Abstract
Recent studies indicate that IL-1α functions intracellularly in pathways independent of its cell surface receptors by translocating to the nucleus and regulating transcription. Similarly, the chromatin-associated protein HMGB1 acts as both a nuclear factor and a secreted proinflammatory cytokine. Here, we show that IL-33, an IL-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines, is an endothelium-derived, chromatin-associated nuclear factor with transcriptional repressor properties. We found that IL-33 is identical to NF-HEV, a nuclear factor preferentially expressed in high endothelial venules (HEV), that we previously characterized. Accordingly, in situ hybridization demonstrated that endothelial cells constitute a major source of IL-33 mRNA in chronically inflamed tissues from patients with rheumatoid arthritis and Crohn's disease. Immunostaining with three distinct antisera, directed against the N-terminal part and IL-1-like C-terminal domain, revealed that IL-33 is a heterochromatin-associated nuclear factor in HEV endothelial cells in vivo. Association of IL-33 with heterochromatin was also observed in human and mouse cells under living conditions. In addition, colocalization of IL-33 with mitotic chromatin was noted. Nuclear localization, heterochromatin-association, and targeting to mitotic chromosomes were all found to be mediated by an evolutionarily conserved homeodomain-like helix–turn–helix motif within the IL-33 N-terminal part. Finally, IL-33 was found to possess transcriptional repressor properties, associated with the homeodomain-like helix–turn–helix motif. Together, these data suggest that, similarly to IL1α and HMGB1, IL-33 is a dual function protein that may function as both a proinflammatory cytokine and an intracellular nuclear factor with transcriptional regulatory properties.
Keywords: heterochromatin, nucleus, endothelium, HMGB1
Cytokines of the IL-1 family play a major role in a wide range of inflammatory, infectious, and autoimmune diseases (1). The three best known members of this family, IL-1α, IL-1β, and IL-18, are highly inflammatory cytokines, and dysregulation of their production or activity can lead to severe pathological events (1, 2). IL-33 is the most recent addition to the IL-1 family (3). IL-33 has been shown to induce T helper (Th) type 2 responses (3) by signaling through the IL-1 receptor-related protein ST2 (IL-1 R4), an orphan member of the IL-1 receptor family (4, 5). Treatment of mice with recombinant IL-33 resulted in blood eosinophilia, splenomegaly, and increased serum levels of IgE, IgA, IL-5, and IL-13 (3). Exposure to IL-33 also caused severe pathological changes in the lungs and gastrointestinal tract, including eosinophilic and mononuclear infiltrates, increased mucus production, and epithelial cell hyperplasia and hypertrophy (3).
IL-33 shares with the other members of the IL-1 family a single structural domain formed from 12 β strands, arranged in a so-called IL-1/FGF β-trefoil fold (6, 7). Similarly to IL-1α (271 aa, 30.6 kDa) and IL-1β (269 aa, 30.7 kDa) (1), IL-33 (270 aa, 30.7 kDa) is synthesized as a 31-kDa protein that lacks a clear signal peptide (3). This IL-33 precursor has been shown to be cleaved by caspase-1 in vitro, and it has been proposed that, similarly to IL-1β and IL-18 (1, 2), IL-33 may require processing by caspase-1 for optimal biological activity (3). However, it remains to be determined whether IL-33 is processed to a mature active form by caspase-1 in vivo.
The observation that the precursor form of each member of the IL-1 family (8), with the exception of the IL-1 receptor antagonist IL-1Ra, lacks a signal peptide suggested persistence of an early evolutionary role of these proteins as intracellular factors (9). Indeed, there is growing evidence for intracellular roles of cytokines and growth factors of the IL-1/FGF family. For instance, IL-1α, which is rarely found in the extracellular compartment but rather is primarily a cell-associated cytokine (1, 10), has been proposed to regulate cell migration, proliferation, senescence, and differentiation through intracrine mechanisms and intracellular pathways independent of its cell-surface membrane receptors (9, 11–16). Nuclear translocation of the IL-1α precursor, mediated by a consensus nuclear localization sequence (NLS) within its N-terminal (Nter) part (17), has been shown to be critical for the intracellular functions of IL-1α (12, 13). Several nuclear targets of the IL-1α precursor have been identified that interact specifically with the acidic Nter propiece but not the C-terminal (Cter) mature form (15, 16). These targets include the histone acetyltransferases p300, PCAF and Gcn5 (16), and the growth suppressor necdin (15). The identification of the histone acetyltransferases as nuclear partners of IL-1α is in agreement with a recent report demonstrating an important role of the IL-1α precursor as an intracrine proinflammatory activator of transcription (9). Together, these observations indicate that IL-1α is a dual-function protein that acts as both a nuclear factor and a proinflammatory cytokine (9). Interestingly, a similar duality of function has been shown for high-mobility group box 1 (HMGB1) protein, an abundant chromatin-associated protein involved in transcriptional regulation that is released by necrotic cells and secreted by activated macrophages during inflammation and functions extracellularly as a potent proinflammatory cytokine (18–20).
