Abstract
Hemolytic uremic syndrome (HUS) is an important cause of acute renal failure in children. Mutations in one or more genes encoding complement-regulatory proteins have been reported in approximately one-third of nondiarrheal, atypical HUS (aHUS) patients, suggesting a defect in the protection of cell surfaces against complement activation in susceptible individuals. Here, we identified a subgroup of aHUS patients showing persistent activation of the complement alternative pathway and found within this subgroup two families with mutations in the gene encoding factor B (BF), a zymogen that carries the catalytic site of the complement alternative pathway convertase (C3bBb). Functional analyses demonstrated that F286L and K323E aHUS-associated BF mutations are gain-of-function mutations that result in enhanced formation of the C3bBb convertase or increased resistance to inactivation by complement regulators. These data expand our understanding of the genetic factors conferring predisposition to aHUS, demonstrate the critical role of the alternative complement pathway in the pathogenesis of aHUS, and provide support for the use of complement-inhibition therapies to prevent or reduce tissue damage caused by dysregulated complement activation.
Keywords: renal disease
Hemolytic uremic syndrome (HUS) (MIM 235400) is characterized by thrombocytopenia, Coomb's test-negative microangiopathic hemolytic anemia and acute renal failure (1). The typical form of HUS follows a diarrheal prodrome and is associated with 0157:H7 Escherichia coli infections (2). Five percent to 10% of HUS patients lack an association with infection and have the poorest long-term prognosis (1). This atypical HUS (aHUS) usually occurs in adults and in very young children and is associated with mutations or polymorphisms in the genes encoding the complement regulators factor H (fH, CFH) (3–8), membrane cofactor protein (MCP, MCP) (9, 10), and factor I (fI, IF) (11, 12).
The molecular mechanisms underlying aHUS are only now beginning to be understood. In contrast to patients with complement deficiencies, aHUS patients present relatively normal complement levels (5). Crucially, functional analysis of several CFH, MCP, and IF aHUS-associated mutations has demonstrated that they affect primarily the control of complement activation on cellular surfaces, whereas complement regulation in plasma is not greatly affected (4, 7, 10, 13). These findings have led to the hypothesis that aHUS results from a defective protection of host cells within the context of a “normal” complement function in the fluid phase (7, 14). From this perspective, a situation that triggers complement activation in the microvasculature will not be properly controlled, and the amplification of the initial damage will result in tissue destruction.
Despite these advances in our understanding of the molecular basis of aHUS, no genetic defect has yet been found in two-thirds of aHUS patients, and incomplete penetrance of the disease in individuals carrying fH, MCP, or fI mutations is relatively frequent, suggesting the existence of additional genetic factors that predispose to aHUS.
To provide further insights into the genetic factors predisposing to aHUS we have examined the hypothesis that genetic variants of complement-activating proteins, resulting in excessive activation of the complement system, are also aHUS risk factors. Like mutations in the complement regulators, genetic variations in complement activators have the potential to disrupt the balance between activation and regulation, dysregulating the complement system and predisposing to pathology when the complement system undergoes activation.
Here, we describe mutations in the gene encoding the complement-activator factor B (fB, BF) and demonstrate these to be associated with aHUS. We report their functional characterization and demonstrate that they are gain-of-function mutations. These data offer a complementary view to the previously described loss-of-function mutations in complement regulators and definitively establish the critical role of the complement alternative pathway in the pathogenesis of HUS. In addition, we analyze the issue of incomplete penetrance of disease in BF mutation carriers and show that, in a large multiple-affected Spanish HUS pedigree, the concurrence of an enhanced activation of the alternative pathway of complement with an impaired protection of host surfaces likely results in unrestricted activation of the alternative pathway at the cellular surfaces and triggers the development of aHUS.
Results and Discussion
Complement Profiles in aHUS Patients Identify a Subgroup with Evidence of Alternative Pathway Activation.
Our registry of aHUS patients includes 74 individuals extensively studied with regard to complement profiles and genomic analyses of the complement-regulatory genes CFH, MCP, IF, and DAF. We found mutations in CFH, MCP, or IF in ≈30% of unrelated aHUS patients in our cohort [see supporting information (SI) Fig. 6a], in concordance with 50% figures from other aHUS cohorts (3–12).
A majority of patients in our aHUS cohort present normal C3, C4, and hemolytic complement levels (SI Fig. 6b), in agreement with reports describing that, in general, HUS patients are not hypocomplementemic (5). We noticed, however, that some of the patients without mutations in the complement-regulatory genes, including patients H21, H49, H55P, H65, and H71, present evidence of alternative pathway activation. They consistently show very low levels of C3 and normal or elevated levels of C4 over multiple determinations (SI Fig. 6b), and analysis of their plasma by Western blot showed the presence of the activation product Ba (SI Fig. 6c). Interestingly, H55P belongs to a large multiple-affected aHUS Spanish kindred in which disease segregated with the MHC region in chromosome 6q24, including the BF gene (15, 16).
