Abstract
We determined the complete mtDNA sequence of the centipede Lithobius forficatus and found that only one of the 22 inferred tRNA genes encodes a fully paired aminoacyl acceptor stem. The other 21 genes encode tRNAs with up to five mismatches in these stems, and some of these overlap extensively with the downstream genes. Because a well-paired acceptor stem is required for proper tRNA functioning, RNA editing in the products of these genes was suspected. We investigated this hypothesis by studying cDNA sequences from eight tRNAs and found the editing of up to 5 nt at their 3′ ends. This editing appears to occur by a novel mechanism with the 5′ end of the acceptor stem being used as a template for the de novo synthesis of the 3′ end, presumably by an RNA-dependent RNA polymerase. In addition, unusual secondary structures for several tRNAs were found, including those lacking a TΨC (T) or a dihydrouridine (D) arm, and having an unusual number of base pairs in the acceptor or anticodon stems.
RNA editing has been defined as “any programmed alteration of RNA primary structure to generate a sequence that could have been directly encoded at the DNA (gene) level” (1). First discovered less than 15 years ago, it is now regarded as a widespread phenomenon occurring in all types of eukaryotic RNA, mainly in mitochondria, less frequently in chloroplasts, and, in a few cases, in the nucleus (2). Four different types of tRNA editing have been found to date (all of them in mitochondria): cytidine to uridine conversion, cytidine or uridine insertion, template-dependent editing of the first three nucleotides at the 5′ ends of tRNAs, and template-independent editing at the 3′ ends of tRNAs (for review, see ref. 3). With a single exception (4), only the last type has been reported so far in animal tRNAs. None of the previously described types of tRNA editing, however, could serve as a potential editing mechanism for the aberrant tRNA structures encoded by the mtDNA of the centipede Lithobius forficatus.
We have sequenced the complete mtDNA of L. forficatus (GenBank accession no. AF309492) and found that all but one of its 22 tRNA genes code for tRNAs with at least one and as many as five mismatches in their aminoacyl acceptor stems (henceforth termed acceptor stems). Some, but not all, of these tRNA genes, if they encode full-length acceptor stems, would also overlap with their downstream genes. Because a well-matched acceptor stem is important for defining tRNA structure (5), directing tRNA processing (6), and assisting tRNA recognition by aminoacyl-tRNA synthetase (7), we suspected that the poorly matched acceptor stems of L. forficatus tRNAs are posttranscriptionally edited. We studied this hypothesis by analyzing cDNA sequences from eight tRNAs and found that a previously unknown type of tRNA editing is present in the mitochondria of L. forficatus.
Materials and Methods
tRNA and DNA Preparation.
Total RNA was prepared by using TRIzol reagent (GIBCO/BRL) from a frozen and homogenized L. forficatus centipede collected in Ann Arbor, MI. To eliminate most of the rRNA, we modified previously published protocols (8, 9) as follows. Total RNA was dissolved in 200 μl of 2 M LiCl/0.1 M potassium acetate, pH 5.0. The mixture was chilled on ice for 5 min and centrifuged for 20 min at 5,000 × g. The supernatant, containing the salt-soluble RNA fraction (mostly tRNA) was transferred to a new tube and RNA was precipitated by the addition of 3 vol of 100% ethanol and centrifugation at 12,000 × g for 15 min. The RNA pellet was washed with 80% ethanol and dissolved in 30 μl of water. The DNA was prepared from the same centipede by using the 2× cetyltrimethylammonium bromide extraction buffer, phenol/chloroform extraction and ethanol precipitation.
tRNA Circularization and cDNA Synthesis.