In this study, we show that IL-33, which we found to be identical to the NF-HEV protein we characterized (21), is a nuclear factor associated with heterochromatin in vivo and mitotic chromosomes in living cells, that possesses potent transcriptional-repressor properties. Our findings suggest that IL-33, similarly to IL-1α, may function as both a proinflammatory cytokine and an intracellular nuclear factor involved in transcriptional regulation.
Results
Endothelial Cells (ECs) Constitute a Major Source of IL-33 mRNA in Chronically Inflamed Tissues from Patients with Rheumatoid Arthritis (RA) and Crohn's Disease.
Using in situ hybridization (ISH), we have shown abundant and preferential expression of NF-HEV/IL-33 mRNA in HEV ECs from human tonsils, lymph nodes, and Peyer's patches (21). In contrast, RT-PCR analysis of human cDNA libraries has revealed expression of IL-33 mRNA in activated dermal fibroblasts, keratinocytes, and activated bronchial smooth muscle cells, but expression in ECs was not reported (3). To define the major cell types responsible for IL-33 mRNA expression in chronically inflamed human tissues, we further characterized IL-33 mRNA expression by ISH. Prominent and specific dot-like signals were detected with the antisense IL-33 ISH probe in blood vessels from human tonsils (Fig. 1A), Crohn's disease intestine (Fig. 1B), and RA synovium (Fig. 1C), whereas the sense probe gave only background signals. Combined ISH for IL-33 and immunohistochemistry (IHC) for postcapillary venule EC marker DARC revealed expression of IL-33 mRNA in both DARC-positive and -negative ECs (Fig. 1C). Semiquantitative RT-PCR confirmed abundant expression of IL-33 mRNA in ECs freshly isolated from human tonsils, RA synovium, and Crohn's intestine (Fig. 1D). In contrast, IL-33 mRNA was not detected in HeLa epithelial cancer cells. Together, these findings indicate that ECs constitute a major source of IL-33 mRNA in chronically inflamed tissues.
Fig. 1.
Expression of IL-33 mRNA in ECs from chronically inflamed human tissues. (A–C) ISH analysis of IL-33 mRNA in human tonsils (A), Crohn's disease intestine (B), and RA synovium (C). Cryosections were hybridized with IL-33 sense and antisense riboprobes. ISH signals were detected with FITC-conjugated anti-biotin antibody (A and C) or incubation with Fast-red (B). In C, ISH (green signal) was combined with IHC for the postcapillary venule marker DARC (red signal), and nuclei were counterstained with DAPI (blue signal). (C Inset) Abundant IL-33 signal (green dots) in a DARC-positive postcapillary venule (red signal). (D) Detection of IL-33 mRNA in ECs freshly purified from human tonsils, Crohn's disease intestine, and RA synovium by using semiquantitative RT-PCR. HeLa epithelial cell line and GAPDH primers were used as controls.
IL-33 Is a Heterochromatin-Associated Nuclear Factor in Vivo.
We have already described NF-HEV/IL-33 as a nuclear factor preferentially expressed in HEV endothelial cells from human tonsils (21). Because IL-33 has been rediscovered as a cytokine ligand for a cell-surface receptor (3), we decided to confirm its nuclear localization using three distinct antisera against the Nter part (Nter, IL-33, amino acids 1–15) and IL-1-like Cter domain (Cter1, Cter2). Immunostaining of human tonsil sections with the three independent antisera demonstrated abundant expression of endogenous IL-33 in the nuclei of HEV ECs double stained with DAPI and HEV-specific antibody MECA-79 (Fig. 2A, D, and G). Nuclear staining of HEV ECs with the anti-IL-33 antibodies was specific because it was abrogated by preincubating the antibodies with the corresponding IL-33 peptides (Fig. 2 B, E, and H) but not control peptides (Fig. 2 A, D, and G). Interestingly, higher magnification revealed that endogenous IL-33 is enriched in multiple nuclear domains (Fig. 2 C, F, and I). The IL-33-containing domains overlapped with high local concentrations of DNA as shown by in situ DAPI (Fig. 2 C, F, and I) or Hoechst (data not shown) staining. Because DAPI and Hoechst are known to bind preferentially heterochromatic AT-rich DNA, this finding indicated association of endogenous IL-33 with heterochromatin. We observed similar heterochromatin staining in the nucleus of HEV ECs with the three independent antisera, indicating that both Nter and Cter parts of IL-33 accumulate in the nucleus of HEV ECs and associate with heterochromatin in vivo.
Fig. 2.
IL-33 is a heterochromatin-associated nuclear factor in HEV ECs in vivo. (A, D, and G) Double staining of human tonsils sections with HEV-specific mAb MECA79 and three distinct IL-33 antisera against the Nter and IL-1-like Cter domains, preabsorbed with a control peptide, demonstrated abundant expression of endogenous IL-33 in the nuclei of HEV ECs in vivo. Nuclei were counterstained with DAPI. (B, E, and H) Nuclear staining of HEV ECs with the three IL-33 antisera was abrogated by preincubation of the antibodies with their corresponding IL-33 peptides. (C, F, and I) Higher magnification revealed that endogenous IL-33 accumulates in nuclear domains that colocalize with dense regions of DAPI staining, indicating association with heterochromatin.
