Biol. 1993;13:668–676. doi: 10.1128/mcb.13.1.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uzan G, Prenant M, Prandini M-H, Martin F, Marguerie G. J Biol Chem. 1991;266:8932–8939. [PubMed] [Google Scholar]
- 12.Block K L, Ravid K, Phung Q H, Poncz M. Blood. 1994;84:3385–3393. [PubMed] [Google Scholar]
- 13.Norris F A, Atkins R C, Majerus P W. J Biol Chem. 1997;272:10987–10989. doi: 10.1074/jbc.272.17.10987. [DOI] [PubMed] [Google Scholar]
- 14.Munday A D, Norris F A, Caldwell K K, Brown S, Majerus P W, Mitchell C A. Proc Natl Acad Sci USA. 1999;96:3640–3645. doi: 10.1073/pnas.96.7.3640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Norris F A, Auethavekiat V, Majerus P W. J Biol Chem. 1995;270:16128–16133. doi: 10.1074/jbc.270.27.16128. [DOI] [PubMed] [Google Scholar]
- 16.Norris F A, Atkins R C, Majerus P W. J Biol Chem. 1997;272:23859–23864. doi: 10.1074/jbc.272.38.23859. [DOI] [PubMed] [Google Scholar]
- 17.Bansal V S, Caldwell K K, Majerus P W. J Biol Chem. 1990;265:1806–1811. [PubMed] [Google Scholar]
- 18.Norris F A, Majerus P W. J Biol Chem. 1994;269:8716–8720. [PubMed] [Google Scholar]
- 19.Cantley L C, Neel B G. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Damen J E, Liu L, Rosten P, Humphries R K, Jefferson A B, Majerus P W, Krystal G. Proc Natl Acad Sci USA. 1996;93:1689–1693. doi: 10.1073/pnas.93.4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Helgason C D, Damen J E, Rosten P, Grewal R, Sorensen P, Chappel S M, Borowski A, Jirik F, Krystal G, Humphries R K. Genes Dev. 1998;12:1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shivdasani R A, Rosenblatt M F, Zucker-Franklin D, Jackson C W, Hunt P, Saris C, Orkin S H. Cell. 1995;81:695–704. doi: 10.1016/0092-8674(95)90531-6. [DOI] [PubMed] [Google Scholar]
- 23.Pear W S, Nolan G P, Scott M L, Baltimore D. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Villeval J L, Cohen-Solal K, Tulliez M, Giraudier S, Guichard J, Burstein S A, Cramer E M, Vainchenker W, Wendling F. Blood. 1997;90:4369–4383. [PubMed] [Google Scholar]
- 25.Crissman H A, Steinkamp J A. Cytometry. 1982;3:84–90. doi: 10.1002/cyto.990030204. [DOI] [PubMed] [Google Scholar]