RNA was ligated by using T4 RNA ligase (GIBCO/BRL) and cDNA was synthesized by using avian myeloblastosis virus reverse transcriptase (Promega) under conditions as described (1). Primers used for reverse transcription were Litho-trnC-R (5′-ataggatccAAACTAGGATTTACCTAATC-3′) for tRNA(C), Litho-trnE-R (5′-ataggatccGTGTGTTGTGATAAATTTTC-3′) for tRNA(E), Litho-trnH-R (5′-ataggatccACAGGCCATTATTCTTC-3′) for tRNA(H), Litho-trnM-R (5′-cacaagcttATGAACCCAGTAGCTTAATTTAGC-3′) for tRNA(M), Litho-trnN-R (5′-ataggatccCAGTGAATAGTCTAGTTCATGAC-3′) or Litho-trnN-RM (5′-ataggatccCAGTGAATAGTCTAGTTC-3′) for tRNA(N), Litho-trnQ-R (5′-ataggatccAAATTATTATGCTAAACATC-3′) for tRNA(Q), Litho-trnR-R (5′-cacaagcttCGAAACTGATTGCAATATATCGC-3′) for tRNA(R), and Litho-trnS1-R (5′-ataggatccGTTAGCGGCTCATGCGC-3′) for tRNA(S). Throughout, tRNAs are designated by the one-letter amino acid code as tRNA(X), with the two leucine (L) and two serine (S) tRNAs differentiated by anticodon sequences as L1 (anticodon = UAG), L2 (UAA), S1 (UCU) and S2 (UGA); the tRNA genes are named trnX. The 5′ end 9 nt in these primers (in lowercase type) were added to create a restriction site for HindIII or BamHI plus three terminal nucleotides.
PCR Amplification, Cloning, and Sequencing.
PCR amplification of cDNA was performed by using a Fisher Biotech Taq DNA polymerase kit and the following primer pairs: Litho-trnC-R and Litho-trnC-F (5′-cacaagcttTTGATTGCAAGTCTTACTCAG-3′) for tRNA(C), Litho-trnE-R and Litho-trnE-F (5′-cacaagcttCACACTTTCTATGTGTTAATGTT-3′) for tRNA(E), Litho-trnH-R and Litho-trnH-F (5′-cacaagcttTTGTGGCGCTATAGGTGTATAAC-3′) for tRNA(H), Litho-trnM-R and Litho-trnM-F (5′-acaggatccTCATACCCCATCGATAGATTCTC-3′) for tRNA(M), Litho-trnN-R or Litho-trnN-RM and Litho-trnN-F (5′-cacaagcttCTGTTAATGAATCCAAAAACTAAG-3′) for tRNA(N), Litho-trnQ-R and Litho-trnQ-F (5′-cacaagtcttGGTGTTATTTGAATCAGTGAGTC-3′) for tRNA(Q), Litho-trnR-R and Litho-trnR-F (5′-ataggatccTTTCGACCTGATAGAAGGGCAC-3′) for tRNA(R), and Litho-trnS1-R and Litho-trnS1-F (5′-cacaagcttCTAACTCATCAAAAACAAACACT-3′) for tRNA(S1). The 5′ end 9 nt in all of these primers were added to create a restriction site for HindIII or BamHI plus three terminal nucleotides. An aliquot of each PCR product was electrophoresed on a 1% agarose gel and visualized; the remainder was extracted with phenol/chloroform and digested with restriction enzymes HindIII and BamHI (Promega). One microliter (≈0.1 μg) of Bluescript (Stratagene) plasmid digested with the same restriction enzymes and 2 μl (≈0.2 μg) of PCR product were ligated in a 10-μl reaction containing 1.5 units of T4 DNA ligase (Promega) for 2 h at room temperature. Stratagene Gold competent cells were transformed, single colonies were harvested, and plasmid DNA was prepared by using the alkaline lysis method. The sequence of each cloned insert was determined on an Applied Biosystems model 377 automated DNA sequencer using a BigDye DNA sequencing kit (Applied Biosystems) and modified T3 primer (5′-GAACAAAAGCTGGAGCTC-3′).
Results and Discussion
Genomic Sequences of L. forficatus tRNAs.
