Abstract
The extent of microbial diversity is an intrinsically fascinating subject of profound practical importance. The term ‘diversity’ may allude to the number of taxa or species richness as well as their relative abundance. There is uncertainty about both, primarily because sample sizes are too small. Non-parametric diversity estimators make gross underestimates if used with small sample sizes on unevenly distributed communities. One can make richness estimates over many scales using small samples by assuming a species/taxa-abundance distribution. However, no one knows what the underlying taxa-abundance distributions are for bacterial communities. Latterly, diversity has been estimated by fitting data from gene clone libraries and extrapolating from this to taxa-abundance curves to estimate richness. However, since sample sizes are small, we cannot be sure that such samples are representative of the community from which they were drawn. It is however possible to formulate, and calibrate, models that predict the diversity of local communities and of samples drawn from that local community. The calibration of such models suggests that migration rates are small and decrease as the community gets larger. The preliminary predictions of the model are qualitatively consistent with the patterns seen in clone libraries in ‘real life’. The validation of this model is also confounded by small sample sizes. However, if such models were properly validated, they could form invaluable tools for the prediction of microbial diversity and a basis for the systematic exploration of microbial diversity on the planet.
Keywords: Bacteria, Archaea, sampling, diversity, species richness, neutral community model
1. Introduction
(a) What is the extent of microbial diversity? Introductory comments
How many different kinds of microbes are there? It is one of those child-like questions in science that exposes the depths of our ignorance and the severe limitations of our most sophisticated measurement tools and intellectual strategies. This alone makes it a question worth tackling. However, it is also a question of profound practical importance. Microbes are required to sustain almost every other form of life on Earth and are especially important to human life: influencing, even dictating, climate, health, agricultural productivity and the fate of pollutants. They are often unanticipated modulators of our activities working in benign (pesticide degradation) or malign (mobilizing arsenic in water) ways. However, microbes in real life rarely, if ever, exist as the pure cultures from which we have learnt so much. They exist as communities of varying and typically unknown complexity. These communities invariably form when the opportunity is offered: be that opportunity a new baby or a new hole in the ground. We do not yet really understand how these communities form. This is in many ways more important and more challenging than estimating the extent of microbial diversity. However, the two questions are inextricably linked and a better understanding of microbial diversity will lead us to a firmer comprehension of microbial community formation.
The question of microbial diversity was not always considered a question, at least not a very interesting or tractable one. Thomas Brock's excellent 1966 text on microbial ecology (Brock 1966) makes only passing (though shrewd) comments. The usually thoughtful and insightful MacArthur & Wilson (1967, p. 182) are sure that ‘higher plants and animals comprise most of the…species on Earth’. These presumably uncontroversial views probably had their root in the almost complete reliance on laboratory cultivation for the detailed characterization of micro-organisms and the historically poor nature of prokaryotic taxonomy. Even as microbial taxonomy improved, one still had to grow an organism in the laboratory to learn all but the most limited information about its properties. That information was gained laboriously by tens of biochemical tests, and new species were ‘discovered’ very slowly and delineated based on poorly constrained criteria.
The extent of microbial diversity is now the subject of polemic. Those who believe that diversity is large have been greatly influenced by the work of Pace and his colleagues (Hugenholtz et al. 1998). They demonstrated that the presence of, and phylogenetic relationships between, microbes in real communities could be inferred from the analysis of sequences of conserved genes (typically 16S rRNA genes) recovered from the environment, independent of the need to grow a pure culture in the laboratory. This strategy is now very widely applied in microbial ecology and whole new phyla have been discovered and the description of new 16S rRNA sequences is commonplace. An insightful and complementary piece of work by Torsvik and her colleagues showed that the rate of re-association of DNA extracted from soil was consistent with the presence of several thousand distinct taxa in a few grams (Torsvik et al. 1990), a finding that led E. O. Wilson (1994) to suggest ‘no one has the faintest idea’ of the extent of microbial diversity. There are other lines of evidence which also suggest that prokaryote diversity is extremely large. There are reports of endemic sequences for example in soil and hot springs and large diversities are possible mathematically (Fulthorpe et al. 1998; Curtis et al. 2002; Whitaker et al. 2003).
Those who believe that global microbial diversity is small have been particularly influenced by the ubiquity of certain protozoan species. For example, small samples from marine and freshwater environments were found to contain the vast majority of the ciliates associated with such environments globally. In an even more remarkable study, Finlay & Clarke (1999) found 32 out of 50 known Paraphysomonas species in just 25 μl of sediment. It was reasoned that this apparent ubiquity was a function of the huge numbers, small size and ease of dispersal of ciliates in particular, and microbes in general. Since there is essentially no obvious barrier to the dispersal of microbes (microbes are no respecters of mountain ranges or oceans) and there are very many of them, microbes can, and do, get everywhere. It has been assumed that high rates of dispersal imply high rates of immigration and thus immigration will also limit speciation, by robbing microbial communities of the isolation required for new taxa to arise allopatrically. Conversely, once formed, it is very hard for a microbial species to go extinct, because they exist at such large numbers. Studying the ecology of protists is still largely based on morphology, rather than nucleic acid sequence data more commonly employed in bacterial ecology (Fenchel & Finlay 2006). It has thus been argued that the analysis of ‘morphospecies’ may obscure differences that exist at the genome level and the same ‘morphospecies’ from different locations may be different ‘genomospecies’ and thus not globally dispersed. However, there is evidence of ubiquity and low diversity in the bacterial world. Hagstrom et al. (2002) have suggested that the rate of reporting of new 16S rRNA sequences from marine bacteria is decreasing year-on-year, suggesting that the majority of taxa which exist have been sampled. Moreover, excellent and authoritative evidence for ubiquity has been produced by the Hugenholtz group using an environmental genomics strategy. They have demonstrated that organisms with virtually the same genome were found in model wastewater treatment plants in the USA and Australia (P. Hugenholtz 2006, personal communication). This is a particularly interesting finding. Neither differences nor similarities in morphology or conserved sequences are infallible guides to relatedness in the microbial world. Thus, it has been possible for the parties in the polemic to suggest that the contrasting findings are a function of the contrasting methods used. While this may in part be true, evidence of ubiquity at the genomic level suggests that more than methodological differences need to be invoked to square the circle. The scale of disagreement is extremely large. Though most papers eschew numbers, ‘moderate’ (Finlay & Clarke 1999) diversity in the context of microbes might be less than 10 000 globally and a few hundred taxa in a sample. On the other hand, ‘high’ diversity might be more than 10 million globally and greater than 5–10 000 locally (Curtis et al. 2002). This implies that we cannot agree, even to within three orders of magnitude, on the extent of diversity. It is as if we could not distinguish the height of Mount Everest and the Eiffel tower.
What then are the causes of the uncertainty? We have already alluded to the use of different methods for inferring differences and similarities among species. In addition, different environments might be expected to have different diversities. Therefore, the diversity of soil and seawater may not be comparable. However, even if all these obvious methodological differences were accounted for, one overriding problem remains: sample size. Thus, a lake or an activated sludge reactor might have 1015–1018 individual bacteria, and yet 16S rRNA clone libraries of more than 1000 are exceptional. This implies examining just one clone or sequence for every 1012–1015 individuals. These are impossibly small samples. The implications of gross under-sampling are only just beginning to be understood. Consider taxa–area relationships (TARs). This probably universal phenomenon is actually difficult to observe in the microbial world. If the same most abundant taxa are found in samples of small and large areas or volumes, then the diversity will look exactly the same, simply because such a small proportion of the microbiota is sampled. TARs therefore only become detectable if environmental, evolutionary or demographic forces affect the structure of the most abundant organisms (Woodcock et al. in press). There may also be a complex relationship between the change in diversity and the change in the number of taxa detected. For example, ‘molecular fingerprinting methods’ will only detect differences in diversity when the true diversity lies in a narrow range and neither very low nor very high (Loisel et al. 2006).
The importance of appropriate sample sizes will seem blindingly obvious to future generations. However, the polemic and confusion that prevails at present, largely as a consequence of sampling limitations, are understandable as we are dealing with a world, the microbial world, that operates at a scale beyond the range of normal human intuition. It is not therefore surprising that we use nebulous terminology, overlook important factors, make mistakes and disagree. However, this is not a situation that can be allowed to persist. The exploration of the microbial world is too important for that. We need to develop effective strategies for authoritatively determining prokaryotic diversity and move on to the greater challenges that lie ahead.
