Abstract
The posttranslational modification of histone and other chromatin proteins has a well recognized but poorly defined role in the physiology of gene expression. With implications for interfering with these epigenetic mechanisms, we now report the existence of a relatively abundant secondary modification of chromatin proteins, the N6-formylation of lysine that appears to be uniquely associated with histone and other nuclear proteins. Using both radiolabeling and sensitive bioanalytical methods, we demonstrate that the formyl moiety of 3′-formylphosphate residues arising from 5′-oxidation of deoxyribose in DNA, caused by the enediyne neocarzinostatin, for example, acylate the N6-amino groups of lysine side chains. A liquid chromatography (LC)–tandem mass spectrometry (MS) method was developed to quantify the resulting N6-formyl-lysine residues, which were observed to be present in unperturbed cells and all sources of histone proteins to the extent of 0.04–0.1% of all lysines in acid-soluble chromatin proteins including histones. Cells treated with neocarzinostatin showed a clear dose–response relationship for the formation of N6-formyl-lysine, with this nucleosome linker-selective DNA-cleaving agent causing selective N6-formylation of the linker histone H1. The N6-formyl-lysine residue appears to represent an endogenous histone secondary modification, one that bears chemical similarity to lysine N6-acetylation recognized as an important determinant of gene expression in mammalian cells. The N6-formyl modification of lysine may interfere with the signaling functions of lysine acetylation and methylation and thus contribute to the pathophysiology of oxidative and nitrosative stress.
Keywords: histone acetylation, oxidative stress, enediyne
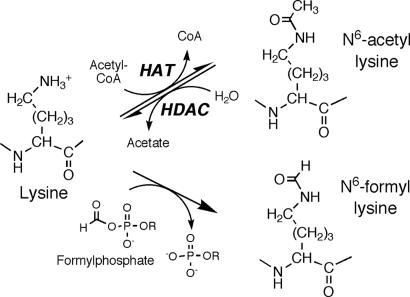
There is a growing appreciation for the complex relationship between posttranslational modification of histone and other chromatin proteins and regulation of most DNA-based processes, including DNA replication, transcription, repair and recombination, and chromosome segregation (1–4). Among the modifications affecting the N and C termini of histone proteins are lysine acetylation, lysine and arginine methylation, serine phosphorylation, ubiquitination, and ADP ribosylation (1–4). Inasmuch as these modifications represent electrophilic adducts of nucleophilic side chains of amino acids, it is reasonable to assume that lysine-rich histone proteins would be subject to chemical reactions with electrophilic oxidation products of lipids and other biomolecules. For example, electrophilic products of lipid peroxidation, such as malondialdehyde and the α,β-unsaturated hydroxyalkenals, have been detected as protein adducts under conditions of oxidative stress (5–7). We now report that histone proteins are subject to a noncanonical secondary modification that arises from oxidative DNA damage: N-formylation of the side-chain amino group of lysine (Fig. 1) by transacylation with the highly reactive 3′-formylphosphate residues arising from 5′-oxidation of deoxyribose in DNA. This endogenous mimic of lysine N6-acetylation has the potential to interfere with the physiology of normal histone modifications.
Fig. 1.
N6-formyl-lysine residues arising from products of oxidative DNA damage are chemical analogs of the biologically important lysine N6-acetylation.
Among the proteins capable of reacting with endogenously generated electrophiles, histone proteins contain an abundance of nucleophilic amino acids that make them likely targets for pathological secondary modification. The primary level of organization of eukaryotic nuclear DNA involves packaging of DNA as nucleosomes, each consisting of a core containing ≈145 bp of DNA wrapped around an octamer of histone proteins H2A, H2B, H3, and H4, with adjacent cores linked by 30–50 bp of DNA and a fifth or linker histone, H1 (8, 9). In general terms, each histone protein is comprised of a globular central domain with lysine- and arginine-rich C and N termini that make extensive contact with the nucleosomal DNA (8, 9) and are the major sites for posttranslational modification by methylation, acetylation, and phosphorylation of amino acid side chains. These modifications are now widely recognized as a major epigenetic mechanism for controlling gene expression (10), and there is an emerging appreciation for the relationship between aberrant modifications and carcinogenesis (1, 11). Although the secondary modification of histones by acetylation of lysine N6-amino groups (Fig. 1) has been known for 40 years (12, 13), it has only recently been identified as a critical determinant of gene expression (14) and a general modulator of p53 and other transcription factors (15, 16). Unlike the N-terminal α-acetylation of proteins that occurs with translation, lysine N6-acetylation is reversible and occurs at highly conserved sites in the N-terminal tails of histone proteins (14). Acetylation is thought to affect gene expression by disrupting electrostatic histone–DNA interactions and by serving as a recognition element for bromodomain modules of various chromatin remodeling proteins and transcription factors (17). By whatever mechanism, lysine acetylation is controlled by two classes of enzymes (Fig. 1): histone acetyltransferases and histone deacetylases (14, 18). Human cells contain several members of each class of protein, with both cytosolic and nuclear forms (14).