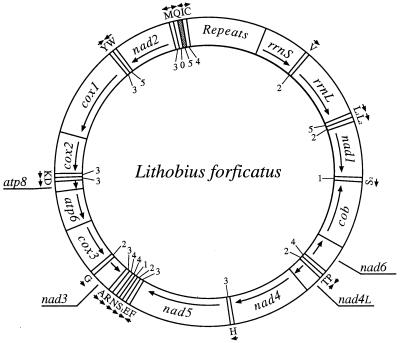
The mitochondrial genome of the centipede L. forficatus is typical for arthropods in terms of its size, nucleotide composition, gene content, and gene arrangement (Fig. 1 and unpublished data). However, all but one of the 22 inferred tRNA genes encode tRNAs with at least one and as many as five mismatches in their acceptor stems (Fig. 1). We analyzed cDNA sequences from eight L. forficatus mitochondrial tRNAs, those for C, E, H, M, N, Q, R, and S1. The genes trnR, trnN, trnS1, and trnE form a contiguous cluster in the mtDNA with trnR and trnN separated by one intervening nucleotide, trnN and trnS1 overlapping by 4 nt, and trnS1 and trnE overlapping by 2 nt (Fig. 2A). trnC, trnH, trnM, and trnQ are located in different parts of the mtDNA and are not adjacent to other tRNA genes on the same strand. trnQ is the only tRNA gene in L. forficatus mtDNA that does not encode any mismatches in the acceptor stem; other tRNA genes studied encode between one and four mismatches, as enumerated in Fig. 1.
Figure 1.
Gene map of L. forficatus mtDNA. Protein and rRNA genes are abbreviated as follows: atp6 and atp8 (genes for subunits 6 and 8 of the F0 ATPase), cox1–cox3 (genes for cytochrome c oxidase subunits 1–3), cob (gene for apocytochrome b), nad1–nad6 and nad4L (genes for NADH dehydrogenase subunits 1–6 and 4L), and rrnS and rrnL (genes for small and large subunit rRNAs). The 22 tRNA genes are identified by the one-letter code for the corresponding amino acid. Two leucine and two serine tRNA genes are differentiated by their anticodon sequence with trnL(uag) marked as L1, trnL(uaa) as L2, trnS(ucu) as S1, and trnS(uga) as S2. The direction of transcription for each gene is shown by an arrow. trnI (shaded area) is the only gene translocated relative to the arthropod primitive gene arrangement exemplified by the mtDNA of the horseshoe crab Limulus polyphemus (10). The numerals near each tRNA gene indicate the number of mismatches in the acceptor stems inferred from mtDNA sequence.
Figure 2.
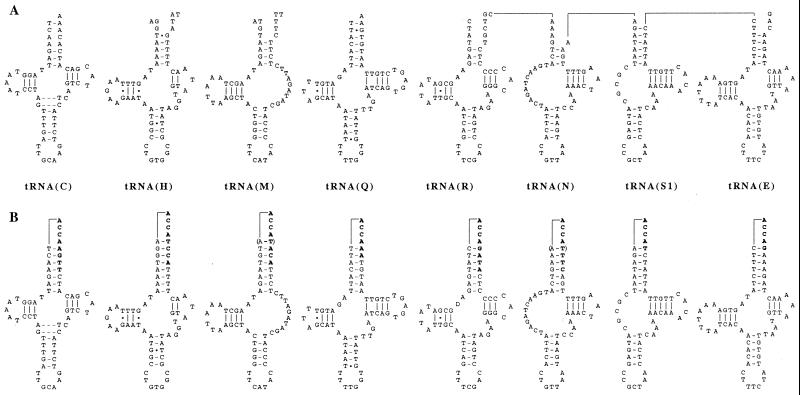
Primary and secondary structures of L. forficatus mitochondrial tRNAs. (A) Inferred from mtDNA sequence. (B) Inferred from cDNA sequences. Edited nucleotides are in boldface type and base pairs that are not present in all clones are in parentheses. Lines connecting tRNAs for R, N, S1, and E in A indicate that this is contiguous genomic sequence. Lines connecting the 5′ and 3′ ends of the tRNAs in B indicate bonds made by the RNA ligase during cDNA preparation.