Identification of BF Mutations in Patients with Alternative Pathway Activation.
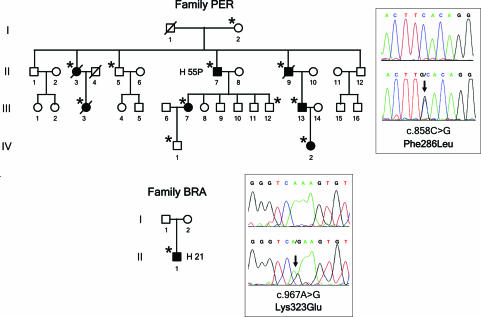
Although an earlier report excluded BF mutations in another aHUS cohort (16), the above observations prompted us to search for mutations in BF in the group of patients showing evidence of alternative pathway activation. Two patients in this subgroup (H21; pedigree BRA and H55P; pedigree PER) carried missense mutations in heterozygosis in the BF gene. Genomic sequencing of BF in H55P revealed a heterozygous mutation (c.858C>G; F286L) in exon 6, encoding the von Willebrand type A domain of fB. The F286L missense mutation segregates with the low levels of C3 that are present in different members of family PER and is inherited by all HUS-affected members of this pedigree (Fig. 1; and see SI Table 1). Genomic sequencing of BF in H21 revealed another heterozygous mutation (c.967A>G; K323E) in exon 7, also encoding a residue within the von Willebrand type A domain of fB. K323E is a de novo mutation absent in H21 parents (Fig. 1). Parental sample integrity in family BRA was confirmed by genotyping markers in the RCA gene cluster in chromosome 1q32. Neither F286L nor K323E mutations were present in a control population of 100 normal individuals.
Fig. 1.
Mutations in the BF gene in two aHUS pedigrees. Pedigrees of families PER and BRA are described. Individuals are identified by numbers within each generation (in Roman numerals). Affected individuals are indicated with filled symbols. Deceased individuals are crossed. Carriers of BF mutations are indicated by an asterisk. For each BF mutation, the chromatogram corresponding to the DNA sequence surrounding the mutated nucleotide in BF is shown for the appropriate HUS patient and for a control sample. The nucleotide and amino acid numbering are referred to the translation start site (A in ATG is + 1; Met is + 1).
Structure–Function Analyses Identify BF Gain-of-Function Mutations in aHUS.
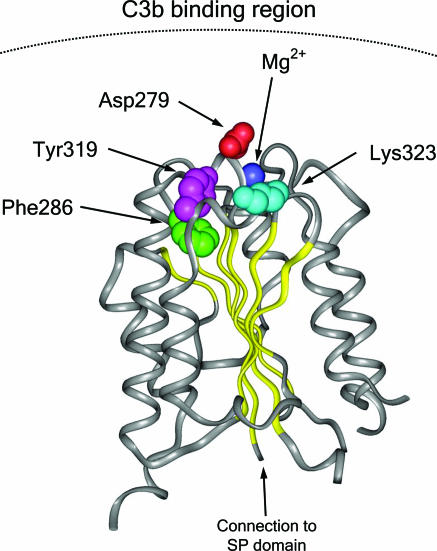
fB is a zymogen that carries the catalytic site of the complement alternative pathway convertase C3bBb. Upon interaction with C3b, fB is cleaved by factor D (fD) into two fragments, Ba and Bb. Ba is released, whereas Bb remains bound to C3b, forming the C3bBb alternative pathway convertase, an active serine protease that cleaves additional C3 into C3b. The Bb fragment of fB comprises two protein domains: a von Willebrand factor type A (vWfA) domain and a serine protease (SP) domain (17). From structural studies, the fB vWfA domain is globular, composed of several parallel β-sheets surrounded by seven α-helices (18, 19). Site-directed mutagenesis of fB residues at the C3b-Bb interface in the fB vWfA domain have identified several residues that are critical for the interaction between C3b and fB and influence the normal dissociation of Bb from C3b, whether it is spontaneous or promoted by the complement regulatory proteins FH, DAF, or complement receptor 1 (CR1). In particular, the mutation D279G, in close proximity to the Mg2+-binding site within the fB vWfA domain, caused resistance to decay acceleration mediated by all three regulators and increased C3b-binding affinity and C3bBb stability (20). Both of the BF mutations F286L and K323E identified here in the aHUS patients alter residues at the C3b–Bb interface in the vWfA domain in close proximity to D279 (Fig. 2). This finding led us to suspect that these mutations might alter stability of the C3bBb convertase and activity of the alternative pathway. The mutation K323E changes a fully exposed residue with a positive charge to one with a negative charge. The mutation F286L influences Y319, another fully exposed residue, modifying the edge-to-face stacking interaction between F286 and Y319. The substitution F286L likely decreases mobility restrictions to Y319 at the C3b–Bb interface (Fig. 2).