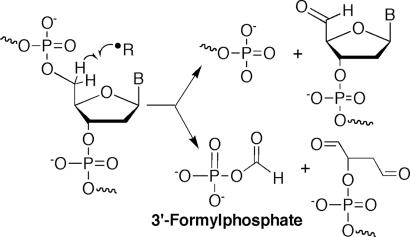
As shown in Fig. 2, oxidation of deoxyribose produces a variety of electrophilic products that have the potential to react with nucleophilic sites in cellular DNA, RNA, and proteins. For example, the base propenal product of 4′-oxidation of deoxyribose in DNA has been shown to be a major cellular source of the mutagenic endogenous DNA adduct, M1G (19). The present work addresses protein adducts formed by the 3′-formylphosphate residue that arises from oxidation of the 5′-position of deoxyribose along with a 5′-(2-phosphoryl-1,4-dioxobutane) species (Fig. 2). The labile phosphate ester of the formylphosphate promotes transfer of the formyl group to primary and secondary amines (20). That the 5′-position of deoxyribose in DNA is the most solvent accessible site in the B-helix suggests that 3′-formylphosphate residues and other 5′-oxidation products will be among the most common features of DNA strand breaks in cells (21).
Fig. 2.
Reactive 3′-formylphosphate residues arise from 5′-oxidation of deoxyribose in DNA.
Given the proximity of lysine-rich histone proteins to DNA in nuclei, one might expect that oxidative DNA damage would result in the reaction of deoxyribose oxidation products with histone proteins to cause noncanonical protein secondary modifications. Here we present evidence for just such a mechanism with N6-formylation of lysines in histone proteins by reaction with the 3′-formylphosphate residues of deoxyribose oxidation in DNA. Further, we observed significant lysine N6-formylation only in histone proteins, which raises the possibility of an endogenous secondary modification with the potential for interference with N6-acetylation of lysine.
Results
Tritium Transfer Studies.
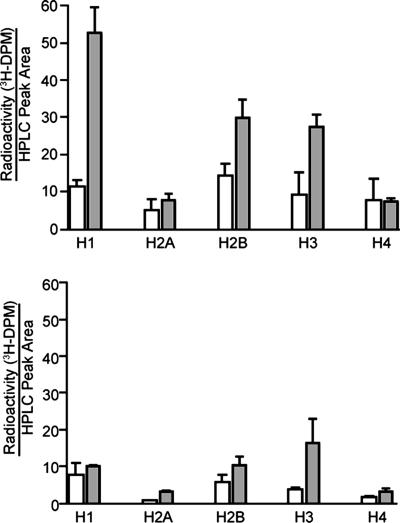
The first approach to establishing the transfer of deoxyribose oxidation products to histones entailed radiolabeling of deoxyribose in cellular DNA followed by treatment of cells with a DNA-oxidizing agent and quantification of the radiolabel in histone proteins by scintillation counting. To this end, DNA in human TK6 cells was labeled with [5′-3H2]- or [methyl-3H]thymidine by addition of the deoxynucleoside to the culture medium, which resulted in incorporation of one radiolabeled thymidine for every 10–30 unlabeled thymidines (data not shown). Isolated nuclei were then treated with neocarzinostatin, an DNA-cleaving enediyne antibiotic that selectively abstracts the 1′-, 4′-, and 5′-hydrogen atoms from deoxyribose in DNA (22). The 3′-formylphosphate species represent ≈10% of the 5′-oxidation products produced by neocarzinostatin (22). After extraction, nuclease digestion and HPLC separation of the chromatin proteins [see supporting information (SI) Fig. 5], tritium in fractions containing the major histone proteins was quantified by scintillation counting. Neocarzinostatin treatment caused an overall 270% increase in radioactivity associated with all acid soluble protein species (data not shown). The radioactivity associated with individual histone classes is depicted in Fig. 3, from which it is apparent that histone H1 sustained the highest level of 3H labeling (41% of total radioactivity; assumes identical spectroscopic properties for all histone classes), with lesser amounts associated with the core histones H2B and H3 (15% and 30%, respectively). When DNA was labeled with [methyl-3H]thymidine, neocarzinostatin treatment caused significantly less labeling of acid soluble chromatin proteins (73% less) than occurred with [5′-3H]thymidine, and there was no apparent association with any particular histone class (Fig. 3).
Fig. 3.
HPLC analysis of neocarzinostatin-induced 3H-labeling of histones from TK6 cells with DNA containing 5′-[3H]thymidine (Upper) or methyl-[3H]thymidine (Lower). Isolated nuclei were treated with neocarzinostatin (30 μM; shaded bars) or methanol vehicle (open bars), followed by histone extraction, nuclease digestion, and HPLC separation, as described in Materials and Methods. Tritium associated with the various histone-containing HPLC fractions was quantified by scintillation counting and reported as disintegrations per minute (DPM) per unit area of UV absorbance signal. Data represent mean ± SD for three independent experiments.