IL-33 Associates with Heterochromatin and Mitotic Chromosomes in Living Cells.
To characterize association of IL-33 with heterochromatin under living conditions, we analyzed the subcellular localization of IL-33 tagged with GFP at the C terminus, in various cell types. Nuclear accumulation and heterochromatin association of ectopically expressed IL-33 was observed in all living cells analyzed, including human HEK-293T (Fig. 3A) and HeLa (data not shown) epithelial cancer cells, and mouse 3T3 fibroblasts (Fig. 3B), whereas GFP alone localized throughout the cell without accumulation in the nucleus (Fig. 3C). In all cell types, IL-33 colocalized with dense regions of Hoechst staining, including the perinucleolar heterochromatin at the nucleolar periphery in human cells and the pericentromeric heterochromatin in mouse cells (Fig. 3 A and B). Nuclear and heterochromatin staining was similarly observed when IL-33 was tagged with GFP at the N terminus (Fig. 3D). We then asked whether IL-33 was still associated with chromatin during mitosis. As expected, GFP alone was distributed throughout the whole cell and mainly excluded from the condensed mitotic chromosomes (Fig. 3E). In contrast, we observed a clear association of IL-33-GFP with mitotic chromatin, as revealed by the bright green fluorescence on the condensed chromosomal arms counterstained with Hoechst (Fig. 3F). Unlike HMGB1 (22), association of IL-33 with mitotic chromosomes was retained after permeabilization and fixation of the cells and immunostaining with IL-33 antibodies (Fig. 3G). Finally, we confirmed association of IL-33 with chromatin, using a biochemical approach (Fig. 3H). Chromatin fractions were prepared from control HEK-293T cells or cells transfected with IL-33 expression vector by digesting nuclei with DNase and extracting proteins with 0.4 M or 0.8 M NaCl. Western blot analysis with the three distinct IL-33 antibodies revealed the presence of full-length IL-33 (≈30 kDa) in both 0.4 M and 0.8 M NaCl chromatin extracts and residual pellet (corresponding to proteins not extracted at 0.8 M NaCl), similarly to the internal control histone H3.
Fig. 3.
IL-33 associates with heterochromatin and mitotic chromatin in living cells. (A–C) Association of IL-33-GFP protein with heterochromatin was observed under living conditions both in human HEK-293T epithelial cells (A) and mouse 3T3 fibroblasts (B). Colocalization of IL-33-GFP with dense regions of Hoechst staining was found at the perinucleolar heterochromatin in human cells and the pericentromeric heterochromatin in mouse cells (A and B, arrowheads), whereas GFP alone localized throughout the cell (C). (D) Heterochromatin association of IL-33 in living cells was also observed with an Nter GFP tag. (E–G) During mitosis, IL-33-GFP was found associated with mitotic chromatin (F), whereas GFP alone was distributed throughout the whole cell (E). Association of IL-33-GFP with mitotic chromosomes was retained in fixed cells (G). Staining of mitotic chromosomes with IL-33 Nter antibody was blocked with the Nter peptide. (H) Western blot analysis of chromatin extracts with the three IL-33 antisera (Nter, Cter1, and Cter2) and control histone H3 antibody. Chromatin fractions were prepared by digestion of nuclei with DNase and extraction with 0.4 M NaCl and 0.8 M NaCl. Residual pellet corresponds to proteins not extracted at 0.8 M NaCl.
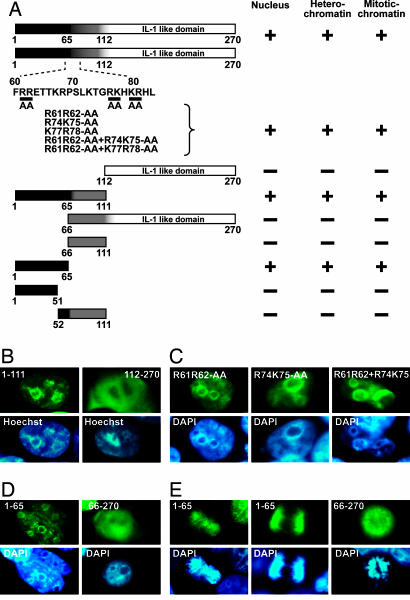
An Evolutionarily Conserved Motif Within the N Terminus Is Necessary and Sufficient for IL-33 Nuclear Localization, Heterochromatin Association, and Targeting to Mitotic Chromosomes.