2. Stratagems for discovering diversity
How then can we determine microbial/prokaryote diversity? Not withstanding our comments about sampling, it is clear that the diversity in a sample exceeds the number of taxa observed by whatever empirical measurement is used. Not least because when prokaryote communities are analysed using 16S rRNA gene clone libraries, a relationship between the clone library size and the number of different taxa observed is almost invariably found. In trying to determine diversity, one can adopt one of a number of strategies, each with their own advantages and disadvantages. The simplest approach is probably to use some form of non-parametric estimator. Alternatively, one can assume some form of distribution, guided by either theoretical reasoning or extrapolation from a dataset. Finally, one can also estimate the diversity and the local distributions by using a calibrated mathematical model of community assembly.
3. Chao's estimators
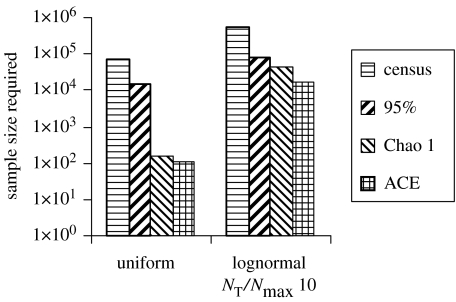
In principle, non-parametric methods are an extremely attractive way to estimate diversity. A suite of such methods, developed by Chao and originally popularized by Colwell (Chao 1984, 1987; Colwell & Coddington 1994), give an estimate of the minimum diversity compatible with the data. These methods are simple and make no assumptions about the underlying distribution. Though the methods themselves are excellent, they can be misinterpreted as giving a true estimate of the diversity, irrespective of the sample size. In reality, if the sample size is too small, then the corresponding estimate of diversity will be too small as well. The minimum sample size required for this class of estimator is of the order of the square root of twice the diversity. However, minimum sample size is extremely sensitive to the underlying distribution. Schloss & Handelsman (in press) have explored the use of non-parametric estimators by simulation. They found that for a sample with a species richness of 5000, it could take anything from 18 000 to 40 000 clones to correctly estimate the true species richness if the bacteria have a lognormal distribution, but a mere 150 clones if the distribution is even (figure 1).
Figure 1.
Non-parametric methods must be used with caution when sample sizes are small. The sample sizes required to correctly characterize a sample with a diversity (i.e. richness) of 5000, undertaking a complete census, a 95% census or using non-parametric methods (Chao 1 and ACE). Note that if the diversity is uniform, non-parametric estimators are very efficient. However, if the diversity is lognormally distributed, then a very large sample is required to obtain the correct answer. The simulations are described in more detail elsewhere (Schloss & Handelsman in press).
Schloss & Handelsman have also undertaken some very interesting analyses of local (Schloss & Handelsman 2005) and global (Schloss & Handelsman 2004) biodiversity data that unequivocally show non-parametric estimates are a function of sample size. They go on to show that we do not yet have sufficient data at a local or global scale to estimate richness using these estimators. The global analysis was undertaken by analysing the sequences in the Ribosornal Database Project II (RDP-II) database of 16S rRNA sequences. While this stratagem is subject to many caveats, in addition to the question of sample size, it does give a lower limit to our estimates of global diversity of about 35 000 (based on species discrimination at 97% sequence identity) and 325 000 (based on species discrimination at 99% sequence identity). We know that the global diversity of prokaryotes is greater than this, but we do not know how much greater. This uncertainty at small and large scale is not the fault of the estimators; they are clever mathematical tools, not magic wands. We simply need to make sure that the sample sizes employed are big enough for the tools to do their job and to answer the questions we ask.
4. Assuming a distribution
One way to try to escape the sample size issue is to assume a particular taxa-abundance distribution from which the samples are taken. The specific nature of the distribution then becomes a critical factor. Ecologists who study larger organisms have commonly observed lognormal distributions within a given group of organisms at a given location. A number of theoretical explanations for such a pattern have been advanced. MacArthur, and later May (MacArthur 1960; May 1974), pointed out that micro-organisms would be subject to exponential growth, but without a large standing population. They suggested that if many different things acted independently on the growth rates of the micro-organisms, then the growth rates would be normally distributed and so the abundance of the organisms would be lognormally distributed (because growth is exponential). They further reasoned that in more extreme environments fewer factors would impinge on growth rates leading to communities with geometric taxa-abundance curves.
In the absence of any reliable data on the relative abundance of bacterial taxa, the lognormal taxa-abundance curve is therefore a plausible place to start. Making a further assumption that the least-abundant bacterium in a sample is represented by a single organism at an abundance of one, it is possible (Curtis et al. 2001) to derive a relationship between ratio of the abundance of the most abundant taxon (Nmax), the total number of individuals (NT) or NT/Nmax ratio and the number of taxa or species richness (figure 2).
Figure 2.
A ‘quick and dirty’ way to estimate diversity by assuming a distribution. (a) The total number of taxa in a community with a lognormal species abundance curve is simply the area under that curve (called the species curve). The individuals curve is the number of species at each abundance (the species curve) multiplied by their abundance (the x-axis). There is therefore a mathematical relationship between the area under a species area curve, the number of individuals NT (the area under the individuals curve), and the maximum and the minimum abundance (Nmax and Nmin). (b) The relationship, over 30 orders of magnitude in population size, for various ratios of NT/Nmax by assuming that Nmin is equal to one (Curtis et al. 2002). As a rule of thumb, soil has a ratio of 10 and seas and lakes have a ratio of 4. There are about 1030 bacteria in the world (Whitman et al. 1998).
This is a ‘quick and dirty’ estimate but has the advantage of requiring relatively little information and the disadvantage of more probably giving an overestimate of species richness at any given level of phylogenetic resolution. Furthermore, by comparing the outcomes of this method and Chao's non-parametric estimators (assuming the sample size is too small) one can get a feel for the range of possible outcomes and possibly use the parametric method to conservatively estimate the sample sizes required to obtain a correct estimate from a non-parametric method. Thus, the NT/Nmax ratio for the Schloss & Handelsman (2005) global dataset is about 67 when taxa are defined at the level of 97% 16S rRNA sequence identity. This implies a global species richness of less than 1010 assuming that there is a single least-abundant organism with an abundance of one. The true value is probably a great deal less than this because the sequence database in general will contain a small number of representative sequences and not reflect the frequency at which particular sequences are recovered in individual studies (which would lower the NT/Nmax values). Thus, if the global NT/Nmax ratio was 10, then under the same assumptions as before the global species richness would be about 108; this probably represents a more plausible upper limit. On the other hand, it is well known that discrimination of taxa based on 16S rRNA sequence identity values is innately conservative which could be used to argue for higher estimates of species richness. These ‘back of an envelope’ estimates must be couched in careful terms and should be regarded as a tentative first pass estimates.
However, there is one ‘bullet proof’ estimator that assumes a distribution, but it can only be used in the unusual situation where all the sequences detected in a 16S rRNA gene clone library are different (Lunn et al. 2004). In this case, one can assume that the underlying distribution is uniform and estimate the probability of observing that sample, given certain levels of diversity in the sample. This approach was used to analyse data from a clone library of 100 singletons obtained from Amazonian soil (Borneman & Triplett 1997). Thus, for 100 singletons it would be very unlikely (p=0.006) if the soil diversity was less than 103, quite unlikely (p=0.6) if the diversity was less than 104 and probable (p=0.95) if the diversity was about 105. The assumption of a uniform distribution is almost certainly wrong. However, if there is any other kind of distribution, the estimated diversity would be even higher. Such samples are of course not common, and there is some doubt about the truly flat nature of the dataset which inspired the work (Schloss & Handelsman in press). Nevertheless, the unequivocal nature of the reasoning makes this work significant.