Development of an LC–Tandem MS (MS/MS) Method to Quantify N6-Formyl-Lysine.
Although informative, the 3H transfer studies could not establish the chemical identity of the deoxyribose species reacting with the histone proteins. To this end, we undertook the development of a sensitive and specific analytical method to quantify the formation of N6-formyl-lysine residues in proteins. The method entails addition of isotopically labeled internal standards (4,4,5,5-[2H], N6-formyl-lysine and -lysine) to solutions of acid-soluble proteins extracted from cells, followed by protease digestion, phenylisothiocyanate (PITC) derivatization of the individual amino acids and resolution of the derivatives by HPLC (SI Fig. 6 Upper), which is similar to the approach taken by Cai and Guengerich (23). Fractions containing the lysine and N6-formyl-lysine PITC derivatives were collected for subsequent LC-MS/MS analysis with atmospheric pressure ionization-electrospray ionization and detection in the positive ion mode with selected reaction monitoring. The HPLC system used in the LC-MS/MS studies provided a 16 min separation of the PITC derivatives of N6-formyl-lysine and arginine, which ensured a lack of contamination of the two in prepurified samples (SI Fig. 6 Lower). The PITC derivatives of arginine, lysine, and N6-formyl-lysine yielded molecular ions (M + 1; unit resolution mode) at m/z 310, 417, and 310, respectively, which were selected in the first quadrupole. Loss of the PITC fragment during decomposition reactions yielded ions at m/z of 175, 282, and 175, respectively. The limit of detection for all three PITC derivatives was 50 fmol, which amounts to a sensitivity of ≈0.2 N6-formyl-lysine per 104 lysines in 50 μg of protein. Calibration curves were all linear (r2 > 0.99; see SI Fig. 7).
Several control studies were performed for various facets of the LC-MS/MS method. First, we established that the HPLC systems provided baseline resolution of the PITC derivatives of lysine, arginine, and N6-formyl-lysine, as shown for the second HPLC system in SI Fig. 6. Second, the calculated exact masses for the PITC derivatives of arginine and N6-formyl-lysine differ by only 0.01 mass units (309.1260 vs. 309.1147), so it was important to verify the identity of the species comigrating with standards on the first (prepurification) and second (LC-MS/MS) HPLC systems and to ensure that the earlier-eluting arginine species did not contaminate the later-eluting N6-formyl-lysine. Both objectives were achieved by high-resolution MS analysis (electrospray ionization–TOF) of fractions collected from the second HPLC separation of PITC derivatives of either synthetic standards or hydrolyzed histone proteins. In both cases, the fraction assigned to PITC-arginine contained a protonated molecular ion (M+H+) with an m/z of 310.1382, whereas that for the PITC-N6-formyl-lysine had an m/z of 310.1279 (SI Fig. 8). These correspond well to calculated masses of 310.1338 and 310.1270, respectively. There was no detectable contamination of the PITC-N6-formyl-lysine signal with PITC-arginine and vice versa (data not shown).
Control studies were also performed to determine the chemical stability of the N6-formyl-lysine species and the potential for its adventitious formation during sample processing. The former was assessed by incubating synthetic N6-formyl-lysine with 0.2 M H2SO4 under the conditions used for histone purification (see Materials and Methods). After PITC derivatization, the acid-treated and control (incubation at pH 7) samples were analyzed by LC-MS/MS, with the results revealing no effect of H2SO4 on the stability of N6-formyl-lysine (data not shown).
The concern for adventitious formation of N6-formyl-lysine arises from the potential for DNA oxidation and thus generation of 3′-formylphosphate residues during nucleus isolation and histone purification. The extent of this artifact was assessed in two ways. First, calf thymus histone proteins were mixed with an equal mass of calf thymus DNA and then subjected to the acid extraction conditions used with isolated nuclei; control samples contained no DNA. Subsequent LC-MS/MS quantification of the PITC derivative of N6-formyl-lysine revealed no differences in the background quantities of N6-formyl-lysine residues in the two samples (7.7 ± 0.9 N6-formyl-lysine per 104 lysine with DNA versus 7.7 ± 0.9 N6-formyl-lysine per 104 lysine without DNA). A second set of control studies was performed by comparing the N6-formyl-lysine levels in histone proteins isolated by different methods, as discussed shortly.
Quantification of N6-Formyl-Lysine in Histone and Other Proteins.
Having established the rigor of the N6-formyl-lysine analytical method, we next undertook an analysis of N6-formyl-lysine residues in histone and other proteins. The quantity of formyl-lysine residues in all cases was normalized to the measured quantity of lysine in each sample (expressed as formyl-lysines per 104 lysines) to correct for differences in the quantities of proteins present in each sample and to allow comparisons between different proteins. The results of analyses performed on different sources of histone proteins and on different types of proteins are presented in Table 1.