Fig. 2.
Structural implications for the HUS-associated BF mutations. Diagram of the von Willebrand type A domain of fB. Insight II (BYOSYM software package; Molecular Simulations, San Diego, CA) was used to draw structure by using PDB files 1Q0P and 1RS0_A. The positions of the residues Phe-286 and Lys-323 that are mutated in the HUS patients are indicated. The position of the Mg2+ ion and that of the Asp-279 and Tyr-319 are also indicated. Notice the edge-to-face stacking of Phe-286 and Tyr-319 residues. Numbering of residues is referred to with the initial methionine as “1” and therefore includes the sequence of the N-terminal signal peptide. Residue Asp-279 was described as Asp-254 by Hourcade et al. (32).
To functionally characterize the effects of the K323E and F286L mutations, we purified fB from plasma of appropriate carriers and normal controls (SI Fig. 7a). We monitored formation and decay of the C3 convertase in real time using surface plasmon resonance (SPR) (21). To assess the effects of mutations, C3b was coated on the biosensor chip surface via the thioester group, and fB from controls or patients was flowed across the surface in the absence or presence of the fB cleaving enzyme fD (2 μg/ml) to form the proenzyme or activated enzyme, respectively.
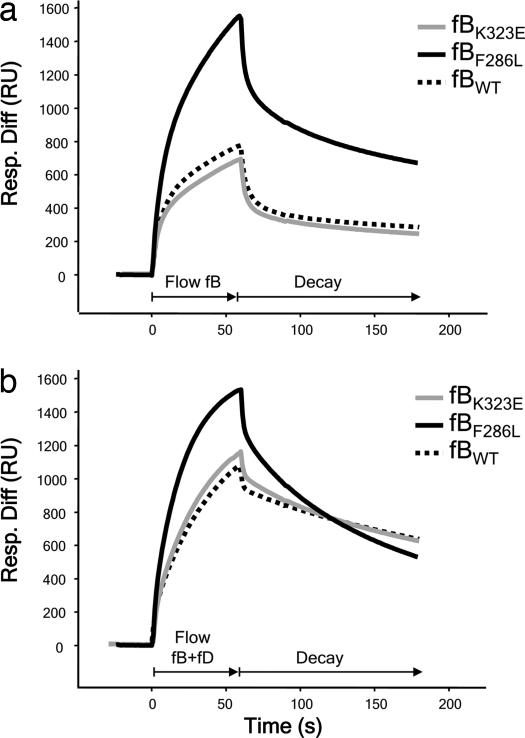
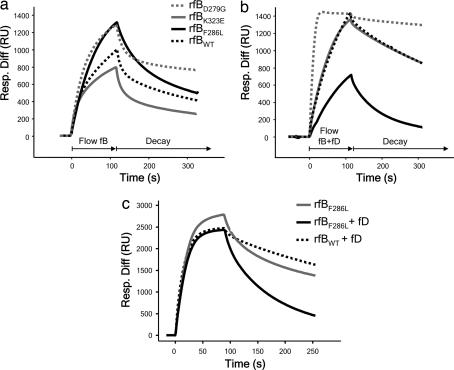
fB isolated from patient H55P demonstrated a much more rapid and efficient formation of the C3bB complex (Fig. 3a). In the presence of fD, the expected enhanced formation of the C3bBb complex was seen for control fB, whereas the kinetics of complex formation for fBF286L were not further enhanced from that obtained in the absence of fD (Fig. 3b). However, the rate of decay of the mutant convertase was increased compared with control. These differences were observed despite the protein purified from patient H55P comprising both normal and mutant fB. To better define the effect of this mutation on convertase formation, we generated recombinant fBWT and fBF286L proteins (SI Fig. 7b). As a positive control, we also generated fBD279G, previously identified in point mutation studies to increase stability of the convertase (20). Convertase formation and decay with recombinant fBs was visualized by SPR. In the absence of fD, high levels of C3bB were formed with recombinant fBF286L, as seen with native fBF286L (Fig. 4a), suggesting that initial binding between C3b and fBF286L was more efficient. However, in the presence of fD, fBF286L appeared to form low levels of active enzyme, a consequence of enhanced rate of decay, particularly apparent at low concentrations of fB (Fig. 4b; 10 μg/ml shown here). As the concentration of recombinant fB was increased, the level of enzyme formed by fBF286L approached that formed by recombinant fBWT (Fig. 4c; 37 μg/ml). From these data, we conclude that, when supply of fB is unlimited (200 μg/ml in plasma), fBF286L will generate abundant, rapidly cycling C3-convertase; in this regard, fBF286L behaves as a gain-of-function mutation that will enhance generation of C3b, dysregulating the alternative pathway. The findings explain the increased levels of fB cleavage and C3 consumption found in carriers of the fBF286L mutation.