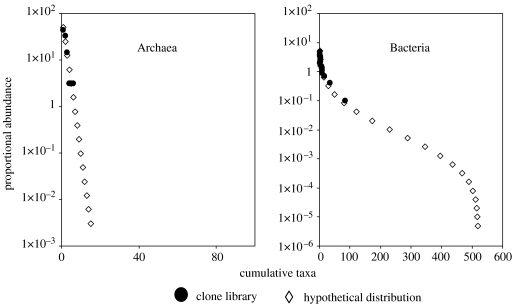
Though lognormal curves appear to be commonly observed in communities of large organisms, we simply do not know if they pertain in the microbial world. Furthermore, MacArthur & May's reasoning may not be infallible. For example, samples of the Archaea and the Bacteria can have apparently different distributions (figure 3) in the same anaerobic digester (Godon et al. 1997). This should imply that the anaerobic digester represents a permissive environment for Bacteria and an extreme environment for methanogens. Similarly, clone libraries of ammonia-oxidizing bacteria (AOB) and the general bacterial community can have radically differing distribution in the same treatment plant engineered to meet all their needs. In retrospect, one can construct a narrative to explain these differences. However, this form of plausible qualitative reasoning is probably of only limited application at present and we should aspire to a more robust approach.
Figure 3.
Different distributions are observed in the same environment. Clone libraries of different groups of organisms (Archaea and Bacteria) in the same environment can appear to have radically different distributions. Hypothetical geometric (Archaea) and lognormal (Bacteria) distributions are shown. In this example, the bacterial and archaeal 16S rRNA gene sequences amplified from the same anaerobic digester exhibit very different distributions. Thus, the distribution of species is not likely to be a simple function of the suitability of the environment (data from Godon et al. 1997).
5. Fitting a taxa-abundance curve to the data
An obvious and superficially attractive alternative to simply assuming a distribution curve is to simply predict the shape of the curve (and therefore the species richness) on the basis of the relative abundance of clones in clone library data (Dunbar et al. 2002; Hong et al. 2006). The major flaw with this approach is that sample sizes are dictated by budgets and technology rather than a rational assessment of the sample size required to undertake the task (but see below). There is, therefore, an implicit assumption that there is enough information in the sample to describe the underlying distribution. This assumption is not necessarily met.
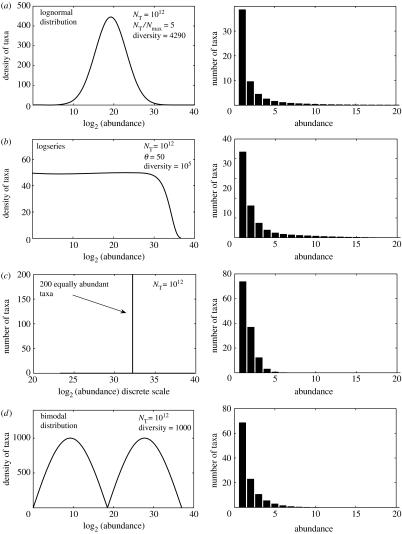
Sloan et al. (in press) have demonstrated that relatively small samples from communities with radically different diversities will look remarkably similar (figure 4) and bear little resemblance to the distribution of taxa in the community from which they were drawn. It follows that one cannot assume that the distribution of sequences observed in a 16S rRNA gene clone library resembles the distribution of the community from which it is drawn. In fairness, some curve fitting papers have emphasized the problem of sample size and given very useful estimates of the sample sizes required for more confident estimates. Thus, Dunbar (Dunbar et al. 2002) made a prediction on the basis of clone libraries of several hundred clones, but showed that, in reality, 16 284–44 000 clones might be required to describe half the taxa present in a diverse soil sample. Schloss & Handelsman (in press) have made analogous calculations suggesting that while a complete census of a diverse soil sample might require over 400 000 clones, a smaller sample of 18 000 clones would yield enough information to make an estimate of the diversity.
Figure 4.
Small samples can be very misleading. To demonstrate the unreliability of small sample sizes, four radically different distributions (on the left) were sampled (200 individuals selected randomly) and the resulting distributions (shown on the right) plotted (Sloan et al. in press). Note that all the sample distributions are superficially alike.
More recently, there has been an increase in both the range of curves examined and the sophistication with which they have been fitted. There has however been no systematic attempt to obtain adequate samples sizes. Gans et al. (2005) have attempted to get round this problem by using DNA–DNA re-association data rather than clone libraries of conserved genes, although this method has its critics. Gans et al. (and others before them) have realized that the pattern of re-association reflects the underlying distribution of similar sequences (and therefore we hope genomic diversity). Only about half the DNA re-associates, but in simulations of data from communities with a diversity of 5000, this was enough to discriminate between distributions. However, when the data were analysed, the best fitting lines were compatible with the presence of over a million distinct genomes. This implies that a far greater proportion of the curve is hidden than perhaps Gans et al. thought when they embarked on the study. This makes the extrapolation more difficult and uncertain. The study has drawn comments about strategies for fitting curves from Hong et al. (2006) and I. Volkov (2005, personal communication). However, the best curve fitting in the world is pointless if there are insufficient data to begin with. The studies reported to date would have done a great service if they simply illustrate this point (Narang & Dunbar 2004).
To persist in curve fitting with insufficient data will yield papers but not knowledge. We need now to accept we require ‘more power’ and that this means more data. Larger sample sizes are technically feasible but are potentially expensive, especially if one considers several environments. However, it is probably not as expensive as the present modus operandi of repeatedly asking questions about diversity and only partially funding the search to find the answer. Moreover, exciting new sequencing technologies could well make very large datasets far more easily available than they have been in the past.
6. ‘A more fruitful approach’
Determining an appropriate distribution and diversity for a given microbial community will be fascinating, because it is difficult and unknown. There is, moreover, tremendous satisfaction to be gained from breaking out of the speculation and polemic that surrounds the field and placing the exploration of the extent of the microbial world on a more solid footing. However, the limits of simply fitting curves, even with enough data, should be acknowledged. In 1957, MacArthur (1957, p. 293), commenting on contemporary debates about taxa-abundance curves, wrote:
One approach… is to fit known statistical distributions of uncertain biological meaning to the data. A far more fruitful approach seems to be… to predict on the basis of simple biological hypotheses.
This critique is doubly relevant for microbial ecology. There will probably be no single ‘one size fits all’ distribution or diversity. The picture will vary between communities, levels of taxonomic resolution and functional groups (functional group is a loose term meaning organisms with the same function, such as the denitrifiers or AOB). We have already alluded to the apparent differences between the Archaea and the Bacteria in the same environment. Hong et al. (2006) noted differing fits with differing levels of phylogenetic resolution. This should remind us that describing the diversity of any given community or functional group is not an end in itself, but a milestone on a search for a deeper understanding of how communities form.
Moreover, calculating prokaryote diversity in particular and microbial diversity in general will often be a laborious and expensive undertaking. We will be better able to justify the expense if we can use such studies to draw meaningful and generalizable inferences. Consequently, we cannot be content to simply plot the lines and say the diversity is X. Rather, we must use this information to test our ability to predict on the basis of simple biological hypotheses.
This in turn will present us with tools to predict how communities form and change, even when we have not had the opportunity to characterize those communities in great detail. If we had such tools, we might be able to ‘sketch out’ the lay of the land over many microbial environments and landscapes using information garnered from small samples that can be analysed cost effectively.
Such estimates could be an invaluable guide to those seeking to investigate, exploit or manipulate a given class of environments. For example, a combined genomics and proteomics investigation of an environment represents a substantial investment in the diversity of a given community or class of community. Predicting the nature, extent and stability of that community will help ensure that the investment is of an appropriate scale and that the information is valid over appropriate spatial and temporal ranges.
7. Criteria and principles for models of microbial community formation
MacArthur's ideas for a better approach eventually bore fruit as ‘The Theory of Island Biogeography’ (MacArthur & Wilson 1967). His co-author Wilson (1998) suggested that a good theory should be judged against the criteria of parsimony, generality, consilience and predictiveness. This is good advice. The ability to predict is particularly important in the context of microbes. Many ecological theories are evaluated by their ability to reproduce a world that has already been well described. Theory is used in lieu of experimentation to gain insights into underlying mechanisms. This can mean using complex models with many parameters, most of which are, necessarily, invented.