Sequence alignment of vertebrate IL-33 sequences revealed two evolutionarily conserved regions [see supporting information (SI) Fig. 6]: the predicted homeodomain-like helix–turn–helix (HTH) motif (amino acids 1–65) within the Nter part (21), and the Cter IL-1-like domain (amino acids 112–270) (3). We observed that fusion of GFP to the IL-1-like domain resulted in an even distribution of the fusion protein throughout the cell, whereas fusion to the Nter part of IL-33 (amino acids 1–111) resulted in nuclear localization and heterochromatin association in living cells (Fig. 4A and B). Essentially identical results demonstrating the critical role of the IL-33 Nter part in nuclear targeting and heterochromatin association were obtained in all cell types analyzed, including human HeLa and 293T epithelial cells and mouse 3T3 fibroblasts. To further define, within the IL-33 Nter part, the molecular determinants responsible for IL-33 localization, we first investigated the role of the candidate bipartite NLS (21). Surprisingly, double and quadruple point mutations in the predicted NLS revealed that it is not required for IL-33 nuclear targeting and heterochromatin association (Fig. 4C). In contrast, deletions targeting the homeodomain-like HTH motif (amino acids 1–65) abrogated nuclear accumulation and heterochromatin association of the IL-33-GFP fusion proteins (Fig. 4 A and D), and this motif alone was sufficient for targeting GFP to the nucleus and heterochromatin (Fig. 4D). Interestingly, the homeodomain-like HTH motif was also found to be sufficient for targeting GFP to mitotic chromatin, whereas its deletion abrogated association of IL-33-GFP fusion proteins with mitotic chromosomes (Fig. 4E). We concluded that the evolutionarily conserved homeodomain-like HTH motif within the Nter part is necessary and sufficient for IL-33 targeting to the nucleus, heterochromatin, and mitotic chromosomes.
Fig. 4.
An evolutionarily conserved homeodomain-like HTH motif within the IL-33 Nter part is necessary and sufficient for nuclear localization, heterochromatin association, and targeting to mitotic chromosomes. (A) Diagram of IL-33 and its deletion- or point-mutant derivatives. The capacity of each mutant to accumulate in the nucleus and associate with heterochromatin and mitotic chromatin was determined. (B–E) The different IL-33 mutants fused to GFP were expressed in HEK-293T cells, and their subcellular localization in interphase (B–D) or mitotic (E) cells was analyzed by fluorescence microscopy. DNA was counterstained with Hoechst 33342 in living cells and with DAPI in fixed cells.
IL-33 Has Transcriptional Repressor Properties Associated with the Homeodomain-Like HTH Motif.
Because one of the hallmarks of heterochromatin is that it constitutes a transcriptionally repressive environment in the nucleus, we next wished to determine whether IL-33 may possess transcriptional-repressor properties. For that purpose, full-length IL-33 or deletion constructs were fused to the Gal4-DNA-binding domain (Gal4-DB) and used in gene reporter assays with a GAL4-responsive luciferase reporter. The full-length human IL-33 protein was found to exhibit significant transcriptional repressor activity (Fig. 5A) that was not observed with a reporter lacking GAL4-responsive elements (Fig. 5B). This effect required tethering of IL-33 to DNA through the GAL4-DB, and it was similar to that of the potent heterochromatin-associated transcriptional repressor histone lysine methyltransferase SUV39H1 (Fig. 5 A and C). Analysis of IL-33 deletion mutants revealed a good correlation between heterochromatin targeting and transcriptional-repression activity, because repression activity was observed for the Nter part of IL-33 (amino acids 1–111) and the homeodomain-like HTH motif (amino acids 1–65), whereas the Cter IL-1-like domain (amino acid 112–270) did not exhibit any significant transcriptional regulatory properties when fused to the GAL4-DB (Fig. 5D). Together, these data indicate that IL-33 possesses potent transcriptional-repressor activity associated with the evolutionarily conserved Nter homeodomain-like HTH motif.
Fig. 5.
IL-33 has potent transcriptional repressor properties. (A and B) Human HEK-293T epithelial cells were transiently transfected with increasing amounts (10, 100, or 500 ng) of expression vectors for GAL4-DB fusions of IL-33 (Gal4-IL33) or Suv39H1 (Gal4-Suv), along with reporter vector pLex-Gal4 in which luciferase gene transcription is under the control of five GAL4-DB-binding sites (A) or with the same reporter vector deleted of the GAL4-DB-binding sites (B). IL-33 not fused to GAL4-DB (IL33) was also tested as a control. Luciferase activities were normalized by cotransfection with pRL-CMV plasmid and expressed relative to the values obtained with empty expression vector pCMV-2N3T (control). Results are means and standard deviations of three independent transfection experiments. (C) Fold repression was calculated by dividing the normalized luciferase activity of cells expressing GAL4-DB alone (Gal4) by the activity of the IL-33 (Gal4-IL33) and Suv39H1 (Gal4-Suv) fusion proteins (500-ng expression vector). (D) The IL-33 Nter part and homeodomain-like HTH motif, but not the IL-1-like Cter part, exhibit transcriptional repressor activity. IL-33 full-length (1–270) or deletion mutants (1–65, 1–111, and 112–270) were expressed as GAL4-DB fusions (10-, 100-, 500-, or 750-ng expression vectors) and tested for their ability to repress the pLex-Gal4 luciferase reporter. Normalized luciferase activities were determined as described in A.