Fig. 3.
SPR analysis of fBs purified from plasma. (a) Formation of C3bB complexes. Control fB (fBWT) and fB purified from H21 (fBK323E) formed similar levels of the proenzyme, whereas fB purified from H55P (fBF286L) formed C3bB complexes much more rapidly and to a much higher level. (b) Formation of C3bBb complexes. fBWT and fBK323E formed similar levels of complexes with C3b. fBF286L formed high and similar levels of complexes with C3b in the absence or presence of fD. Resp. Diff, response difference; RU, resonance units.
Fig. 4.
SPR analysis of recombinant fB proteins. (a) Formation of C3bB complexes. fBWT and fBK323E formed similar levels of the proenzyme, whereas fBF286L formed C3bB complexes much more rapidly and to a much higher level, similar to those formed by the fBD279G mutant described by Hourcade et al. (20). (b) Formation of C3bBb. At 10 μg/ml, fBWT and fBK323E formed similar levels of complexes with C3b. fBF286L formed lower levels of complexes with C3b in the presence than in the absence of fD as a consequence of accelerated rate of spontaneous decay of the cleaved fB. (c) Formation of C3bBb. At 37 μg/ml, fBF286L formed high levels of proenzyme, but when fD was included in the incubation, levels of activated enzyme formed by either recombinant fBWT or fBF286L were comparable. Resp. Diff, response difference; RU, resonance units.
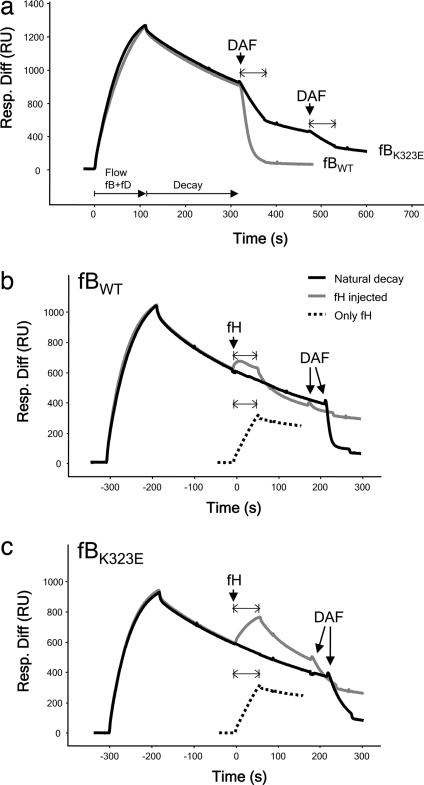
Similar experiments were carried out to test the K323E mutation. fB was isolated from the plasma of patient H21 and used to form convertase on the C3b-coated chip. In contrast to fB from patient H55P, enzyme formation and decay of H21 fB was indistinguishable from control fB, either in the presence or absence of fD (Fig. 3). Similarly, when recombinant proteins were analyzed, enzyme formation by fBWT and fBK323E were comparable (Fig. 4). To further explore the defect in fBK323E, we used SPR to compare the ability of DAF and fH to decay the cleaved, activated enzyme. Convertase was formed on the C3b surface by flowing recombinant fBWT or fBK323E in the presence of fD. Enzyme was allowed to decay naturally for several minutes, and then soluble DAF (600 nM) was flowed across the surface. fBWT was rapidly released from the C3b surface, whereas C3bBb formed from fBK323E was clearly more resistant to accelerated decay (Fig. 5a); some Bb remained, even after a second injection of soluble DAF. Similarly, this enzyme was more resistant to accelerated decay by fH (Fig. 5b). In contrast to DAF, which rapidly dissociates from the convertase after decay (21), fH binding to C3b is stable. During injection of fH (40 nM) onto the convertase surface, an overall change in amount of protein bound to the surface was evident, a balance between fH binding and Bb release. To visualize the change due to fH binding only, a separate injection of fH was carried out on the C3b surface in the absence of any fB (Fig. 5b; dashed black line). During fH injection onto recombinant fBWT convertase, the balance between fH binding and Bb release resulted in an initial small increase in mass. To distinguish between bound fH and residual Bb, DAF was then injected. No further decay was seen, confirming that fH had decayed the convertase. In contrast, when DAF was flowed over an identical convertase that had not been decayed by fH, efficient decay was seen (Fig. 5b). When fH was flowed over the fBK323E convertase, a much larger mass change, the balance between fH binding and Bb decay, was seen, indicating that decay was impaired; injection of DAF after fH caused further decay, confirming presence of residual Bb (Fig. 5c). Taken together, these data demonstrate that each of the mutant fB proteins cause increased alternative pathway activation. Formation of the C3bB proenzyme by mutant fBF286L is much enhanced, which will produce more active enzyme in vivo, whereas fBK323E forms a C3bBb enzyme more resistant to decay by DAF and fH, also causing increased enzyme activity in vivo. This finding of gain-of-function mutations in the BF gene and their association with aHUS illustrates the important contribution of the alternative pathway C3 convertase to the pathogenesis of aHUS.