In the microbial world it is different. We must use the parameters to predict the microbial world. It follows that the parameters must be as near to reality as possible and therefore inferable from either first principles or reasonable measurements. This in turn reinforces the need for parsimony, since this minimizes the number of parameters. This implies a theory based on simple truths.
In searching for these truths, one might be tempted to look at certain putatively ubiquitous phenomena such as TARs or lognormal taxa-abundance curves. These phenomena are good indicators that universal principles are at work. They are not universal principles themselves. TARs have been, rather unfortunately, described as ‘one of the few laws in ecology’ (Pounds & Puschendorf 2004). This statement confuses a phenomenon, the taxa–area curve, with the underlying principles behind it, which might include demographic considerations, selection or evolution. This is not academic hair splitting. Since the microbial world stretches over 30 orders of magnitude (i.e. from one cell to all the cells on the planet), scale is one of the more important challenges in microbial ecology. It will often be easier to confidently upscale simple principles than to extrapolate certain phenomena. The Earth orbits the Sun, but a pea will not orbit a grapefruit: the law of gravity explains both.
8. The simplest possible models
An almost infinite number of factors impinge upon microbial life. However, the simplest possible truths about an open microbial system are that organisms reproduce, die and immigrate. MacArthur & Wilson (1967) used birth, death and immigration as the founding principle of the theory of island biogeography. Somewhat defensively noting in the preface (p. 5) that crude theory ‘if it can account for, say, 85% of the variation in some phenomenon of interest, it will have served its purpose well’, they suggested that the diversity observed on an island represented a balance between immigration from some source community and local extinction. Intriguingly they sought, and found, their first qualitative evidence for this conception in microbial communities. However, though the theory of island biogeography is very simple, it is too complicated for microbial ecologists. This is because it is predicated on a complete, or almost complete, census of local and source diversity, a question that we found we could not satisfactorily answer, as described earlier. Nevertheless, by suggesting that the composition of a local community is a function of immigration and some source community, they do suggest a strategy for predicting local diversity.
More recently, stochastic models of community assembly have been proposed (independently) by Bell & Hubbell (Bell 2000, 2001; Hubbell 2001). These models are conceptually analogous to the original theory of island biogeography and have been termed neutral as they implicitly assume, on average, the equivalence of species. Hubbell's publication is the more ambitious and wide ranging of the two. It is intuitively appealing as it has just three parameters, a source community, an immigration parameter and the number of individuals in the local community (figure 5), and yet, at least superficially, appears able to generate the paradoxical range of diversities and community abundance patterns found in the real world. Thus, the Bacteria and the Archaea in the same environment to which they are both well adapted could have radically different distributions if one occurred at higher numbers or with a higher source diversity.
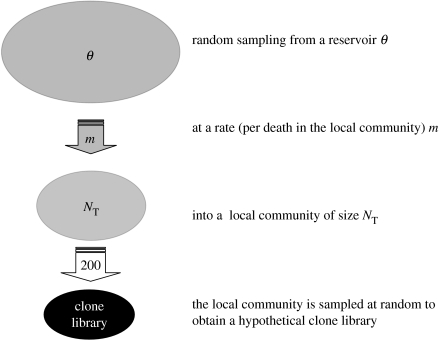
Figure 5.
A schematic of the model used to estimate diversity. When an individual dies in the local community, it is replaced from outside the community with a probability m, or from within the community with a probability 1−m (after Hubbell 2001; Sloan et al. 2006, in press).
Hubbell's model is also unusable in its original format as far as microbial ecologists are concerned. The most important problem is that it was based on a discrete Markov chain and thus becomes computationally intractable for number of individuals greater than 104: you would find a larger community in a one-tenth of a thimble of seawater. The second problem was that the original model was corroborated, using two fitted parameters (the source term and the immigration term), to known taxa-abundance curves. This is unfortunate, in the microbial world we are still unsure about the nature of the source diversity or indeed the immigration term and we do not yet have a single reliable taxa-abundance curve. Finally, the model conceptualizes the source diversity as being a log series; this may or may not be true for a given microbial source community. The model for the evolution of the source community in Hubbell's model assumes that variation is generated through simple point mutations. This might be an adequate model for certain genes used to characterize microbial diversity. However, it might not be an adequate model for the whole organism as horizontal gene transfer (HGT) appears to be a major force in the speciation of prokaryotes.
In summary, a simple neutral community model is an attractive option for exploring microbial diversity. However, such a model must be able to cope with large numbers, be calibrated without recourse to a taxa-abundance curve, or more than one independent parameter, and not be wholly reliant on Hubbell's conception of the source term. One such approach is described in Sloan et al. (2006).
9. A neutral community model for microbes
To develop a stochastic model that may cope with very large numbers, Sloan et al. (2006) have derived a continuous form of Hubbell's discrete model. They did this by drawing on methods widely used in the study of neutral evolution and originally developed by physicists to scale up random walks. The conceptual basis of the model is identical to that of Hubbell. It is predicated on the idea that, over a small period of time, the number of individuals in a community can either increase by one organism, decrease by one organism or not change. The probability of each of these possibilities can be expressed in terms of the number of organisms (NT), the probability that a death can be replaced by an organism from outside the local community (m) and the proportional abundance of the species in the source community (pi). Based on these probabilities, it is possible to derive an equation that describes the rate of change of the probability that the species will have a particular relative abundance, xi. The relative abundance is assumed to be a continuous random variable for large microbial populations. The steady-state solution of the equation gives an expression for the probability density function for the relative abundance of ith species. It is possible to confer a slight advantage or disadvantage over other taxa (which introduces a fourth advantage parameter that can be used to represent competition) or have a purely neutral system (in which case the advantage parameter is zero). When the advantage parameter is zero, then xi is beta distributed,
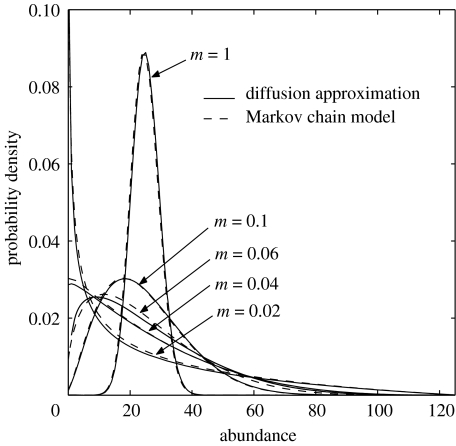
This is not the only analytical solution to Hubbell's model (Houchmandzadeh & Vallade 2003; Vallade & Houchmandzadeh 2003; Volkov et al. 2003; McKane et al. 2004), but it is the simplest and makes identical predictions to those of the discrete model (figure 6), even at very low NT values. For any given mean proportional abundance in the source community, the probability distribution spreads out or tightens up as the value of the number of individuals (NT) multiplied by the invasion rate (m) or NTm decreases or increases (figure 7).
Figure 6.
Predictions of the Sloan et al. (2006, in press) model. Probability density functions, ϕi for the local abundance of a species that makes up 20% of the source community. m=1 means the space vacated by every death is filled by an immigrant and the source and local communities are highly coupled. Therefore, ϕi forms a tight bell-shaped distribution with mean relative abundance 20%. As m drops, the local community becomes increasingly more isolated and the internal neutral dynamics act to increase the skew and variance of the distributions, making low abundances more probable, but increasing the uncertainty or variability. As m continues to drop, the mode of the distribution becomes zero (m<0.04) and the likelihood of the species being absent increases.
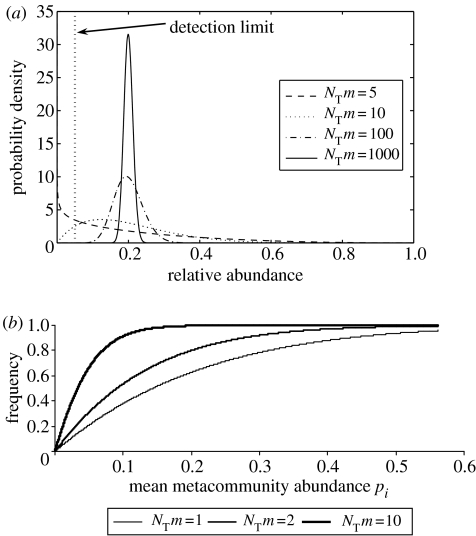
Figure 7.