Discussion
IL-33 is a recently described member of the IL-1 family that signals through the IL-1 receptor-related protein ST2 (3). IL-33 was found to be a potent inducer of Th2 responses and Th2-associated cytokines IL-4, IL-5, and IL-13, suggesting that IL-33 may play an important role in asthma and other allergic-type diseases (3, 23). In this study, we show that IL-33 localizes to the nucleus, associates with heterochromatin and mitotic chromosomes, and exhibits potent transcriptional-repressor properties. To the best of our knowledge, interleukins have not been shown previously to exhibit such a localization profile in living cells and/or human tissues in vivo. In addition, we identify an evolutionarily conserved domain within the IL-33 Nter part that is necessary and sufficient for IL-33 nuclear localization, association with heterochromatin and mitotic chromosomes, and transcriptional-repression activity. This domain is predicted to exhibit structural homology with the homeodomain and other HTH DNA-binding domains (21), but has no similarity with the Nter part of other IL-1 family cytokines. Together, our data provide strong evidence that IL-33 is a “dual-function” cytokine that may function as both an intracellular nuclear factor and a potent proinflammatory cytokine. A similar duality of function has been shown for IL-1α and chromatin-associated factor HMGB1 (9, 11–16, 18–20).
By acting as a dual-function cytokine, IL-33 may be more similar to IL-1α than to IL-1β. Indeed, IL-33 (270 aa) and IL-1α (271 aa) have a similar size, share the IL-1/FGF fold at their C terminus, translocate to the nucleus through signals located within their Nter parts, and exhibit transcriptional regulatory properties. IL-33 also shares similarity with HMGB1 by its capacity to associate with chromatin in interphase and mitosis. HMGB1 has been shown to be passively released by necrotic cells (19) or secreted by activated macrophages after hyperacetylation of lysine residues (24). It remains to be seen whether similar mechanisms may contribute to IL-33 release during inflammation. The mature form of IL-33 has been proposed to be secreted after maturation by caspase-1 (3). Surprisingly, the predicted cleavage site for caspase-1 was not conserved in the canine, bovine, and porcine IL-33 orthologues (SI Fig. 6), casting some doubts about the proposed maturation of IL-33 by caspase-1 in vivo (3). In agreement with this observation, we found no evidence for IL-33 processing in vivo, i.e., differential localization of N- and Cter parts, for both endogenous IL-33 in HEV ECs (Fig. 2 A, D, and G) and ectopic IL-33 in HEK-293T epithelial cells (Fig. 3 A, D, and G). Membrane-associated IL1α biological activity has been demonstrated in many studies (1, 10, 25), and it remains possible that IL-33 may similarly function as a membrane-associated cytokine. In any case, our results clearly show that IL-33 is a heterochromatin-associated nuclear factor in vivo, and future studies will therefore be required to determine how the IL-1-like cytokine domain is released and/or presented to the ST2 receptor expressed on target cells.
Our data show that ECs constitute a major source of IL-33 mRNA and protein in vivo. ISH analysis in human chronically inflamed tissues revealed abundant expression of IL-33 mRNA in blood vessel ECs from RA synovium and Crohn's disease intestine, suggesting that IL-33 may play an important role at the level of the endothelium during chronic inflammation. IHC with three distinct antisera directed against the Nter part and IL-1-like Cter domain, indicate that IL-33 is abundantly expressed by HEVs in human tonsils. Analyses have shown that IL-33 mRNA constitutes one of the major transcripts preferentially expressed by HEV ECs (21, 26). Together, these expression data indicate that IL-33 is likely to play important roles in HEV ECs. Like HMGB1, IL-33 may act at the chromatin level and function as a nonhistone chromosomal protein involved in the assembly of nucleoprotein complexes on DNA and the maintenance or establishment of chromatin structure (27, 28). Similarly to the heterochromatin-associated lymphocyte-specific transcription factor Ikaros, which plays a key role in the transcriptional regulation of genes required for lymphocyte development (29), IL-33 may regulate gene-expression programs required for development and/or maintenance of HEV ECs in vivo. We found that IL-33 has transcriptional repression properties and localizes to heterochromatin, a nuclear domain linked to gene silencing (30). IL-33 may therefore repress gene expression in HEV ECs. However, as proposed for Ikaros (31), IL-33 could also function as a potentiator of gene expression by squelching transcriptional repressors at heterochromatin, thus decreasing their local concentrations on specific promoters and allowing activators to bind more efficiently. Ikaros has been shown to associate with histone deacetylases and chromatin remodeling complexes (32, 33), and it will be important in future studies to determine whether IL-33 also associates with histone-modifying and chromatin-remodeling complexes.
So far, HEV ECs constitute the only cell type that has been shown to express endogenous IL-33 at both the mRNA and protein level. Unfortunately, these cells can not be grown in culture because they rapidly dedifferentiate outside the lymphoid tissue microenvironment (34). We looked for another cell culture model, but we were not able to detect endogenous IL-33 protein expression in any cultured cell lines, even after stimulation with proinflammatory mediators. These later observations suggest that in vivo studies will be required to further define the biological roles of IL-33 and the molecular mechanisms regulating its production.