Analysis of cDNA.
For all tRNAs analyzed, at least some cDNAs have sequence at the 3′ end of the acceptor stems that is different from that encoded by the mtDNA and perfectly matches the 5′ end of the stems (Fig. 2B). In all of these cases, the discriminator nucleotide (always A) and, in all but one, the trinucleotide CCA are also present at the 3′ end of the tRNA, indicating that these cDNA sequences represent mature tRNAs (Table 1). Because the rest of the cDNA sequence matches that of the mtDNA perfectly and because no import of tRNAs has been reported for arthropod mitochondria, we conclude that these differences between genomic and cDNA sequences are the result of RNA editing.
Table 1.
cDNA sequence variation in circularized Lithobius mitochondrial tRNAs
| tRNA | No. clones | cDNA sequence |
|---|---|---|
| tRNA (C) | 7 | TCAGTCTACGACATCTTGAACCATCAAGATT |
| 1 | TCAGT–––tatcacacagcagtg–––AGATT | |
| 1 | TCAG–––––––––actag–––––––––GATT | |
| tRNA (E) | 10 | TGTTAAAAACTAGAAAGACCACTTTCTATAGTGAAA |
| 1 | TGTTA––––––AGAAAGACCACTTTCTATAGTGAAA | |
| tRNA (H) | 4 | TAACTTTTACCTACCAAGGTAAAATAGTTTAAGAAG |
| 2 | TAACTTTTACC-ACCA––––––––––––––––GAAG | |
| 6 | TAACTTTTGA–––––––GGTAAAATAGTTTAAGAAG | |
| 1 | TAACTTTTGAT––––––GATAAAATAGTTTAAGAAG | |
| tRNA (M) | 2 | TCTCTTACATACCAATGTAAGATAAGCTA |
| 4 | TCTCTTACA-ACCA-TGTAAGATAAGCTA | |
| 3 | TCTCTTCTTTTT–––TGTAAGATAAGCTA | |
| tRNA (N) | 4 | TAAGTTTTGACTTTACCAAAAGTCATGAAC |
| 1 | TAAGTTTT––––––––––AAAGTCATGAAC | |
| 1 | TAAGTTTTGACTTTACCAAAAGTCATGAAC | |
| 1 | TAAGTTTTGACTT-ACCA-AAGTCATGAAC | |
| 1 | TAAGTTTTGACTCTACCA-AAGTCATGAAC | |
| 4 | TAAGTTTTGAA–TGCTGCAAAGTCATGAAC | |
| 1 | TAAGTTTTGAAAATGCTGCAAAGTCATGAAC | |
| tRNA (Q) | 6 | AGTCTGTTAATGTAAACCATTACATTAGATG |
| 1 | AGTCTGTTAATGTAAAC––TTACATTAGATG | |
| 1 | AGTCTGTTA–––––tttt––––––TTAGATG | |
| tRNA (R) | 4 | GGGCACCCCCTCATAGACCACTATGAGADGCGA |
| 2 | GGGCACCCCCCTAAA––––––––––––AAGCGA | |
| 2 | GGGCACCAAAAA–––––––––––––––AAGCGA | |
| 1 | GGGCACCCCCAAAA–––––––––––––AAGCGA | |
| 1 | GGGCACCCCC––––––––––––––––––AGCGA | |
| tRNA (S1) | 12 | CACTTGTTATTATCTACCAAGATAATCGCG |
Underlined nucleotides are part of the primer sequence. Nucleotides in boldface are those inferred to be edited at the 3′end of the acceptor stem. Nucleotides in italics are inferred to be added by some alternative (nontemplated) editing process(es). Nucleotides in lowercase are inferred to be artifacts of the experimental procedure. D indicates nucleotide variation (A, T, or G) among the four edited cDNA clones for tRNA (R), possibly caused by posttranscriptional modification at this tRNA position (26).
Characteristics of tRNA Editing.