How the value of NTm and m can be determined. (a) For a given mean source community abundance, the frequency with which an organism is observed is related to NTm, the total number of individuals multiplied by the immigration parameter. At high NTm values, the local distribution is tightly clustered around the mean meta-community distribution (in this case 0.2). As the NTm values drop, the distribution widens and eventually the mode of the curve falls below the detection limit and the organism is no longer observed. There is therefore a relationship between the mean source community abundance NTm and the frequency with which an organism is observed. (b) The expected frequency–relative abundance relationships observed in a local community for differing NTm values. Thus, for a given dataset, the NTm values can be found by fitting a line to these data; where the value of NT is known, m can be easily inferred.
10. Parameter estimation
Though the development of analytical solutions, especially simple ones, represents a helpful step forward, plausible values for the parameters NT, pi and m are required to put the model to work.
NT, the total number of individuals in a sample might be the number of clones examined in a clone library, or the number of bacteria in a sample from which 16S rRNA gene fragments are subsequently amplified and then analysed using a microbial community fingerprinting technique, such as denaturing gradient gel electrophoresis (DGGE). Where subsets of the bacterial community are examined using molecular fingerprinting techniques, it may be necessary to use fluorescent in situ hybridization (FISH) or some other quantitative method to determine the number of individuals comprising that subset of the community. It is worth noting that though the model itself is stochastic, the parameter NT is not. Rather, the number of individuals in a particular functional group is a function of the efficiency with which that particular group converts energy to biomass. Where this is known, the number of individuals can be estimated a priori.
The proportional abundance in the meta-community, pi, can also be estimated. Hubbell (2001) pointed out that each local community is, in effect, a sample of the community from which it is formed. Thus, by sampling many local communities sourced from the same meta-community, one can build-up a picture of the proportional abundance of that organism in the source community. This may be achieved by analysis of a large number of 16S rRNA gene clone libraries or microbial community fingerprints from similar environments. The sample sizes should be approximately the same and the samples themselves should be independent, ideally from different communities in similar environments.
The parameter m is the probability that a death within the community replaced from outside the community (1−m is the probability that a death is replaced by growth/reproduction of a member of the local community). Authoritative commentators have suggested that m, the immigration parameter, is impossible to measure (Maurer & McGill 2004). Fortunately, there are at least two ways by which m can be inferred. Firstly, as the value of NTm increases or decreases the variance of the local distribution will also increase and decrease. For any given molecular method, there is a detection limit (d), a proportional abundance a microbe must exceed to be detected. For example, to be detected in a clone library an organism must comprise at least 1/the number of clones, while in DGGE an organism must be at least 1% of the sample (Muyzer et al. 1993). Since the NTm value controls the variance of the local distribution, it controls the proportion of communities in which an organism is present above the detection limit and thus the frequency with which it is observed. Therefore, for an organism of mean abundance pi, the probability that the local abundance of the organism (xi) exceeds the detection limit (d) can be related to the value of NT, which can be measured and related to m by the following equation:
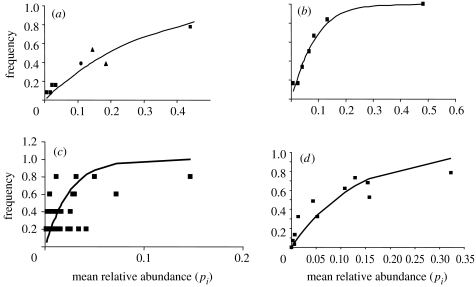
The predicted relationship between pi and NTm is shown in figure 7. The prediction can be evaluated with data on community composition from a number of sites obtained using methods based on 16S rRNA or some other conserved gene. Such a survey will provide an estimate of both pi and the frequency with which a particular organism is observed in several independent samples. Since NT and d are already known, m can be found using a simple spreadsheet-based routine. Figure 8 shows the relationship between pi and frequency for a range of quite different environments; further examples can be found in Sloan et al. (2006). We suspect that this pattern is widespread, perhaps even universal in the microbial world.
Figure 8.
Data and theory compared. The theoretical (solid line) and observed (squares or triangles) relationship between the mean relative abundance of taxa and the frequency with which they appear in a population of fixed size. Each of the points represents a different taxon. (a) Clone libraries of different ammonia monooxygenase (AMO) genes at 13 different domestic sewage works, NTm=1.41 (Wagner & Loy 2002). The triangles represent putative salt tolerant taxa. (b) Clone libraries of 16S rRNA genes from different AOB at six sites from the Humber Estuary, NTm=14. (c) Clone libraries of 16S rRNA genes from five experimental aquatic microcosms, NTm=10.3 (Horner-Devine et al. 2003). (d) 16S RNA sequences for 16 different bacterial taxa that are considered to be specific to freshwater environments sampled from 96 different lakes (Zwart et al. 2003), NTm=1.36. It is interesting to note that though both panels (c) and (d) pertain to lakes, the fit is much better in the larger dataset. This might be for two reasons: first, a larger dataset from larger systems will give a better indication of pi but also because prior to the analysis of (d) we removed data that represented three cyanobacterial lineages, leaving only data from one functional group of putative heterotrophs whereas (c) contains all the bacterial sequences. Other examples and a full description are given in Sloan et al. (2006, in press).
The absence of a species definition might be considered a barrier to theoretical microbial ecology. However, the parameter m is the probability of an individual immigration and should therefore be independent of the species definition. Data from Horner-Devine et al. (2003) offer some insight into this issue as it was recorded at different levels of phylogenetic resolution (95, 97 and 99% 16S rRNA sequence identity used to discriminate taxa), which might crudely correspond to genus, species and subspecies discrimination. The estimate of m obtained using data at different levels of phylogenetic resolution is similar (0.13, 0.13 and 0.2, respectively). This suggests that the model parameters are not constrained by, or an artefact of, species-specific, niche-adaptation considerations. This observation may have a bearing on ecology in general as frequency abundance relationships were first observed by Darwin and niche-based and stochastic rationales have been offered (Brown 2000).
The immigration parameter m can also be inferred without recourse to complex mathematical models. In colony forming organisms, each colony represents an immigration event. The number of individuals (not involved in the immigration) in a colony represents reproduction events in the local community. Where a single organism immigrates, the value of m is the ratio of the total number of colonies to the number of individuals in those colonies. This is also the minimum possible value for m, since more than one organism could form a colony (and thus decrease the ratio of immigration events to increase in numbers by reproduction), but less than one organism cannot initiate a colony and increase that ratio. AOB in wastewater treatment occur in just such microcolonies and a reanalysis of recently published data (Coskuner et al. 2005) permits us to estimate immigration rates by this method. We found rates of 7×10−3 (s.d., 0.0017; n=12) and no evidence of genus-specific immigration parameters from data obtained by the specific quantification of Nitrosomonas spp. (m=0.006) and Nitrosospira spp. (m=0.007; Curtis et al. in preparation).
11. Scaling of immigration rates
The immigration parameter is not likely to be constant; Sloan et al. (in press) demonstrate, albeit using almost complete censuses of tree communities, that immigration scales with the size of the community and potentially its physical attributes. Moreover, the immigration parameter identified using small random samples like clone libraries from a neutrally assembled community is confounded by the variance introduced by sampling effects. This means that the value of m that one perceives in a small sample can be much higher than the migration into a larger sample, or the community as a whole. Sloan et al. (in press) derived a relationship between the effective immigration that can be seen in a sample of size Ns and the true immigration rate m in a neutral community of size NT. The relationship proposed should be used with caution. It may only pertain in successively larger samples of relatively well-mixed communities and will not indicate migration into a discrete community or over a landscape. It does indicate that the immigration rate will decline as the samples get larger and larger; this in itself is significant.
By estimating the value of m using differing methods working at differing scales, we can see how m varies with the number of individuals in a well-mixed environment (wastewater treatment plants; figure 8). The observed migration rates do scale in the manner that Sloan et al. (in press) suggested. The data has a slope of −0.876 (standard error of 0.082) while the tentative theory suggested a slope of −1. We can probably cautiously extrapolate migration rates within the system on the basis of the observed and predicted relationships and suggest that for 1018 individuals (i.e. all bacteria in the plant) the value of m might be 1.5×10−16. We do not know what happens to migration at the boundary of the system, but it presumably drops below this value and may further decline or plateau.