Fig. 5.
fBK323E convertase decay by sDAF or fH monitored by SPR. (a) Decay by sDAF. C3bBb was allowed to decay spontaneously for several minutes before injection of sDAF. Bb from fBWT was removed from the surface by accelerated decay, whereas Bb formed from fBK323E demonstrated a much slower rate of accelerated decay. (b) Decay of fBWT by fH. After a period of spontaneous decay, fH or buffer (“natural decay”) was injected over the surface. Mass of protein bound reflected a balance between fH binding to C3b and Bb release. To visualize the binding of fH to C3b in isolation, fH was injected over the C3b surface (“only fH”). To confirm efficiency of fH decay, DAF was flowed and released no more Bb. (c) Decay of fBK323E by fH. Injects were as described above. Bb from fBK323E was much less efficiently decayed by fH as evidenced by the increased protein mass at the surface during the inject and the presence of residual Bb revealed by DAF-mediated decay. Resp. Diff, response difference; RU, resonance units.
Polymorphisms in the BF gene have recently been associated with age-related macular degeneration (AMD) (22), a disease that has been shown to strongly associate with polymorphisms in the CFH gene and where the macular pathology is driven by complement activation (23–26). Two BF polymorphisms were identified that were protective for AMD, one of which had been shown to reduce the activity of fB measured in hemolytic assays and the other in the signal peptide, was suggested to modulate secretion of fB. The observed protection in each case was ascribed to a reduced activity of the complement alternative pathway. This report provides timely support for our suggestion that genetic changes that alter the function of fB may influence pathology driven by alternative pathway activation.
Disease Penetrance in Carriers of BF Mutations Is Modulated by Other Complement-Associated Risk Alleles.
Within the HUS pedigree reported here, we have identified a total of 11 individuals carrying the F286L BF mutations, but only 7 of them have developed HUS thus far, and the majority of the healthy carriers are old enough to assume that they are not at risk (Fig. 1; and see SI Table 2). Incomplete penetrance of the disease has also been reported for the aHUS-associated mutations in the CFH, MCP, and IF genes (7, 9–11). Previously we, and others, have reported that two relatively frequent CFH and MCP SNPs haplotype blocks are strongly associated with aHUS (27, 28). We have proposed that concurrence of different susceptibility alleles greatly influences predisposition to aHUS and provides an explanation for the incomplete penetrance of aHUS in carriers of mutations in the complement regulator genes (28, 29). To investigate whether penetrance of the disease in BF mutations carriers is influenced by other aHUS genetic risk factors, we genotyped the CFH and MCP aHUS-associated SNP haplotype blocks and found that, among carriers of the F286L mutation, all of the HUS patients presented the MCPggaac risk allele, whereas among the healthy carriers (I-2, II-5, III-12, and IV-1) only the youngest, and probably still at risk, individual IV-1 harbored the HUS-associated MCPggaac risk allele. Of note, the single affected individual with the K323E mutation also presented the MCPggaac risk allele. These data strongly suggest that the HUS phenotype associated with the BF gain-of-function mutations is modulated by common complement regulator gene variants, such as the MCPggaac allele, associated with increased risk of developing HUS (28).
Complement Dysregulation Is the Unifying Event in aHUS Patients.
The efficiency of the complement system as an innate defense mechanism against microbial infections depends on a fine control that avoids the wasteful consumption of its components and restricts its activation to the surface of microorganisms, thus preventing nonspecific damage to host tissues. Genetic and functional analyses of the complement-regulatory proteins fH, MCP, and fI have shown that this critical control of complement activation may be impaired in aHUS patients (4, 7, 10). The identification of gain-of-function mutations in BF associated with aHUS reinforces this view and illustrates that dysregulation of the complement system may also result from an abnormally increased activity of the alternative pathway enzymes. These findings implicate other complement components in the same pathway as BF, such as C3 and CFP (encoding properdin), as potential candidate genes contributing to aHUS susceptibility.