In total, 28 nt are inferred to be edited in eight tRNAs studied (22 forming base pairs at the 3′ end of the acceptor stem and six at the discriminator nucleotide position). The observed editing is not limited to a subset of nucleotides and always creates a perfect match between the edited nucleotide and its complement in the 5′ end of the acceptor stem. Even the U⋅G pair in the completely matched acceptor stem of tRNA(Q) is replaced with a Watson-Crick U-A pair. Therefore, we infer that the editing is template-dependent and uses the 5′ end sequence of the acceptor stem as a template.
By contrast, the editing found in several other species of animals (11-14) is hypothesized to occur by a template-independent mechanism, most likely posttranscriptional polyadenylation. Indeed, in all of those cases but one, only adenosines are added, even when this recreates a mismatch in the acceptor stem. The single exception is platypus tRNA(S) where the 3′ end sequence in the acceptor stem is edited to CCCA (15). This editing also recreates one of the mismatches in the acceptor stem and, therefore, also appears to be template-independent. The mechanism for the latter editing might include the participation of tRNA nucleotidyltransferase, which is otherwise used for 3′ end CCA addition and/or repair in various organisms (16). Consistent with this idea, the repair of adenosines and cytidines in the tRNA acceptor stem has also been recently reported in human and mouse mitochondrial in vitro systems (17).
Alternative Editing Found in Some tRNAs.
Interestingly, the type of editing found in other animal species seems also to occur in L. forficatus mitochondria. Indeed, alternatively edited cDNA sequences were found for several tRNAs. The proportion of these sequences increases with the number of mismatches in the acceptor stem (Table 1). The alternative editing includes polyadenylation and, in two cases, C insertions, exactly the type of editing observed in other animal mitochondrial tRNAs. However, in L. forficatus, this alternative editing does not correct mismatches in the acceptor stem and is usually associated with cDNAs of aberrant length or/and sequence. Thus, it seems likely that these are the products of a different editing system, possibly one involved in adding/repairing the discriminator nucleotide and 3′ end CCA.
The representation of mature tRNAs in the cDNA pool seems also to be influenced by the primer design. Two reverse transcription primers were designed for tRNA(N), and the representation of cDNAs corresponding to mature tRNAs was different between them (Table 1). This may be due to the presence of modified nucleotides, which may interfere with reverse transcription when using certain primers, as suggested (11).
A Candidate Enzyme for tRNA Editing in L. forficatus.
The inferred mode of L. forficatus mitochondrial tRNA editing would require a 5′-to-3′ RNA-dependent RNA polymerase (RdRp). RdRp is encoded by most RNA viruses (except retroviruses), including mitochondria-associated double-stranded RNAs that are widespread in eukaryotes (18). A functional viral RdRp enzyme has been found in mitochondria of the fungus Ophiostoma novo-ulmi (19) and a gene homologous to viral RdRp has been incorporated into the Arabidopsis thaliana mitochondrial genome (20). Proteins homologous to RdRps have also been found in the nuclear genomes of a variety of eukaryotes; in some cases, they are part of the gene-silencing viral-defense mechanism (21). Thus, the enzyme involved in tRNA editing could be encoded by the nuclear genome and targeted to mitochondria or have a viral origin. If a viral enzyme is participating in the tRNA editing observed in L. forficatus, the virus, or at least the RdRp-encoding portion of its genome, would be indispensable for this animal.
Implication for tRNA Processing.
The inability of the unedited tRNA gene sequences to form paired acceptor stems raises questions about RNA processing. According to the generally accepted tRNA punctuation model for animal mitochondria (22), the secondary structures of tRNAs serve as the signals for processing of the polycistronic transcript. Their enzymatic removal not only creates tRNAs, but also liberates the intervening mRNAs. Most of the tRNA processing enzymes are rather specific in their requirements (23, 24) and most use the paired acceptor stem of tRNA as at least a part of the template. Consistently, it was demonstrated in plants that the editing of the tRNA acceptor stem has to precede the tRNA 3′ end excision (25). In animal mitochondrial tRNAs, however, the processing of the 3′ end seems to precede its editing and, therefore, was hypothesized to occur by a passive mechanism, such as the cleavage at the 5′ end of the properly folded downstream pre-tRNA present in the same polycistronic transcript (11). Indeed, in all cases but one, the edited part of the tRNA acceptor stem is encoded by the portion of the gene that overlaps with the downstream tRNA gene (for review, see ref. 3) and thus is removed when the transcript of the downstream gene is cleaved at the 5′ end. An alternative view would be that the edited portion of the acceptor stem is not encoded at all in these tRNA genes, which are, consequently, immediately adjacent to their downstream tRNA genes.