12. An aside about speciation and migration rates
In considering the implications of ubiquitous taxa, it has been assumed that dispersal rates are so high that evolution is slowed down by immigration, leading to a few very abundant taxa. The implicit assumption is that immigration rates are sufficiently high to permit immigration to outweigh evolution. This is an assumption we can now consider in more detail by comparing plausible immigration rates with plausible speciation rates. Mutations are thought to occur at a rate of about 0.003 per genome per replication. This corresponds to a rate of about 10−10 per base pair per replication in a 6 Mb genome (Drake 1991); speciation cannot occur faster than this, except perhaps by HGT. The probability of a mutation being observed in a specific gene will be perhaps 7×10−7 substitutions per replication for a 1.5 kb gene. It appears that mutation in specific genes could exceed migration in as few as 106–107 organisms. This is less than the number of individuals found in a few millilitres of seawater or lake water, or a few drops from a wastewater treatment bioreactor. This is consistent with recent observations of sequence variation in bacteria in the open sea, which could be interpreted as evidence of the accumulation of neutral mutations and periodic selective sweeps (Thompson et al. 2005). Speciation will of course be slower than mutation and influenced by the selection pressures present at a particular time. However, if migration rates are a function of NT, then they could be very low indeed (see below). There therefore seems no reason why evolution should not occur in a system subject to immigration provided that the probability of a cell lost from the system by death being replaced by an immigrant decreases as the number of individuals increases, but the probability of that death being replaced by reproduction and replacement with a genetically different individual does not. The two processes of selection and evolution should exist in some form of dynamic equilibrium in the microbial world and similarities and differences between the genes in differing communities should contain information about the relative importance, perhaps even the rate, of these two mechanisms. There is the possibility of some link here between ecology and the work of population geneticists. For example, Slatkin & Maddison (1989) have developed methods for inferring immigration from phylogenies. They concluded that when the value of the effective population size Ne multiplied by the immigration rate m exceeds one, then there is enough gene flow to prevent the divergence of neutral genetic loci. Roberts & Cohan (1995) studied communities of Bacillus subtilis and Bacillus mojaviensis and found that Nem values did indeed exceed one. However, it is not certain that the Nem of Slatkin & Maddison (1989) is comparable to the NTm presented here. A more formal linkage between bacterial population genetics and microbial neutral community models may be possible, but would probably require a thorough re-examination of both. This would be a very worthwhile exercise, not only because of the insights one might gain about immigration, but also because community models and the population biology models should, ideally, be consilient.
13. Simple scenarios
Now that we have a model for community assembly and some plausible parameters, it is possible to begin an exploration of the diversity of the microbial world from a more ‘MacArthurian’ perspective. That is to invoke the rules and parameters described earlier in an attempt to gain insight into the extent of microbial diversity. The extent of the source diversity does of course remains unknown, local diversity remains unmeasured and a formal validation of the model is therefore unwarranted.
However, we do have a tentative method for estimating immigration rates, we do know how many individuals are present in certain functional groups and we do know how many different sequences are found in a clone library of a given size. It is therefore possible to run the model to generate a hypothetical local community which we can sample to generate an in silico clone library to crudely compare with observed clone library species richness.
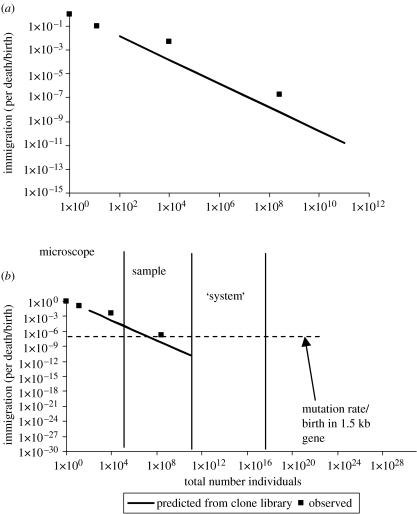
The parameters we have already obtained pertain to β-proteobacterial AOB and the bacterial community as a whole (greater than 95% heterotrophs) in aerobic wastewater treatment plants in the British midlands. The two groups are of contrasting species richness and evenness. The AOB are of low richness and evenness with a 50 clone clone library comprising five distinct sequences (at the 97% level) with one sequence comprising 40–45 clones. By contrast, a clone library of 542 sequences from the whole bacterial community might comprise 242 different sequences, with the most abundant clone comprising only about 10% of the total clone library (J. C. Baptista 2005, unpublished data). If the wastewater treatment reactor was 1000 m3, then one might expect the local community to have an NT value of about 1018 for the whole bacterial community and 1016 for the AOB (Coskuner et al. 2005). The data and methods described earlier suggest that this might lead to immigration rates of 10−12 and 1.5×10−16, respectively (figure 9).
Figure 9.
The scaling of immigration with NT. From left to right are shown a single cell, through a clone library to a FISH study and a DGGE gel. The solid line is the scaling predicted within a system from the clone library data. The data is shown at (a) a local scale and (b) a global scale, there being approximately 1030 bacteria in the world. It is apparent that a plausible extrapolation of the observed data could bring the rate of immigration below a plausible rate of speciation. This would permit speciation in an open system.
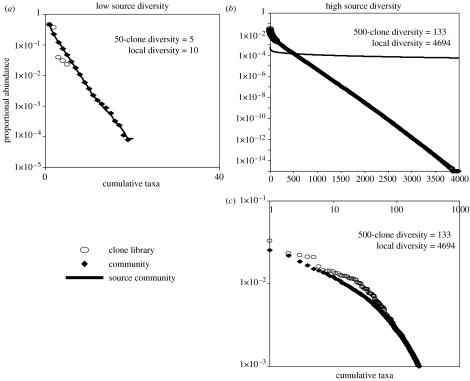
To attempt to reproduce sample diversities comparable to those observed using the parameters we have inferred, we must select possible source terms which can be sampled (Sloan et al. 2006, in press) to produce ‘local diversities’ which can be sampled an appropriate number of times to give a ‘clone library’ (figure 5). Thus, AOB might have a very low source diversity, perhaps 100–200 species capable of growing in a wastewater treatment plant in a global community of 1027. By contrast, the general bacterial or ‘heterotroph’ source community might be very large indeed, perhaps containing just over 105 different taxa in a global community of 1029 bacteria. We used these high and low diversities as source terms in the model. The in silico clone library of 50 clones from the low diversity simulation yielded five types of ‘AOB’ (approx. 20% of the putative source diversity), the most abundant sequence comprised 50% of the sample (figure 10). By contrast, about 500 clones from the high diversity simulation yielded just ‘133 heterotroph’ taxa, though the true local species richness might be 4500. The AOB and general ‘heterotroph’ species richness in the simulated clone libraries were, respectively, very close to, and of the same order as, those observed in real life. We believe that this is tentative and qualitative evidence that simple models using parameters derived from real systems can reproduce patterns observed when real analyses are done.
Figure 10.
Model predictions are qualitatively consistent with the patterns seen in ‘real life’. A sample of 50 ammonia-oxidizing bacteria (AOB) clones and 500 bacterial clones in one of the wastewater treatment plants featured in figure 9 found taxa diversities of 7 and 202, respectively, in the two samples. Here, we examine the ability of the Sloan et al. model to qualitatively reproduce that finding. We used known NT values and, hypothetical source diversities of 200 (for the low source diversity group corresponding with the AOB) and 100 000 (for the high source diversity corresponding with the general bacterial population); immigration parameters were estimated using the extrapolation procedure mentioned in this paper (10−12 for the AOB and 1.5×10−16 for the general bacterial population). (a) Represents the AOB, the diversity is similar to that found in the samples. (b) Represents the high source diversity group, the most abundant members of which are shown in (c) for greater clarity. The clone and local diversity are obviously distinct. Slightly less diversity was found in the model than observed in practice, suggesting some small errors in the source term or the immigration parameter. This is neither proof nor disproof of the model, but an encouraging indication that a more formal validation may be worthwhile undertaking.