Table 1.
Quantification of N6-formyl-lysine in histone and other proteins
| Identity and source of protein | N6-formyl-lysines per 104 lysines |
|---|---|
| Acid soluble protein from TK6 nuclei | 9.1 ± 2.6 |
| Calf thymus total histone* | 7.7 ± 0.9 |
| Chicken blood total histone† | 11 ± 0.5 |
| Acid soluble protein from TK6 cells | 4.2 ± 1.5 |
| Total protein from TK6 nuclei | 12 ± 3 |
| Proteinase K (4% lys)‡ | <0.2 |
| Bovine serum albumin (10% lys) | <0.2 |
| Human collagen (5% lys) | <0.2 |
| Human IgG (6% lys) | <0.2 |
*Acid-soluble protein containing all histone classes.
†Acid-soluble/TCA-precipitated proteins from erythrocytes.
‡Lysine content as mol percent of total amino acids.
One set of studies presented in Table 1 entails a comparison of different sources of histone proteins and different histone purification methods. The standard for comparison here is total acid-soluble protein isolated from TK6 cell nuclei (mainly histone proteins; ref. 8), which contained nine N6-formyl-lysine residues per 104 lysines. This is similar to 8 and 11 N6-formyl-lysine residues per 104 lysines for commercial calf thymus and chicken blood histones, respectively; these commercial histone preparations were also purified by acid extraction and are similar in histone content to our TK6 nuclear acid extracts. To assess the effects of the nucleus isolation procedure and the acid extraction step, N6-formyl-lysine residues were quantified in acid-soluble protein from intact TK6 cells (four residues per 104 lysines) and in total protein from isolated nuclei (12 residues per 104 lysines), respectively. The lower value for intact TK6 cells likely arises as a result of dilution of the nuclear proteins with other cellular acid soluble proteins.
A second set of studies compared the levels of N6-formyl-lysine in different protein bovine serum albumin species. As shown in Table 1, human IgG, collagen, and bovine serum albumin all had levels of N6-formyl-lysine below the limit of detection of the assay (0.2 N6-formyl-lysine per 104 lysines). Further, we observed that the proteinase K used to hydrolyze the various proteins contributed negligibly to the level of N6-formyl-lysine (<0.2 residues per 104 lysines; Table 1).
Deoxyribose Oxidation-Induced Lysine N-Formylation in Cells.
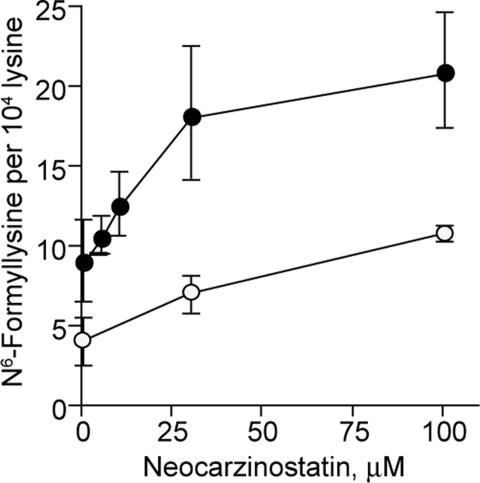
The role of deoxyribose oxidation in the level of N6-formyl-lysine in cells was investigated by treating human TK6 cells and nuclei with neocarzinostatin, a deoxyribose-specific DNA oxidizing antibiotic that causes formation of the 3′-formylphosphate residue hypothesized to lead to lysine N-formylation in cellular histone proteins (24). As shown in Fig. 4, there were clear dose–response relationships between neocarzinostatin concentration and N6-formyl-lysine content in histones from both whole cells and isolated nuclei treated with the antibiotic.
Fig. 4.
N6-formyl-lysine formation in TK6 nuclei (closed circles) and cells (open circles) treated with neocarzinostatin.
Discussion
We have presented evidence consistent with a previously undescribed protein secondary modification, the N6-formylation of lysine, that appears to arise from products of DNA oxidation in cells. A variety of control experiments and alternative approaches to establishing the presence of the N6-formyl-lysine species attest to the authenticity of the lesion as an endogenous protein secondary modification present in histone and other nuclear proteins at relatively high levels. Although the discovery of this protein modification is important in terms of understanding the pathophysiology of oxidative and nitrosative stresses in cells, that the N6-formylation of lysine appears to be limited to histone and possibly other nuclear proteins and is analogous to lysine N6-acetylation suggests a possible role for this lesion in affecting gene expression.