In conclusion, these data demonstrate the critical role of the alternative complement pathway in the pathogenesis of aHUS and expand our understanding of the genetic factors conferring predisposition to aHUS. Furthermore, they support the hypothesis that susceptibility to aHUS involves the concurrence of multiple factors (genetic and environmental) contributing to dysregulation in the alternative pathway. This hypothesis provides an explanation for the incomplete penetrance of the disease in carriers of aHUS-associated mutations. The involvement of genetic factors may also be relevant in explaining the severe or fatal outcome of some individuals with the more common diarrhea-associated typical HUS. Lastly, our findings provide further support for the use of complement inhibition therapies in aHUS and perhaps also in severe forms of typical HUS to prevent or reduce tissue damage caused by dysregulated complement activation.
Patients, Materials, and Methods
Patients.
This study includes a total of 74 aHUS patients selected on the basis of a clinical history of HUS with non-diarrhea-associated origin. Of special relevance are patients H55P, H55, H85, and H112, all members of a large multiple-affected Spanish pedigree, referred to as family PER, and H21, the propositus in family BRA; specific clinical details are provided in SI Text. All protocols included in these studies have been approved by national and/or local institutional review boards, and all subjects gave their informed consent.
Complement Analyses.
C3, C4, fH, and fI levels were measured in serum or plasma as described (5, 30, 31). FB plasma levels were determined by ELISA using a polyclonal goat anti-human fB antibody (New Scientific Company, Cormano, Milan, Italy) as the capture antibody and a monoclonal mouse anti-human fB (JC1, in house) as the detection antibody. To assess complement function and regulation, we used the standard hemolytic assays CH50 and AP50 and a fH-dependent hemolytic assay (13). Expression levels of the membrane regulators MCP and DAF in peripheral blood lymphocytes (PBLs) were determined by flow cytometry as described (28).
Genotyping.
Patients and relatives were screened for mutations and polymorphisms in CFH, MCP, IF, and BF genes. DNA from these individuals was extracted from PBLs or from buccal mucosa cells by using standard procedures. Each exon of the CFH, MCP, and IF genes was amplified from genomic DNA by using specific primers derived from the 5′ and 3′ intronic sequences as described (5, 10, 11). All BF exons were amplified from genomic DNA in three amplicons of 1,800, 2,500, and 2,000 bp (see SI Text). Automatic sequencing was performed in an ABI 3730 sequencer using a dye terminator cycle sequencing kit (Applied Biosystems, Foster City, CA). Genotyping of HUS-associated CFH and MCP SNPs was performed by resequencing or by allelic discrimination using TaqMan probes (Applied Biosystems).
FB Purification.
FB from patients and controls was purified in one chromatographic step as follows. Two to 5 ml of plasma-EDTA were directly applied to a Sepharose column coated with the JC1 mouse monoclonal anti-human fB antibody and previously equilibrated with Tris·HCl (20 mM, pH 7.4) containing NaCl (50 mM). After extensive washes with equilibration, acetate (0.1 M, pH4, containing NaCl 500 mM) and borate (0.1 M, pH 8, containing NaCl 500 mM) buffers, the proteins bound to the column were eluted with 1 ml of glycine (100 mM, pH 2.5), followed by equilibration buffer. Twenty microliters of Tris·HCl (2M, pH 8) were added to the 500-μl eluted fractions to equilibrate pH. Those fractions containing fB were pooled and dialyzed against Hepes (10 mM, pH 7.4) containing 50 mM NaCl.
Recombinant fB Mutants.
The full-length human fB cDNA from clone IRAUp969B085D6 (RZDP) was introduced in the eukaryote expression vector pCI-Neo (Promega, Madison, WI), and the F286L, K323E, and D279G mutations introduced by site-directed mutagenesis using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. All of the constructs were completely sequenced to confirm that no additional mutations had been introduced. Recombinant fB proteins were expressed in stably transfected, cloned CHO cells cultured in Ham's F-12 medium containing 1% FBS and 0.5 mg/ml G418 sulfate (Geneticin; GIBCO-BRL, Carlsbad, CA). FB recombinant proteins were purified from the cell supernatants by affinity chromatography, as described above.
SPR Analysis of Convertase Formation and Decay.
Interactions of fB with C3b were analyzed on a BIAcore 3000 (BIAcore, Uppsala, Sweden) at 25°C. C3b was deposited on the surface of a CM5 chip as described (21). In brief, human C3, fB, and fD (Complement Technology, Tyler, TX) were flowed to seed the chip surface with small amounts of C3bBb; successive cycles of C3, followed by factors B and D, resulted in dense coating of the chip with C3b coupled in the “native” conformation via the thioester to hydroxyl groups on the carboxy-methyl dextran surface. The surface was stabilized by a short injection of 50 mM diethylamine, pH 11. fB proteins (37 μg/ml, fB purified from patient plasma; 10 μg/ml, recombinant fB) were then flowed over the C3b surface at 20 μl/min in HBS/Mg2+ (10 mM Hepes, pH 7.4/50 mM NaCl/1 mM MgCl2) with or without 2 μg/ml fD. Interactions were analyzed by using BIAcore BIAevaluation software, and data from a blank reference cell were subtracted to correct for changes in refractive index of flowed solutions.