The presence of similarly overlapping (abbreviated) tRNA genes in L. forficatus suggests that the tRNA processing also should precede tRNA editing in this organism. However, because the editing occurs in the transcripts of both overlapping and nonoverlapping genes, the latter often followed by a protein gene, the excision of unedited pre-tRNAs from polycistronic transcript should be an active process and the enzymes involved should recognize the unusual secondary structures formed by these sequences. The replacement of the U⋅G pair with a U-A pair in otherwise matched acceptor stem of tRNA(Q) supports this point of view and also suggests that the editing of 3′ ends of tRNAs may be even more extensive than inferred, with some matching identities between cDNA and genomic sequence being coincidental.
Unusual tRNA Secondary Structures.
In addition to the tRNA editing, three types of unusual secondary structures for L. forficatus mitochondrial tRNAs have been revealed by the analysis of cDNA data:
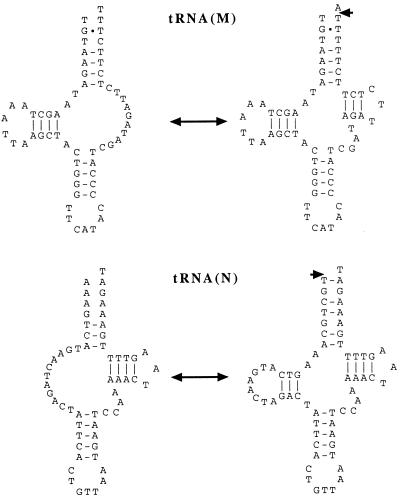
(i) Three of the eight tRNAs analyzed deviate from the classical cloverleaf secondary structure: tRNA(M) has a TV-replacement loop instead of a T-arm and variable (V) loop, whereas tRNA(S1) and tRNA(N) have D-arm replacement loops (Fig. 2). Although mitochondrial tRNA(S1) lacks the D-arm in all metazoan species examined (26), the deviation in the structure of the two other tRNAs are unusual but not unprecedented. The processing of such tRNAs from a presumed polycistronic transcript would present additional problems, because the D-arm and T-arm are generally needed for tRNA recognition by 5′ and 3′ tRNA end processing enzymes (23, 24). Interestingly, the genes for tRNA(M) and tRNA(N), along with some adjacent nucleotides, have the potential to encode secondary structures that are more like the typical cloverleaf form (Fig. 3) and RNA processing has been observed at their 5′ and 3′ ends (Table 1). However, no cDNAs were found that indicated acceptor-stem editing or 3′ CCA addition to such tRNAs, so it is unclear if these could serve any function in transcript processing.
Figure 3.
Alternative foldings for the unedited L. forficatus tRNAs with unusual secondary structures. Arrows indicate the deduced processing at the boundaries of these structures as observed in cDNA.