The radiolabel transfer studies established that a deoxyribose-derived species was responsible for the tritium labeling of the histone proteins. Although one of the two 5′-tritium atoms on the thymidine used to label the DNA was removed by neocarzinostatin during the oxidation reaction, the other tritium atom remained associated with the formyl residue of the resulting 3′-formylphosphate and was involved in the histone labeling. This conclusion is supported by the reduction in radiolabeling of histones when the tritium label resides on the thymidine base rather than the deoxyribose moiety (Fig. 3). The labeling of histone proteins observed with the tritiated thymine base is consistent with some form of covalent DNA–protein crosslinking in addition to the N-formylation, such as the formation of Schiff bases between the amino groups of histone lysines and the ketone and aldehyde groups in many of the deoxyribose oxidation products (e.g., nucleoside-5′-aldehyde and 2-phosphoryl-1,4-dioxobutane residues shown in Fig. 2; ref. 24). Further support for the conclusion that a deoxyribose-derived species was responsible for the tritium labeling of the histone proteins comes from the observed preferential radiolabeling of histone H1 in the neocarzinostatin-treated samples. The intercalative binding mode of neocarzinostatin limits its DNA cleaving activity to the linker region between adjacent nucleosome cores (25), the region associated with binding of histone H1. Thus, neocarzinostatin-induced 3′-formylphosphate residues form only in the vicinity of histone H1 and bias radiolabel transfer to this linker histone. This observation serves as proof of concept and holds no biological relevance, because histone H1 has not been observed to be subject to lysine acetylation (8).
Although radiolabeling studies established transfer of an electrophile from deoxyribose to histone proteins, rigorous identification of the histone adduct as an N6-formyl-lysine residue was achieved by LC-MS/MS and electrospray ionization–TOF analyses. The concern about the similar masses of arginine and N6-formyl-lysine was obviated by the chromatographic resolution of the PITC derivatives of these species and by control experiments indicating a lack of arginine contamination of the N6-formyl-lysine signal. Further concerns about adventitious formation of the N6-formyl-lysine and its stability during histone purification were also quelled by control studies. Thus, the LC-MS/MS method to quantify N6-formyl-lysine was determined to be rigorous and relatively sensitive, and served to identify the presence of this species in cells.
Subsequent application of the analytical method to quantify N6-formyl-lysine in various proteins revealed results consistent with the radiolabeling studies. Neocarzinostatin was again found to cause N6-formyl-lysine formation in histone proteins from treated cells. However, the most intriguing result was the observation of N6-formyl-lysine residues only in acid soluble proteins from cells and nuclei, which points to histone and other chromatin proteins as a major source of N6-formyl-lysine in cells. The absence of N6-formyl-lysine in proteins as diverse as proteinase K, Ig, albumin, and collagen further suggests that N6-formyl-lysine residues arise solely in chromatin proteins, a conclusion consistent with the proposed mechanism of formation by the 3′-formylphosphate arising from 5′-oxidation of deoxyribose in DNA. The paucity of N6-formyl-lysine residues in nonchromatin proteins cannot be an analytical artifact because of the lower proportion of lysine in these proteins because, as noted in Table 1, there is at most a 4-fold difference in lysine content between the histone proteins (average 15% lys) and nonhistone proteins, yet the formyl-lysine content differed by 46-fold. One test of this model would be to perform the neocarzinostatin reactions with nuclei under anaerobic conditions in the presence of the radiation sensitizer, misonidazole. This should lead to a large increase in the proportion of 3′-formylphoshate species among products of 5′-oxidation chemistry (Fig. 2; reviewed in ref. 24) and a consequent increase in the level of lysine N formylation.
The biological relevance of N6-formyl-lysine in histones and other chromatin proteins lies in its chemical similarity to N6-acetylation and the potential for disrupting signaling mediated by N6-acetylation and -methylation in the control of gene expression. Future studies may establish the location of N6-formyl-lysines in the core histone proteins relative to sites of acetylation and methylation, as well as recognition of the N6-formyl groups by histone deacetylases. However, relatively high steady-state level of lysine N6-formylation observed in histones from a variety of sources (0.04–0.1%), coupled with the relatively slow turnover of histone adducts (half-life matching the kinetics of cell turnover; ref. 26), raises the probability that this apparently endogenous protein modification could interfere with normal chromatin protein function.
Materials and Methods
Materials.
All chemicals and reagents were of the highest purity available and were used without further purification unless noted otherwise. [5′-3H2]thymidine or [methy-3H3]thymidine were obtained from Moravek Biochemicals (Brea, CA). 4,4,5,5, -[2H]lysine was obtained from Cambridge Isotope Laboratories (Cambridge, MA). Calf thymus total histone proteins (Type IIs), type VI human collagen, and human IgG bovine serum albumin were obtained from Roche Diagnostic (Indianapolis, IN), and chicken blood total histone proteins (core histones) were obtained from Upstate Biotechnology (Charlottesville, VA). Human lymphoblastoid TK6 cells were provided by William Thilly (Massachusetts Institute of Technology). Neocarzinostatin holoantibiotic was purchased from Kayaku [Tokyo, Japan (no longer available from Kayaku; currently produced by Sigma, St. Louis, MO)]. Calf thymus DNA, arginine, lysine, and N6-formyl-lysine were purchased from Sigma. DNA and protein samples were dissolved at a concentration of 1 mg/ml in Chelex- (Bio-Rad, Hercules, CA) treated 50 mM potassium phosphate buffer, pH 7.4, containing 1 mM diethylenetriaminepentaacetic acid and dialyzed four times against 4 liters of Chelex-treated 50 mM potassium phosphate buffer, pH 7.4. Distilled and deionized water was further purified with a Milli-Q system (Millipore, Bedford, MA) and was used in all experiments.