To analyze DAF-mediated decay of convertase formed by recombinant fB, the chip surface was coated with C3b as described above and fB (10 μg/ml) with fD (2 μg/ml) were flowed across to form active enzyme. Soluble DAF (600 nM) was flowed across the surface for 60 s, and decay was monitored in real time. To analyze decay by fH, convertase was formed by using 20 μg/ml fB and 2 μg/ml fD. After convertase formation, either buffer (natural decay) or fH (40 nM) was injected, and decay was monitored in real time. To remove residual Bb from the surface, soluble DAF was injected at 2 μM. Because the overall change in mass at the chip surface after injection of fH was a combination of fH binding and Bb decay, fH was also injected over the surface in the absence of convertase formation to assess the mass change that was due to fH binding alone.
Supplementary Material
Acknowledgments
We are grateful to all members of families PER and BRA and the collaborating clinicians for their participation in this study. We also thank the members of the DNA sequencing laboratory at the Centro de Investigaciones Biológicas (CIB) and Dr. M. García-Lacoba of the Bioinformatics Department (CIB) for invaluable help. This work was supported by Spanish Ministerio de Educación y Cultura Grant SAF2005-00913; Spanish Fondo de Investigaciones Sanitarias Grants G03/054, G03/011, FIS 03/0621, and FIS 01/A046; and Wellcome Trust Grants 068823/Z and 068599. J.E.-G. was supported by European Molecular Biology Organization Long-Term Fellowship LTF 522-2006.
Abbreviations
- aHUS
atypical HUS
- HUS
hemolytic uremic syndrome
- SPR
surface plasmon resonance
- vWfA
von Willebrand factor type A.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0603420103/DC1.
References
- 1.Moake JL. N Engl J Med. 2002;347:589–600. doi: 10.1056/NEJMra020528. [DOI] [PubMed] [Google Scholar]
- 2.Karmali MA. Mol Biotechnol. 2004;26:117–122. doi: 10.1385/MB:26:2:117. [DOI] [PubMed] [Google Scholar]
- 3.Caprioli J, Bettinaglio P, Zipfel PF, Amadei B, Daina E, Gamba S, Skerka C, Marziliano N, Remuzzi G, Noris M. J Am Soc Nephrol. 2001;12:297–307. doi: 10.1681/ASN.V122297. [DOI] [PubMed] [Google Scholar]
- 4.Manuelian T, Hellwage J, Meri S, Caprioli J, Noris M, Heinen S, Jozsi M, Neumann HPH, Remuzzi G, Zipfel PF. J Clin Invest. 2003;111:1181–1190. doi: 10.1172/JCI16651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pérez-Caballero D, González-Rubio C, Gallardo ME, Vera M, López-Trascasa M, Rodríguez de Córdoba S, Sánchez-Corral P. Am J Hum Genet. 2001;68:478–484. doi: 10.1086/318201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Richards A, Buddles M, Donne R, Kaplan B, Kirk E, Venning M, Tielemans C, Goodship J, Goodship T. Am J Hum Genet. 2001;68:485–490. doi: 10.1086/318203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sánchez-Corral P, Pérez-Caballero D, Huarte O, Simckes AM, Goicoechea E, López-Trascasa M, Rodríguez de Córdoba S. Am J Hum Genet. 2002;71:1285–1295. doi: 10.1086/344515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Warwicker P, Goodship THJ, Donne RL, Pirson Y, Nicholls A, Ward RM, Turnpenny P, Goodship JA. Kidney Int. 1998;53:836–844. doi: 10.1111/j.1523-1755.1998.00824.x. [DOI] [PubMed] [Google Scholar]
- 9.Noris M, Brioschi S, Caprioli J, Todeschini M, Bresin E, Porrati F, Gamba S, Remuzzi G. Lancet. 2003;362:1542–1547. doi: 10.1016/S0140-6736(03)14742-3. [DOI] [PubMed] [Google Scholar]