Materials and Methods
Plasmid Constructions.
Plasmid pEGFP.N3 (BD Biosciences Clontech, Mountain View, CA) encoding EGFP was modified by deletion of Kozak and ATG sequences of EGFP. IL-33 and IL-33 deletion mutants were amplified by PCR using the human NF-HEV/IL-33 cDNA (21) as a template; all sense primers contained an EcoRI site, a Kozak sequence (CCACC) and an ATG start codon, whereas the stop codon in antisense primers was replaced with a BamHI site. The PCR fragments, thus obtained, were digested with EcoRI and BamHI and cloned in-frame upstream of EGFP in pEGFP.N3 modified vector. Point mutants of IL-33 were generated by two successive rounds of PCR using pEGFP.N3-IL-33 as a template. IL-33 cDNA was also cloned in plasmid pEGFP.C2 (BD Biosciences Clontech) to express IL-33 with an Nter GFP tag and in plasmid pEGFP.N3 with a stop codon before GFP to express IL-33 with no tag. All primer sequences are available upon request.
Mammalian Cell Culture and Fluorescence Microscopy.
Human HeLa and HEK293T epithelial cell lines and mouse 3T3 fibroblasts were grown in DMEM supplemented with 10% FCS and 1% penicillin-streptomycin (all from Invitrogen, Carlsbad, CA). For live cell imaging, cells seeded (150 × 103 cells per well) in Chamber Slide System 2 wells (Lab-Tek; Nunc, Roskilde, Denmark) 24 h before transfection were transfected with 2 μg of plasmid DNA by using a phosphate calcium precipitation method. Two days after transfection, DNA was stained for 10 min at 37°C in culture medium containing 5 μg/ml Hoechst 33342 and washed once, and living cells were observed by fluorescence microscopy on an inverted fluorescence microscope (Eclipse TE300; Nikon Corp, Tokyo, Japan).
In Situ Hybridization.
Tissue samples for ISH and IHC experiments have been described (35). ISH was performed as described (34) by using sense and antisense IL-33 riboprobes (GenBank accession no. NM_033439, IL-33 cDNA, nucleotides 216–828), labeled with the DIG RNA Labeling Kit (Roche, Indianapolis, IN). Hybridized probe was detected with rabbit anti-DIG HRP-conjugated antibody (1/15; DAKO, Carpenteria, CA) by using the biotin-tyramide amplification system (GenPoint kit; DAKO) and Fast red substrate (Sigma, St. Louis, MO). For combined ISH/IHC, ISH signal was revealed with FITC-conjugated goat anti-biotin antibody (1/100; Vector Laboratories, Burlingame, CA) and sections were further incubated with DARC mAb [10 μg/ml, mAb Fy6; kindly provided by Y. Colin (Institut National de la Transfusion Sanguine, Paris, France)]and Cy3-conjugated goat anti-mouse IgG (1/1,000; GE Healthcare, Nottinghamshire, U.K.). Sections were counterstained with DAPI and viewed on a Nikon Eclipse TE300 fluorescence microscope.
IHC.
IHC experiments with IL-33 antibodies were performed on 5-μm sections from Bouin-fixed, paraffin-embedded human tonsils. Antigen retrieval was done in a microwave oven in citrate buffer, pH6.5 (4 × 5 mn). The sections were blocked in 5% goat serum and incubated overnight at 4°C with IL-33 Nter rabbit polyclonal antibody (1/200; Eurogentec, Seraing, Belgium) adsorbed with IL-33 Nter peptide (amino acid 1–15) or control peptide for 1 h at 37°C. Two other IL-33 antisera were also used, IL-33 Cter1 and IL-33 Cter2 rabbit polyclonal antibodies (1/200, nos. 210–447 and 210–933; Alexis Biochemicals, Lausen, Switzerland), together with their specific IL-33 Cter peptide (no. 522–098; Alexis Biochemicals). The sections were successively incubated with biotinylated goat anti-rabbit (1/200; Jackson ImmunoResearch, West Grove, PA) and Cy3-coupled streptavidin (1/200; Zymed, San Francisco, CA), followed by MECA79 mAb (20 μg/ml; Pharmingen, San Diego, CA) and Cy2 goat anti-rat IgM (1/200; Jackson ImmunoResearch). Sections were finally counterstained with DAPI and mounted with Moviol.
Semiquantitative RT-PCR.
Isolation of microvascular ECs from the different tissues and RNA analysis by semiquantitative RT-PCR was performed as described (34, 36) with the following primer pairs: 5′-CACCCCTCAAATGAATCAGG-3′ and 5′-GGAGCTCCACAGAGTGTTCC-3′ for IL-33, 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′ for the internal control GAPDH.
Preparation of Chromatin Fractions.