Cell Culture, 3H-Labeling of DNA, and Nucleus Isolation.
Human lymphoblastoid TK6 cells were cultured at 37°C and 5% CO2 in RPMI medium 1640 with 10% FCS/2 mM l-glutamine/100 units/ml penicillin/100 μg/ml streptomycin. Radiolabeling of DNA in cells and nuclei was accomplished by overnight growth of TK6 cells (2–3 × 105 ml) in medium containing 10 μCi/ml (1 Ci = 37 GBq) of [5′-3H2]thymidine or [methyl-3H3]thymidine. All subsequent procedures were performed on ice or at 4°C. After harvesting, cells were washed twice in PBS (140 mM NaCl/2.7 mM KCl/10 mM Na2HPO4/1.8 mM KH2PO4, pH 7.4) buffer and then once in buffer A (20 mM Hepes/120 mM NaCl/4 mM MgCl2/10 mM KCl/10 mM 2-mercaptoethanol/2 mM EDTA, pH 7.4) containing an EDTA-free protease inhibitor mixture (Roche), with centrifugation at 200 × g for 5 min at 4°C. Nuclei were prepared by Nonidet P-40 lysis and sucrose cushion sedimentation in the presence of protease inhibitors, as described (25). The nuclei were washed once in buffer A containing 0.3 M sucrose, resuspended to a final concentration of ≈7.5 × 106 nuclei/ml in buffer A containing 0.3 M sucrose, and used immediately. To characterize radiolabeling of DNA, nuclear DNA was purified by proteinase K digestion and phenol-chloroform extraction, as described (25), and aliquots were subjected to scintillation counting.
Purification of Histone Proteins.
Histone proteins were isolated from TK6 cell nuclei by sulfuric acid extraction as described in detail (all steps were carried out on ice or at 4°C; ref. 27). Briefly, a nuclear pellet was resuspended in 0.4 M H2SO4 for 4 h, and acid-soluble proteins in a postcentrifugation supernatant (10,000 × g, 20 min) were recovered by overnight precipitation with 20% trichloroacetic acid and centrifugation (16,000 × g, 30 min). The pellets containing all histone proteins were washed once in ice-cold acetone containing 1% HCl and then washed once with ice-cold acetone. The pellet was dried under vacuum and stored at −80°C. Protein concentration was determined by the Bradford method (28) using commercial total histone protein as the standard. The integrity of the histone proteins in the acid soluble extract was evaluated by 8–16% gradient gel SDS/PAGE with Coomassie staining (27).
Individual histone proteins were purified by reversed-phase HPLC by using an Agilent (Palo Alto, CA) HP Series 1100 HPLC system. The dried acid-extracted total histone proteins were dissolved in 25% acetonitrile containing 0.1% trifluoroacetric acid and injected onto a Vydac (Hesperia, CA) 201TP54 C18 reversed-phase column (4.6 × 250 mm, 5-μm particles, 300-Å pores) eluted with 0.1% trifluoracetic acid and a linear 23–60% acetonitrile gradient at a flow rate of 1 ml/min for 80 min. With protein elution monitored by UV absorbance at 206 nm, protein-containing fractions were collected, dried under vacuum and stored at −80°C.
Treatment of Cells and Nuclei with Neocarzinostain.
DNA damage reactions were performed by adding aliquots of methanolic solutions of neocarzinastatin to final concentrations of 5–100 μM to suspensions of nuclei or cells (both at ≈7.5 × 106 ml) in buffer C (0.3 M sucrose in 20 mM Hepes/120 mM NaCl/4 mM MgCl2/10 mM KCl/2 mM EDTA and EDTA-free protease inhibitor mixture, pH 7.4, containing 10 mM glutathione to activate the neocarzinostatin; ref. 25). The reaction was allowed to proceed in the dark at 37°C for 30 min, followed by acid extraction of total histone proteins, as described above.
Synthesis of 4,4,5,5-[2H]N6-Formyl-Lysine Internal Standard.
4,4,5,5 -[2H]N6-formyl-lysine was prepared by formylation of 4,4,5,5 -[2H]lysine with acetic anhydride in 98% formic acid, as described (29). The resulting crystals had mass spectrometric and NMR properties identical to published values (data not shown; ref. 29).
PITC Derivitization of Amino Acids.