- 10.Richards A, Kemp EJ, Liszewski MK, Goodship JA, Lampe AK, Decorte R, Muslumanogglu MH, Kavukcu S, Filler G, Pirson Y, et al. Proc Natl Acad Sci USA. 2003;100:12966–12971. doi: 10.1073/pnas.2135497100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fremeaux-Bacchi V, Dragon-Durey MA, Blouin J, Vigneau C, Kuypers D, Boudailliez B, Loirat C, Rondeau E, Fridman WH. J Med Genet. 2004;41:e84. doi: 10.1136/jmg.2004.019083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kavanagh D, Kemp EJ, Mayland E, Winney RJ, Duffield JS, Warwick G, Richards A, Ward R, Goodship JA, Goodship THJ. J Am Soc Nephrol. 2005;16:2150–2155. doi: 10.1681/ASN.2005010103. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez-Corral P, Gonzalez-Rubio C, Rodriguez de Cordoba S, Lopez-Trascasa M. Mol Immunol. 2004;41:81–84. doi: 10.1016/j.molimm.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 14.Rodriguez de Cordoba S, Esparza-Gordillo J, Goicoechea de Jorge E, Lopez-Trascasa M, Sanchez-Corral P. Mol Immunol. 2004;41:355–367. doi: 10.1016/j.molimm.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Carreras L, Romero R, Requesens C, Oliver AJ, Carrera M, Clavo M, Alsina J. J Am Med Assoc. 1981;245:602–604. [PubMed] [Google Scholar]
- 16.Kavanagh D, Kemp EJ, Richards A, Burgess RM, Mayland E, Goodship JA, Goodship THJ. Mol Immunol. 2006;43:856–859. doi: 10.1016/j.molimm.2005.06.041. [DOI] [PubMed] [Google Scholar]
- 17.Smith CA, Vogel CW, Muller-Eberhard HJ. J Exp Med. 1984;159:324–329. doi: 10.1084/jem.159.1.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bhattacharya AA, Lupher JML, Staunton DE, Liddington RC. Structure (London) 2004;12:371–378. doi: 10.1016/j.str.2004.02.012. [DOI] [PubMed] [Google Scholar]
- 19.Ponnuraj K, Xu Y, Macon K, Moore D, Volanakis JE, Narayana SVL. Mol Cell. 2004;14:17–28. doi: 10.1016/s1097-2765(04)00160-1. [DOI] [PubMed] [Google Scholar]
- 20.Hourcade DE, Mitchell LM, Oglesby TJ. J Immunol. 1999;162:2906–2911. [PubMed] [Google Scholar]
- 21.Harris CL, Abbott RJM, Smith RA, Morgan BP, Lea SM. J Biol Chem. 2005;280:2569–2578. doi: 10.1074/jbc.M410179200. [DOI] [PubMed] [Google Scholar]
- 22.Gold B, Merriam JE, Zernant J, Hancox LS, Taiber AJ, Gehrs K, Cramer K, Neel J, Bergeron J, Barile GR, et al. Nat Genet. 2006;38:458–462. doi: 10.1038/ng1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Edwards AO, Ritter R, III, Abel KJ, Manning A, Panhuysen C, Farrer LA. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 24.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, et al. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, et al. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 26.Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, et al. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Caprioli J, Castelletti F, Bucchioni S, Bettinaglio P, Bresin E, Pianetti G, Gamba S, Brioschi S, Daina E, Remuzzi G, et al. Hum Mol Genet. 2003;12:3385–3395. doi: 10.1093/hmg/ddg363. [DOI] [PubMed] [Google Scholar]
- 28.Esparza-Gordillo J, Goicoechea de Jorge E, Buil A, Berges LC, Lopez-Trascasa M, Sanchez-Corral P, Rodriguez de Cordoba S. Hum Mol Genet. 2005;14:703–712. doi: 10.1093/hmg/ddi066. [DOI] [PubMed] [Google Scholar]
- 29.Esparza-Gordillo J, Goicoechea de Jorge E, Garrido CA, Carreras L, Lopez-Trascasa M, Sanchez-Corral P, Rodriguez de Cordoba S. Mol Immunol. 2006;43:1769–1775. doi: 10.1016/j.molimm.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 30.Esparza-Gordillo J, Soria JM, Buil A, Almasy L, Blangero J, Fontcuberta J, Rodriguez de Cordoba S. Immunogenetics. 2004;56:77–82. doi: 10.1007/s00251-004-0660-7. [DOI] [PubMed] [Google Scholar]
- 31.Gonzalez-Rubio C, Ferreira-Cerdan A, Ponce IM, Arpa J, Fontan G, Lopez-Trascasa M. Arch Neurol. 2001;58:1923–1928. doi: 10.1001/archneur.58.11.1923. [DOI] [PubMed] [Google Scholar]
- 32.Hourcade DE, Mitchell L, Kuttner-Kondo LA, Atkinson JP, Medof ME. J Biol Chem. 2002;277:1107–1112. doi: 10.1074/jbc.M109322200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.