For preparation of nuclear chromatin extracts, 3 × 107 control 293T cells or cells transfected with IL-33 (with no tag) expression vector were washed in PBS and resuspended in 4 ml of chromatin fractionation buffer (0.15 M NaCl/10 mM MgCl2/10 mM CaCl2/1 mM PMSF/15 mM Tris, pH 7.5/0.1% Tween 20). Cells were ruptured by using Ultra-Turrax (Labortechnik; Staufen, Germany) in the presence of NP-10 to a final concentration of 0.1%. After centrifugation at 800 × g (10 min at 4°C), nuclei were digested with DNase 1 (0.2 μg/μl) for 10 min at 30°C and pelleted by a brief centrifugation. Chromatin fractions were prepared by adding NaCl, to a final concentration of 400 mM, to the nuclear pellets resuspended in chromatin fractionation buffer. After 30 min at 4°C, the nuclei were centrifuged at 21,000 × g for 10 min, and the supernatant (chromatin fraction 0.4 M) was saved. Chromatin fraction 0.8 M was similarly prepared by adding NaCl to a final concentration of 0.8 M NaCl. The final pellet was saved as residual pellet. Anti-histone H3 antibody [kindly provided by D. Trouche (Laboratoire de Biologie Moléculaire des Eucaryotes–Centre National de la Recherche Scientifique, Toulouse, France) was used to validate the chromatin fractions.
Reporter Assay.
Gal4-IL-33 expression vectors were generated by inserting the corresponding IL33 fragments, generated by PCR, into pCMVGT vector downstream of the Gal4-DB (amino acids 1–147). Cotransfection of HEK-293T cells with 10, 100, 500, or 750 ng of pCMVGT constructs, 700 ng of firefly luciferase reporter vector (pLex-Gal4), and 50 ng of Renilla luciferase construct (pRL-CMV; Promega, Madison, WI) was performed by using JetPEI (Polyplus-transfection, San Marcos, CA). The amount of CMV promoter was kept constant (750 ng) by using pCMV-2N3T empty vector. pCMVGT, pCMVGT-SUV39H1 (Gal4-Suv), and pLex-Gal4 vectors were provided by D. Trouche (37). After 24 h, firefly and Renilla luciferase activities were assayed with the Dual Luciferase Assay System (Promega). All transfections were normalized to Renilla luciferase activity and repeated at least three times.
Supplementary Material
Acknowledgments
We thank D. Trouche for plasmid vectors and histone H3 antibody; Y. Colin for DARC mAb; P. Brousset (Institut National de la Santé et de la Recherche Médicale U563, Toulouse, France) for paraffin-embedded tissues; F. Viala for iconography; and Prof. F. Amalric for stimulating discussions. This work was supported by Ligue Nationale Contre le Cancer (Equipe Labellisée), Agence Nationale de la Recherches Programme Blanc “Cuboïdale,” Région Midi-Pyrénées, and Migration and Inflammation European Network of Excellence Grant FP6-502935. V.C. was supported by the Association pour la Recherche sur le Cancer.
Abbreviations
- Cter
C-terminal
- EC
endothelial cell
- HEV
high endothelial venules
- HTH
helix–turn–helix
- ISH
in situ hybridization
- IHC
immunohistochemistry
- NLS
nuclear localization sequence
- Nter
N-terminal
- RA
rheumatoid arthritis.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606854104/DC1.
References
- 1.Dinarello CA. Blood. 1996;87:2095–2147. [PubMed] [Google Scholar]
- 2.Dinarello CA. Eur Cytokine Netw. 2000;11:483–486. [PubMed] [Google Scholar]
- 3.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, et al. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Coyle AJ, Lloyd C, Tian J, Nguyen T, Erikkson C, Wang L, Ottoson P, Persson P, Delaney T, Lehar S, et al. J Exp Med. 1999;190:895–902. doi: 10.1084/jem.190.7.895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Townsend MJ, Fallon PG, Matthews DJ, Jolin HE, McKenzie AN. J Exp Med. 2000;191:1069–1076. doi: 10.1084/jem.191.6.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Priestle JP, Schar HP, Grutter MG. EMBO J. 1988;7:339–343. doi: 10.1002/j.1460-2075.1988.tb02818.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang JD, Cousens LS, Barr PJ, Sprang SR. Proc Natl Acad Sci USA. 1991;88:3446–3450. doi: 10.1073/pnas.88.8.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunn E, Sims JE, Nickliny MJ, O'Neill LA. Trends Immunol. 2001;22:533–536. doi: 10.1016/s1471-4906(01)02034-8. [DOI] [PubMed] [Google Scholar]