Derivatization of amino acids with PITC was accomplished as described in detail (23). Briefly, acid extracted histone proteins (with [2H]-labeled internal standards added at this point) were dissolved in 200 μl of 250 mM potassium 3-(N-morpholino) propanesulfonic acid buffer (K-Mops buffer, pH 7.5 or adjusted to 7.5 with NaOH), digested once with proteinase K (7 mg/ml; 37°C; 14 h) followed by the addition of more proteinase K (7 mg/ml) and incubation for 7 h. In some instances, individual amino acids (arginine, lysine, or N6-formyl-lysine and deuterium-labeled internal standards) were also dissolved in K-Mops buffer without proteinase K. In either case, the mixtures were dried under vacuum overnight and redissolved in 200 μl of coupling buffer (acetonitrile/pyridine/triethylamine/H2O with vol/vol ratio of 20/10/4/6). PITC was then added to a final concentration of 2.4 M and the samples incubated at 37°C for 30 min, followed by drying under vacuum and storage at −20°C.
Quantification and Characterization of PITC-Derivatized Amino Acids.
Specific amino acid derivatives in the PITC-treated mixtures were quantified by using an Agilent 1100 Series LC-MSD mass spectrometer. The dried residues were dissolved in 200 μl of 100 mM ammonium acetate (pH 7.4)/acetonitrile (3/7 vol/vol) of which 5 μl was injected onto a Vydac 201TP54 C18 reversed-phase column (4.6 × 250 mm, 5-μm particles, 300-Å pores). Elution was carried out by using a gradient starting with 90% solution A (10 mM ammonium acetate containing 0.1% acetic acid, pH 4.4) and 10% solution B (10 mM ammonium acetate/acetonitrile in a vol/vol ratio of 3/7, pH 7.4) to 100% solution B at a flow rate of 0.4 ml/min over 70 min.
The HPLC-resolved PITC derivatives of arginine, lysine, and N6-formyl-lysine were quantified by isotope dilution MS by using single or triple quadrupole mass spectrometers with atmospheric pressure ionization–electrospray (API-ES) ionization and detection in the positive ion mode with selected ion monitoring. Single quadrupole MS (Agilent 1100) parameters were as follows: drying gas (N2) flow, 12 liters/min at 300°C; nebulizing gas pressure, 35 psi; capillary potential, 3,000 V; fragmentor potential, 75 V; electron multiplier potential, 2,500 V; quadruple temperature, 99°C. LC-MS/MS analyses entailed the use of a PE SCIEX API 3000 tandem quadrupole mass spectrometer coupled to an Agilent 1100 HPLC system. HPLC prepurified N6-formyl-lysine samples were injected onto an Agilent ZORBA 300SB C8 Column (1 × 150 mm, 3.5-μm particles, 300-Å pores) eluted by using 8% acetonitrile containing 0.1% acetic acid at a flow rate of 0.06 ml/min in 30 min. The mass spectrometer was operated in unit resolution mode with the following parameters: nebulizer gas, 8 liters/min; curtain gas, 8 liters/min; collision gas, 4 liters/min; ion spray, 4,250 V; drying gas temperature, 450°C; declustering potential, 200 V; focusing potential, 10 V; entrance potential, 10 V; collision energy, 20 V; collision cell exit potential, 15 V; deflector, −250 V; channel electron multiplier, 2,500 V. In some cases, PITC-treated amino acid samples were resolved by using the same HPLC system, and specific fractions were analyzed on an Agilent G1969A LC-MSD TOF high-resolution MS system.
Calibration curves for the LC-MS and LC-MS/MS analyses of N6-formyl-lysine and lysine were obtained by adding a fixed amount of deuterium-labeled internal standards ([2H4]formyl-lysine, 57.2 pmol/μl; [2H4]lysine: 43.9 pmol/μl) to variable quantities of unlabeled standards (N6-formyl-lysine: 0–1,831 pmol/μl and lysine: 0–1,406 pmol/μl) in K-Mops buffer, pH 7.5. These mixtures were subjected to PITC derivatization and then analyzed by LC-MS or LC-MS/MS to determine the ratio of peak areas for unlabeled and labeled standards. Examples of daily calibration curves are shown in SI Fig. 7. The quantity of N6-formyl-lysine was calculated relative to total lysines present in the acid soluble protein fraction by dividing values for N6-formyl-lysine by the sum of N6-formyl-lysine and lysine. This approach was also applied to the nonhistone proteins.
Supplementary Material
Acknowledgments
We thank Dr. John Wishnok and Ms. Elaine Plummer for expert assistance with chromatography and MS. This work was supported by National Cancer Institute Grant CA103146 and National Institute of General Medical Sciences Grant GM59790. LC-MS/MS and electrospray ionization–TOF analyses were performed in the Bioanalytical Facilities Core of the Massachusetts Institute of Technology Center for Environmental Health Sciences, which is supported by National Institute on Environmental Health Sciences Center Grant ES002109.
Abbreviations
- LC
liquid chromatography
- MS/MS
tandem MS
- PITC
phenylisothiocyanate.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0606775103/DC1.