(ii) Three tRNAs (those for H, M, and N) have other than the typical 7 bp in their acceptor stems in at least some cDNA clones. All previously studied histidine tRNAs have an additional nucleotide, almost universally a G, at their 5′ ends (27), either encoded in the sequence or added posttranscriptionally (28). This nucleotide is located opposite to the discriminator nucleotide in the acceptor stem and may or may not be complementary to it. The inferred L. forficatus tRNA(H) also has an additional nucleotide at the 5′ end, an unusual A, potentially encoded by the mtDNA. However, this tRNA differs from all other histidine tRNAs by also having an additional (discriminator) nucleotide at the 3′ end of an 8-bp acceptor stem (Fig. 2B). Interestingly, two cDNA clones for tRNA(M) also are from mature tRNAs with 8-bp acceptor stems. Although this can be an error of tRNA excision, it is also possible that the alternative structures found for this tRNA are differentially used as an initiator tRNA (aminoacylated with formyl-methionine) and an elongator tRNA (aminoacylated with methionine). If confirmed, this observation may provide an answer to the long-standing question of how a single trnM gene present in most mtDNAs can produce two different products, tRNA(M) and tRNA(fM). In addition to mature tRNAs with 8-bp acceptor stems, one cDNA clone from a mature tRNA(N) with 6 bp in the acceptor stem was found.
(iii) tRNA(C) is inferred to have an unusual secondary structure with 7 bp in the anticodon stem. Similar structures have been described for mammalian mitochondrial tRNA(S) (29), fungal mitochondrial tRNA(C), echinoderm mitochondrial tRNA(T), and some others (30). The L. forficatus trnC codes for two T⋅T mismatches in the acceptor stem. These two mismatches were confirmed to be present in the cDNA from the mature transcript product of this tRNA gene (data not shown). Either two T⋅T mismatches or a single mismatch and bulged T are also present in the anticodon stem of mitochondrial tRNA(C) of Neurospora crassa (30).
Evolutionary Considerations.
The editing observed in L. forficatus tRNAs is more extensive than any previously reported. It appears to occur in all mitochondrial tRNA genes and represents a further step in the degeneration of tRNA genes observed in mitochondria (31). It is likely that, once evolved, this editing would persist in the lineage, because it is difficult to imagine a mechanism that would correct the sequences of all tRNA genes simultaneously and, thus, allow for its loss. Thus, the presence of this type of tRNA editing may be a nearly irreversible phylogenetic character. However, its usefulness for phylogenetic studies would depend on the probability of it arising independently in different lineages, which itself depends on the underlying mechanism. So far, in addition to L. forficatus, we have observed tRNA genes that encode sequences with the potential for similar editing only among species of onychophorans (unpublished data). Further studies are needed to confirm whether this represents an independent origin or a phylogenetically useful character.
Acknowledgments
We thank K. G. Helfenbein and J. V. Moran for comments on the manuscript. This work was supported by National Science Foundation Dissertation Improvement Grant DEB 9972712 to W.M.B. and D.V.L., National Science Foundation Grant DEB 9807100 to W.M.B. and J.L.B., and a University of Michigan predoctoral fellowship to D.V.L.
Abbreviation
- RdRp
RNA-dependent RNA polymerase
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF309492).
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250402997.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250402997
References
- 1.Price D H, Gray M W. Curr Genet. 1999;35:23–29. doi: 10.1007/s002940050428. [DOI] [PubMed] [Google Scholar]