- 9.Werman A, Werman-Venkert R, White R, Lee JK, Werman B, Krelin Y, Voronov E, Dinarello CA, Apte RN. Proc Natl Acad Sci USA. 2004;101:2434–2439. doi: 10.1073/pnas.0308705101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaplanski G, Farnarier C, Kaplanski S, Porat R, Shapiro L, Bongrand P, Dinarello CA. Blood. 1994;84:4242–4248. [PubMed] [Google Scholar]
- 11.Maier JA, Voulalas P, Roeder D, Maciag T. Science. 1990;249:1570–1574. doi: 10.1126/science.2218499. [DOI] [PubMed] [Google Scholar]
- 12.Maier JA, Statuto M, Ragnotti G. Mol Cell Biol. 1994;14:1845–1851. doi: 10.1128/mcb.14.3.1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McMahon GA, Garfinkel S, Prudovsky I, Hu X, Maciag T. J Biol Chem. 1997;272:28202–28205. doi: 10.1074/jbc.272.45.28202. [DOI] [PubMed] [Google Scholar]
- 14.Stevenson FT, Turck J, Locksley RM, Lovett DH. Proc Natl Acad Sci USA. 1997;94:508–513. doi: 10.1073/pnas.94.2.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu B, Wang S, Zhang Y, Feghali CA, Dingman JR, Wright TM. Proc Natl Acad Sci USA. 2003;100:10008–10013. doi: 10.1073/pnas.1737765100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buryskova M, Pospisek M, Grothey A, Simmet T, Burysek L. J Biol Chem. 2004;279:4017–4026. doi: 10.1074/jbc.M306342200. [DOI] [PubMed] [Google Scholar]
- 17.Wessendorf JH, Garfinkel S, Zhan X, Brown S, Maciag T. J Biol Chem. 1993;268:22100–22104. [PubMed] [Google Scholar]
- 18.Wang H, Bloom O, Zhang M, Vishnubhakat JM, Ombrellino M, Che J, Frazier A, Yang H, Ivanova S, Borovikova L, et al. Science. 1999;285:248–251. doi: 10.1126/science.285.5425.248. [DOI] [PubMed] [Google Scholar]
- 19.Scaffidi P, Misteli T, Bianchi ME. Nature. 2002;418:191–195. doi: 10.1038/nature00858. [DOI] [PubMed] [Google Scholar]
- 20.Lotze MT, Tracey KJ. Nat Rev Immunol. 2005;5:331–342. doi: 10.1038/nri1594. [DOI] [PubMed] [Google Scholar]
- 21.Baekkevold ES, Roussigne M, Yamanaka T, Johansen FE, Jahnsen FL, Amalric F, Brandtzaeg P, Erard M, Haraldsen G, Girard JP. Am J Pathol. 2003;163:69–79. doi: 10.1016/S0002-9440(10)63631-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pallier C, Scaffidi P, Chopineau-Proust S, Agresti A, Nordmann P, Bianchi ME, Marechal V. Mol Biol Cell. 2003;14:3414–3426. doi: 10.1091/mbc.E02-09-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinarello CA. Immunity. 2005;23:461–462. doi: 10.1016/j.immuni.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Bonaldi T, Talamo F, Scaffidi P, Ferrera D, Porto A, Bachi A, Rubartelli A, Agresti A, Bianchi ME. EMBO J. 2003;22:5551–5560. doi: 10.1093/emboj/cdg516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kurt-Jones EA, Fiers W, Pober JS. J Immunol. 1987;139:2317–2324. [PubMed] [Google Scholar]
- 26.Palmeri D, Zuo FR, Rosen SD, Hemmerich S. J Leukoc Biol. 2004;75:910–927. doi: 10.1189/jlb.0903408. [DOI] [PubMed] [Google Scholar]
- 27.Thomas JO, Travers AA. Trends Biochem Sci. 2001;26:167–174. doi: 10.1016/s0968-0004(01)01801-1. [DOI] [PubMed] [Google Scholar]
- 28.Agresti A, Bianchi ME. Curr Opin Genet Dev. 2003;13:170–178. doi: 10.1016/s0959-437x(03)00023-6. [DOI] [PubMed] [Google Scholar]
- 29.Georgopoulos K. Nat Rev Immunol. 2002;2:162–174. doi: 10.1038/nri747. [DOI] [PubMed] [Google Scholar]
- 30.Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- 31.Koipally J, Heller EJ, Seavitt JR, Georgopoulos K. J Biol Chem. 2002;277:13007–13015. doi: 10.1074/jbc.M111371200. [DOI] [PubMed] [Google Scholar]
- 32.Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, et al. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- 33.Koipally J, Renold A, Kim J, Georgopoulos K. EMBO J. 1999;18:3090–3100. doi: 10.1093/emboj/18.11.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lacorre DA, Baekkevold ES, Garrido I, Brandtzaeg P, Haraldsen G, Amalric F, Girard JP. Blood. 2004;103:4164–4172. doi: 10.1182/blood-2003-10-3537. [DOI] [PubMed] [Google Scholar]
- 35.Middleton J, Americh L, Gayon R, Julien D, Mansat M, Mansat P, Anract P, Cantagrel A, Cattan P, Reimund JM, et al. J Pathol. 2005;206:260–268. doi: 10.1002/path.1788. [DOI] [PubMed] [Google Scholar]
- 36.Patterson AM, Gardner L, Shaw J, David G, Loreau E, Aguilar L, Ashton BA, Middleton J. Arthritis Rheum. 2005;52:2331–2342. doi: 10.1002/art.21222. [DOI] [PubMed] [Google Scholar]
- 37.Vandel L, Nicolas E, Vaute O, Ferreira R, Ait-Si-Ali S, Trouche D. Mol Cell Biol. 2001;21:6484–6494. doi: 10.1128/MCB.21.19.6484-6494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.