References
- 1.Hake SB, Xiao A, Allis CD. Br J Cancer. 2004;90:761–769. doi: 10.1038/sj.bjc.6601575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Strahl BD, Briggs SD, Brame CJ, Caldwell JA, Koh SS, Ma H, Cook RG, Shabanowitz J, Hunt DF, Stallcup MR, Allis CD. Curr Biol. 2001;11:996–1000. doi: 10.1016/s0960-9822(01)00294-9. [DOI] [PubMed] [Google Scholar]
- 3.Martin C, Zhang Y. Nat Rev Mol Cell Biol. 2005;6:838–849. doi: 10.1038/nrm1761. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Fischle W, Cheung W, Jacobs S, Khorasanizadeh S, Allis CD. Novartis Found Symp. 2004;259:3–17. [PubMed] [Google Scholar]
- 5.Carbone DL, Doorn JA, Kiebler Z, Petersen DR. Chem Res Toxicol. 2005;18:1324–1331. doi: 10.1021/tx050078z. [DOI] [PubMed] [Google Scholar]
- 6.Montine TJ, Neely MD, Quinn JF, Beal MF, Markesbery WR, Roberts LJ, Morrow JD. Free Radic Biol Med. 2002;33:620–626. doi: 10.1016/s0891-5849(02)00807-9. [DOI] [PubMed] [Google Scholar]
- 7.Sayre LM, Zelasko DA, Harris PL, Perry G, Salomon RG, Smith MA. J Neurochem. 1997;68:2092–2097. doi: 10.1046/j.1471-4159.1997.68052092.x. [DOI] [PubMed] [Google Scholar]
- 8.van Holde KE. Chromatin. New York: Springer; 1989. [Google Scholar]
- 9.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 10.Jaskelioff M, Peterson CL. Nat Cell Biol. 2003;5:395–399. doi: 10.1038/ncb0503-395. [DOI] [PubMed] [Google Scholar]
- 11.Kelly WK, Marks PA. Nat Clin Pract Oncol. 2005;2:150–157. doi: 10.1038/ncponc0106. [DOI] [PubMed] [Google Scholar]
- 12.Allfrey VG, Pogo BG, Littau VC, Gershey EL, Mirsky AE. Science. 1968;159:314–316. doi: 10.1126/science.159.3812.314. [DOI] [PubMed] [Google Scholar]
- 13.Gershey EL, Haslett GW, Vidali G, Allfrey VG. J Biol Chem. 1969;244:4871–4877. [PubMed] [Google Scholar]
- 14.Kuo MH, Allis CD. BioEssays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 15.Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 16.Bell SD, Botting CH, Wardleworth BN, Jackson SP, White MF. Science. 2002;296:148–151. doi: 10.1126/science.1070506. [DOI] [PubMed] [Google Scholar]
- 17.Zeng L, Zhou MM. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 18.Grozinger CM, Schreiber SL. Chem Biol. 2002;9:3–16. doi: 10.1016/s1074-5521(02)00092-3. [DOI] [PubMed] [Google Scholar]
- 19.Zhou X, Taghizadeh K, Dedon PC. J Biol Chem. 2005;280:25377–25382. doi: 10.1074/jbc.M503079200. [DOI] [PubMed] [Google Scholar]
- 20.Chin D-H, Kappen LS, Goldberg IH. Proc Natl Acad Sci USA. 1987;84:7070–7074. doi: 10.1073/pnas.84.20.7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Balasubramanian B, Pogozelski WK, Tullius TD. Proc Natl Acad Sci USA. 1998;95:9738–9743. doi: 10.1073/pnas.95.17.9738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dedon PC, Jiang Z-W, Goldberg IH. Biochemistry. 1992;31:1917–1927. doi: 10.1021/bi00122a004. [DOI] [PubMed] [Google Scholar]
- 23.Cai H, Guengerich FP. Chem Res Toxicol. 2000;13:327–335. doi: 10.1021/tx000003p. [DOI] [PubMed] [Google Scholar]
- 24.Dedon PC, Goldberg IH. Chem Res Toxicol. 1992;5:311–332. doi: 10.1021/tx00027a001. [DOI] [PubMed] [Google Scholar]
- 25.Yu L, Goldberg IH, Dedon PC. J Biol Chem. 1994;269:4144–4151. [PubMed] [Google Scholar]
- 26.Ozbal CC, Velic I, SooHoo CK, Skipper PL, Tannenbaum SR. Cancer Res. 1994;54:5599–5601. [PubMed] [Google Scholar]
- 27.Dedon PC, Soults JA, Allis CD, Gorovsky MA. Mol Cell Biol. 1991;11:1729–1733. doi: 10.1128/mcb.11.3.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bradford MM. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 29.Hofmann K, Stutz E, Spühler G, Yajima H, Schwartz ET. J Am Chem Soc. 1960;82:3727–3732. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.