- 2.Brennicke A, Marchfelder A, Binder S. FEMS Microbiol Rev. 1999;23:297–316. doi: 10.1111/j.1574-6976.1999.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 3.Price D H, Gray M W. In: Modification and Editing of RNA. Grosjean H, Benne R, editors. Washington, DC: Am. Soc. Microbiol.; 1998. pp. 289–305. [Google Scholar]
- 4.Janke A, Pääbo S. Nucleic Acids Res. 1993;21:1523–1525. doi: 10.1093/nar/21.7.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dirheimer G, Keith G, Dumas P, Westhof E. In: tRNA: Structure, Biosynthesis, and Function. Söll D, RajBhandary U, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 93–126. [Google Scholar]
- 6.Martin N C. In: tRNA: Structure, Biosynthesis, and Function. Söll D, RajBhandary U, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 127–140. [Google Scholar]
- 7.McClain W H. In: tRNA: Structure, Biosynthesis, and Function. Söll D, RajBhandary U, editors. Washington, DC: Am. Soc. Microbiol.; 1995. pp. 335–347. [Google Scholar]
- 8.Avital S, Elson D. Biochim Biophys Acta. 1969;179:297–307. doi: 10.1016/0005-2787(69)90038-0. [DOI] [PubMed] [Google Scholar]
- 9.Buck M, Connick M, Ames B N. Anal Biochem. 1983;129:1–13. doi: 10.1016/0003-2697(83)90044-1. [DOI] [PubMed] [Google Scholar]
- 10.Staton J L, Daehler L L, Brown W M. Mol Biol Evol. 1997;14:867–874. doi: 10.1093/oxfordjournals.molbev.a025828. [DOI] [PubMed] [Google Scholar]
- 11.Yokobori S, Pääbo S. Proc Natl Acad Sci USA. 1995;92:10432–10435. doi: 10.1073/pnas.92.22.10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tomita K, Ueda T, Watanabe K. Nucleic Acids Res. 1996;24:4987–4991. doi: 10.1093/nar/24.24.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokobori S, Pääbo S. J Mol Biol. 1997;265:95–99. doi: 10.1006/jmbi.1996.0728. [DOI] [PubMed] [Google Scholar]
- 14.Reichert A, Rothbauer U, Mörl M. J Biol Chem. 1998;273:31977–31984. doi: 10.1074/jbc.273.48.31977. [DOI] [PubMed] [Google Scholar]
- 15.Yokobori S I, Pääbo S. Nature (London) 1995;377:490. doi: 10.1038/377490a0. [DOI] [PubMed] [Google Scholar]
- 16.Deutscher M P. Enzymes. 1982;15:183–215. [Google Scholar]
- 17.Reichert A S, Mörl M. Nucleic Acids Res. 2000;28:2043–2048. doi: 10.1093/nar/28.10.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koga R, Fukuhara T, Nitta T. Plant Mol Biol. 1998;36:717–724. doi: 10.1023/a:1005907310553. [DOI] [PubMed] [Google Scholar]
- 19.Cole T E, Hong Y, Brasier C M, Buck K W. Virology. 2000;268:239–243. doi: 10.1006/viro.1999.0097. [DOI] [PubMed] [Google Scholar]
- 20.Hong Y, Cole T E, Brasier C M, Buck K W. Virology. 1998;246:158–169. doi: 10.1006/viro.1998.9178. [DOI] [PubMed] [Google Scholar]
- 21.Cogoni C, Macino G. Nature (London) 1999;399:166–169. doi: 10.1038/20215. [DOI] [PubMed] [Google Scholar]
- 22.Ojala D, Montoya J, Attardi G. Nature (London) 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 23.Rossmanith W. J Mol Biol. 1997;265:365–371. doi: 10.1006/jmbi.1996.0750. [DOI] [PubMed] [Google Scholar]
- 24.Nashimoto M, Tamura M, Kaspar R L. Biochemistry. 1999;38:12089–12096. doi: 10.1021/bi9911942. [DOI] [PubMed] [Google Scholar]
- 25.Kuzmann A, Brennicke A, Marchfelder A. Proc Natl Acad Sci USA. 1998;95:108–113. doi: 10.1073/pnas.95.1.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garey J R, Wolstenholme D R. J Mol Evol. 1989;28:374–387. doi: 10.1007/BF02603072. [DOI] [PubMed] [Google Scholar]
- 27.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Williams J B, Cooley L, Söll D. Methods Enzymol. 1990;181:451–462. doi: 10.1016/0076-6879(90)81143-i. [DOI] [PubMed] [Google Scholar]
- 29.Yokogawa T, Watanabe Y, Kumazawa Y, Ueda T, Hirao I, Miura K, Watanabo K. Nucleic Acids Res. 1991;19:6101–6105. doi: 10.1093/nar/19.22.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinberg S, Leclerc F, Cedergren R. J Mol Biol. 1997;266:269–282. doi: 10.1006/jmbi.1996.0803. [DOI] [PubMed] [Google Scholar]
- 31.Lynch M. Mol Biol Evol. 1996;13:209–220. doi: 10.1093/oxfordjournals.molbev.a025557. [DOI] [PubMed] [Google Scholar]