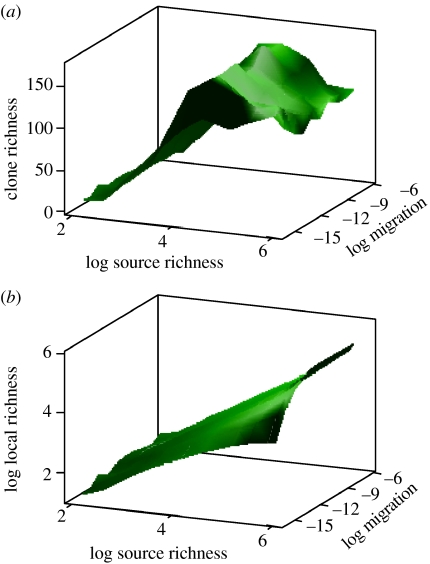
This may sound like convincing evidence for a source diversity (i.e. richness) of over a 100 000 for heterotrophs: it is not. The local community is so grossly under sampled that source richness values varying between 104 and 106 (or more) distinct taxa are superficially indistinguishable even with ‘large’ clone libraries (figure 11) for any migration parameter less than 10−8. Nevertheless, it is apparent that relatively large source diversities are required to generate the clone library diversities commonly observed in the natural world, presumably, because the low immigration rates act as a sort of ‘barrier’ or ‘filter’ which must be overcome if moderately diverse local communities are to form. However, further insights into the source diversity might be gleaned from the proportional abundance of the taxa detected. These are early days with simple models. We should however be able to get a much clearer picture of the un-sampled diversity through an intelligent mixture of model calibration, simulation and samples of an appropriate size. If such an approach could be validated, there seems to be no reason why we should not, in principle, start to systematically infer local and source diversities of microbial communities throughout the world and thus start to systematically map the microbial diversity of the planet.
Figure 11.
The validation of the model is impossible with small sample sizes. Using the neutral model to simulate the effect of migration and source community diversity in a microbial community of 1018 individuals (e.g. a wastewater treatment plant) on (a) the number of taxa in a library of 500 clones and (b) the true local richness in a community of 1018. The number of taxa in the sample soon plateaus, which means we can infer relatively little about the local or source diversity except that both probably exceed 103 and 104, respectively. Note how the clone library species richness actually dips slightly at higher diversities, probably owing to a change in evenness in the underlying local community.
Preliminary evidence from the study of taxa–area curves looks hopeful. In particular, the radical differences in observed taxa–area curves for microbes can be easily reproduced using such a model (Woodcock et al. in press). Thus, the very flat or none existent observed taxa–area curves seen in some studies are to be expected in most environments examined using small samples. Taxa–area curves will be more easily observed in low diversity communities with a relatively low number of individuals. Moreover and more intriguingly, Woodcock et al. (in preparation) have recently shown that the model can be calibrated in one insular community (treeholes) and used to predict the diversity measured using DGGE in others. If this finding could be reproduced, it would make the systematic mapping and prediction of microbial diversity a great deal more tractable.
14. The future
Contemporary studies of microbial diversity are severely handicapped by extremely small sample sizes. The data, and thus our perception, of the microbial world are a function of methods and budgets, rather than a rational assessment of the sample size required. The inadequate nature of the samples commonly obtained from the environment must go some way to explain the polemic that surrounds microbial diversity. Moreover, unless and until sufficiently large samples are obtained in studies of microbial diversity, definitive statements about microbial diversity should only be made with caution. At present, we do not know quite how large the samples will have to be, and it may become apparent that ‘adequate’ sample sizes will be beyond the reach of most laboratories. Predictive mathematical models of microbial diversity could help in both cases. We have described one such model and how it can be calibrated here. The predictions of the model could be used to design an intensive sampling programme to determine the diversity of selected local communities and thus give us valuable information about the wider communities from which they are drawn. This in turn would allow laboratories to make predictions about microbial community diversity by calibrating the model using small samples. A routine and generally accepted method for predicting microbial community diversity, complemented by occasional deep sampling, would open the way for a systematic survey of the extent of the diversity of the microbial world. The exploration of the microbial world could become a routine, accumulative and very powerful long-term endeavour underpinning the knowledge and economic base of the sponsoring nation.
Footnotes
One contribution of 15 to a Discussion Meeting Issue ‘Species and speciation in micro-organisms’.
References
- Bell G. The distribution of abundance in neutral communities. Am. Nat. 2000;155:606–617. doi: 10.1086/303345. doi:10.1086/303345 [DOI] [PubMed] [Google Scholar]
- Bell G. Ecology—neutral macroecology. Science. 2001;293:2413–2418. doi: 10.1126/science.293.5539.2413. doi:10.1126/science.293.5539.2413 [DOI] [PubMed] [Google Scholar]
- Borneman J, Triplett E.W. Molecular microbial diversity in soils from eastern Amazonia: evidence for unusual microorganisms and microbial population shifts associated with deforestation. Appl. Environ. Microbiol. 1997;63:2647–2653. doi: 10.1128/aem.63.7.2647-2653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock T. Prentice-Hall; Englewood Cliffs, NJ: 1966. Principles of microbial ecology. [Google Scholar]
- Brown J.H. University of Chicago Press; Chicago, IL: 2000. Macroecology. [Google Scholar]
- Chao A. Nonparametric-estimation of the number of classes in a population. Scand. J. Stat. 1984;11:265–270. [Google Scholar]
- Chao A. Estimating the population-size for capture recapture data with unequal catchability. Biometrics. 1987;43:783–791. doi:10.2307/2531532 [PubMed] [Google Scholar]
- Colwell R.K, Coddington J.A. Estimating terrestrial biodiversity through extrapolation. Phil. Trans. R. Soc. B. 1994;345:101–118. doi: 10.1098/rstb.1994.0091. [DOI] [PubMed] [Google Scholar]
- Coskuner G, Ballinger S.J, Davenport R.J, Pickering R.L, Solera R, Head I.M, Curtis T.P. Agreement between theory and measurement in quantification of ammonia-oxidizing bacteria. Appl. Environ. Microbiol. 2005;71:6325–6334. doi: 10.1128/AEM.71.10.6325-6334.2005. doi:10.1128/AEM.71.10.6325-6334.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, T. P., Sloan, W. & Scannell, J. 2001 The estimation of prokaryotic diversity and its limits. In Nineth Int. Symp. on Microbial Ecology, Amsterdam.
- Curtis T.P, Sloan W, Scannell J. Estimating prokaryotic diversity and it limits. Proc. Natl Acad. Sci. USA. 2002;99:10 494–10 499. doi: 10.1073/pnas.142680199. doi:10.1073/pnas.142680199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis, T. P., Baptista, J. C. & Sloan, W. T. In preparation. Estimating migration rates in microbial communities.
- Drake J.W. A constant rate of spontaneous mutation in DNA-based microbes. Proc. Natl Acad. Sci. USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. doi:10.1073/pnas.88.16.7160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunbar J, Barns S.M, Ticknor L.O, Kuske C.R. Empirical and theoretical bacterial diversity in four Arizona soils. Appl. Environ. Microbiol. 2002;68:3035–3045. doi: 10.1128/AEM.68.6.3035-3045.2002. doi:10.1128/AEM.68.6.3035-3045.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenchel T, Finlay B.J. The diversity of microbes: resurgence of the phenotype. Phil. Trans. R. Soc. B. 2006;361:1965–1973. doi: 10.1098/rstb.2006.1924. doi:10.1098/rstb.2006.1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay B.J, Clarke K.J. Ubiquitous dispersal of microbial species. Nature. 1999;400:828. doi:10.1038/23616 [Google Scholar]
- Fulthorpe R.R, Rhodes A.N, Tiedje J.M. High levels of endemicity of 3-chlorobenzoate-degrading soil bacteria. Appl. Environ. Microbiol. 1998;64:1620–1627. doi: 10.1128/aem.64.5.1620-1627.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. doi:10.1126/science.1112665 [DOI] [PubMed] [Google Scholar]
- Godon J.J, Zumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl. Environ. Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagstrom A, Pommier T, Rohwer F, Simu K, Stolte W, Svensson D, Zweifel U.L. Use of 16S ribosomal DNA for delineation of marine bacterioplankton species. Appl. Environ. Microbiol. 2002;68:3628–3633. doi: 10.1128/AEM.68.7.3628-3633.2002. doi:10.1128/AEM.68.7.3628-3633.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S.H, Bunge J, Jeon S.O, Epstein S.S. Predicting microbial species richness. Proc. Natl Acad. Sci. USA. 2006;103:117–122. doi: 10.1073/pnas.0507245102. doi:10.1073/pnas.0507245102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner-Devine M.C, Leibold M.A, Smith V.H, Bohannan B.J.M. Bacterial diversity patterns along a gradient of primary productivity. Ecol. Lett. 2003;6:613–622. doi:10.1046/j.1461-0248.2003.00472.x [Google Scholar]
- Houchmandzadeh B, Vallade M. Clustering in neutral ecology. Phys. Rev. E. 2003;68:061912. doi: 10.1103/PhysRevE.68.061912. doi:10.1103/PhysRevE.68.061912 [DOI] [PubMed] [Google Scholar]
- Hubbell S.P. Princeton University Press; Princeton, NJ: 2001. The unified neutral theory of biodiversity and biogeography. [DOI] [PubMed] [Google Scholar]
- Hugenholtz P, Goebel B.M, Pace N.R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J. Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel P, Harmand J, Zemb O, Latrille E, Lobry C, Delgenes J.P, Godon J.J. Denaturing gradient electrophoresis (DGE) and single-strand conformation polymorphism (SSCP) molecular fingerprintings revisited by simulation and used as a tool to measure microbial diversity. Environ. Microbiol. 2006;8:720–731. doi: 10.1111/j.1462-2920.2005.00950.x. doi:10.1111/j.1462-2920.2005.00950.x [DOI] [PubMed] [Google Scholar]
- Lunn M, Sloan W.T, Curtis T.P. Estimating bacterial diversity from clone libraries with flat rank abundance distributions. Environ. Microbiol. 2004;6:1081–1085. doi: 10.1111/j.1462-2920.2004.00641.x. doi:10.1111/j.1462-2920.2004.00641.x [DOI] [PubMed] [Google Scholar]
- MacArthur R. On the relative abundance of bird species. Proc. Natl Acad. Sci. USA. 1957;43:293–295. doi: 10.1073/pnas.43.3.293. doi:10.1073/pnas.43.3.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur R. On the relative abundance of species. Am. Nat. 1960;874:25–36. doi:10.1086/282106 [Google Scholar]
- MacArthur R, Wilson E. Princeton Univerity Press; Princeton, NJ: 1967. The theory of island biogeography. [Google Scholar]
- Maurer B.A, McGill B.J. Neutral and non-neutral macroecology. Basic Appl. Ecol. 2004;5:413–422. doi:10.1016/j.baae.2004.08.006 [Google Scholar]
- May R.M. Patterns of species abundance and diversity. In: Cody M.L, Diamond J.M, editors. Ecology and evolution of communities. Harvard University Press; Cambridge, MA: 1974. pp. 81–120. [Google Scholar]
- McKane A.J, Alonso D, Sole R.V. Analytic solution of Hubbell's model of local community dynamics. Theor. Popul. Biol. 2004;65:67–73. doi: 10.1016/j.tpb.2003.08.001. doi:10.1016/j.tpb.2003.08.001 [DOI] [PubMed] [Google Scholar]
- Muyzer G, Dewaal E.C, Uitterlinden A.G. Profiling of complex microbial-populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16s ribosomal-RNA. Appl. Environ. Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang R, Dunbar J. Modeling bacterial species abundance from small community surveys. Microb. Ecol. 2004;47:396–406. doi: 10.1007/s00248-003-1026-7. doi:10.1007/s00248-003-1026-7 [DOI] [PubMed] [Google Scholar]
- Pounds J.A, Puschendorf R. Ecology—clouded futures. Nature. 2004;427:107–109. doi: 10.1038/427107a. doi:10.1038/427107a [DOI] [PubMed] [Google Scholar]
- Roberts M.S, Cohan F.M. Recombination and migration rates in natural populations of Bacillus subtilis and Bacillus mojavensis. Evolution. 1995;49:1081–1094. doi: 10.1111/j.1558-5646.1995.tb04435.x. doi:10.2307/2410433 [DOI] [PubMed] [Google Scholar]
- Schloss P.D, Handelsman J. Status of the microbial census. Microbiol. Mol. Biol. Rev. 2004;68:686–691. doi: 10.1128/MMBR.68.4.686-691.2004. doi:10.1128/MMBR.68.4.686-691.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D, Handelsman J. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 2005;71:1501–1506. doi: 10.1128/AEM.71.3.1501-1506.2005. doi:10.1128/AEM.71.3.1501-1506.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D, Handelsman J. Toward a census of bacteria in soil. PLoS Comput. Biol. 2006;2:786–793. doi: 10.1371/journal.pcbi.0020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Maddison W.P. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–613. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan W.T, Lunn M, Woodcock S, Head I.M, Nee S, Curtis T.P. Quantifying the roles of immigration and chance in shaping prokaryote community structure. Environ. Microbiol. 2006;8:732–740. doi: 10.1111/j.1462-2920.2005.00956.x. doi:10.1111/j.1462-2920.2005.00956.x [DOI] [PubMed] [Google Scholar]
- Sloan, W. T., Woodcock, S., Head, I. M., Lunn, M. & Curtis, T. P. In press. Using environmental genomic data to identifying patterns in the structure of microbial communities. Microb. Ecol. [DOI] [PubMed]
- Thompson J.R, Pacocha S, Pharino C, Klepac-Ceraj V, Hunt D.E, Benoit J, Sarma-Rupavtarm R, Distel D.L, Polz M.F. Genotypic diversity within a natural coastal bacterioplankton population. Science. 2005;307:1311–1313. doi: 10.1126/science.1106028. doi:10.1126/science.1106028 [DOI] [PubMed] [Google Scholar]
- Torsvik V, Goksoyr J, Daae F.L. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 1990;56:782–787. doi: 10.1128/aem.56.3.782-787.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallade M, Houchmandzadeh B. Analytical solution of a neutral model of biodiversity. Phys. Rev. E. 2003;68:061902. doi: 10.1103/PhysRevE.68.061902. doi:10.1103/PhysRevE.68.061902 [DOI] [PubMed] [Google Scholar]
- Volkov I, Banavar J.R, Hubbell S.P, Maritan A. Neutral theory and relative species abundance in ecology. Nature. 2003;424:1035–1037. doi: 10.1038/nature01883. doi:10.1038/nature01883 [DOI] [PubMed] [Google Scholar]
- Wagner M, Loy A. Bacterial community composition and function in sewage treatment systems. Curr. Opin. Biotechnol. 2002;13:218–227. doi: 10.1016/s0958-1669(02)00315-4. [DOI] [PubMed] [Google Scholar]
- Whitaker R.J, Grogan D.W, Taylor J.W. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. doi:10.1126/science.1086909 [DOI] [PubMed] [Google Scholar]
- Whitman W.B, Coleman D.C, Wiebe W.J. Prokaryotes: the unseen majority. Proc. Natl Acad. Sci. USA. 1998;95:6578–6583. doi: 10.1073/pnas.95.12.6578. doi:10.1073/pnas.95.12.6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson E.O. Penguin; Baltimore, MD: 1994. The diversity of life. [Google Scholar]
- Wilson E.O. Vintage; New York, NY: 1998. Consilience: the unity of knowledge. [Google Scholar]
- Woodcock S, Curtis T.P, Head I.M, Lunn M, Sloan W.T. Taxa–area relationships for microbes: the unsampled and the unseen. Ecol. Lett. 2006;9:805–812. doi: 10.1111/j.1461-0248.2006.00929.x. [DOI] [PubMed] [Google Scholar]
- Woodcock, S., van der Gast, C. J., Bell, T., Lunn, M., Curtis, T. P., Head, I. M. & Sloan, W. T. In preparation. Neutral assembly of bacterial communities. [DOI] [PubMed]
- Zwart G, et al. Rapid screening for freshwater bacterial groups by using reverse line blot hybridization. Appl. Environ. Microbiol. 2003;69:5875–5883. doi: 10.1128/AEM.69.10.5875-5883.2003. doi:10.1128/AEM.69.10.5875-5883.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]