Abstract
We present results from extensive molecular dynamics simulations of collapse transitions of hydrophobic polymers in explicit water focused on understanding effects of lengthscale of the hydrophobic surface and of attractive interactions on folding. Hydrophobic polymers display parabolic, protein-like, temperature-dependent free energy of unfolding. Folded states of small attractive polymers are marginally stable at 300 K and can be unfolded by heating or cooling. Increasing the lengthscale or decreasing the polymer–water attractions stabilizes folded states significantly, the former dominated by the hydration contribution. That hydration contribution can be described by the surface tension model, ΔG = γ(T)ΔA, where the surface tension, γ, is lengthscale-dependent and decreases monotonically with temperature. The resulting variation of the hydration entropy with polymer lengthscale is consistent with theoretical predictions of Huang and Chandler [Huang DM, Chandler D (2000) Proc Natl Acad Sci USA 97:8324–8327] that explain the blurring of entropy convergence observed in protein folding thermodynamics. Analysis of water structure shows that the polymer–water hydrophobic interface is soft and weakly dewetted, and is characterized by enhanced interfacial density fluctuations. Formation of this interface, which induces polymer folding, is strongly opposed by enthalpy and favored by entropy, similar to the vapor–liquid interface.
Keywords: dewetting, folding, hydration entropy, hydrophobic hydration, hydrophobic interaction
Hydrophobic interactions are one of the major contributors to biological self-assembly in solution, including protein folding and aggregation, micelle and membrane formation, and biomolecular recognition (1–5). Recent work in this area has focused on the lengthscale dependencies of hydrophobic hydration and interactions (4, 6–9). In particular, a recent theory by Lum, Chandler, and Weeks (6) highlighted the different physical mechanisms of solvation of small and large hydrophobic solutes in water. Small solutes are accommodated in water through molecular-scale density fluctuations (10, 11), whereas solvation of larger solutes requires formation of an interface similar to that between a liquid and a vapor (4, 6, 12). This change in physics is also reflected in thermodynamic (entropy vs. enthalpy dominated hydration) (9) and structural (wetting vs. dewetting of the solute surface) (4, 12, 13) aspects of hydration. Similarly, interactions between larger hydrophobic solutes in water (14–18) are characteristically distinct from those between their molecular counterparts (19, 20).
The differences between the hydration and interactions of small and large solutes characterize many-body effects in hydrophobic phenomena. Effects of similar origin are also at work in association of small hydrophobic solutes into a larger aggregate (21, 22) and are quantified by the n-particle potential of mean force (PMF) (23–26). For n > 3, however, the dimensionality of the system makes calculations of n-particle PMFs computationally prohibitive.
To this end, ten Wolde and Chandler (27) performed studies of a hydrophobic polymer in water, a many-body model system of potential relevance to protein folding. Their calculations, using a coarse-grained model of water, showed that a sufficiently long hydrophobic polymer readily collapses in water driven by a significant free energy difference (≈30 kBT for their model polymer). Their simulations also predicted that the essentially wet extended states of the polymer go through dewetted transition states at a smaller radius of gyration before collapsing into an ensemble of compact globular states. Although molecular simulation studies of conformational equilibria of solvophobic oligomers in explicit water and in other solvents have been performed in the past (28–33), a systematic study of how attractive interactions and lengthscales affect the thermodynamics of collapse or folding-unfolding transitions has not been reported.
Motivated by the work of ten Wolde and Chandler (27), we present results from extensive molecular dynamics simulations that address the dependence of folding of hydrophobic polymers in water on lengthscales and attractive interactions. We show that compact folded states of smaller polymers with attractive polymer–water interactions are only marginally stable and display thermodynamic characteristics of both warm and cold denaturation. Increasing the polymer lengthscale makes the hydration contribution to folding more favorable, and stabilizes folded states. Reducing the polymer–water attractions also stabilizes folded states. We explore the suitability of area- and volume-based models to describe the hydration contribution to folding. Entropy obtained from temperature dependence of the hydration contribution to folding varies with the polymer lengthscale, explaining the origin of blurring of the “entropy convergence” in protein folding using arguments proposed by Huang and Chandler (7). Structural signatures, such as water density and its fluctuations in the vicinity of the polymer, indicate that the polymer–water hydrophobic interface is soft and weakly dewetted and allows enhanced density fluctuations, collectively inducing a collapse into compact folded conformations.
Results and Discussion
To observe collapse transitions potentially important to understanding protein folding thermodynamics, the model polymer needs to be sufficiently long, such that it can adopt compact globular as well as extended states. Preliminary studies indicate that 6-, 8-, or 12-mer chains are too short to form compact states. In contrast, somewhat longer, 25-mer hydrophobic oligomers can fold into tightly wound helical states. We therefore focus on the collapse transitions of such 25-mer hydrophobic chains in water.
To study the lengthscale dependence, we studied two versions of the 25-mer, a smaller lengthscale polymer (denoted C25) comprising methane-sized monomers (m) (with σmm = 0.373 nm and m m bond length of 0.153 nm, and m
m bond length of 0.153 nm, and m m
m m bond angle of 111.0°) and a larger lengthscale polymer (denoted CG25) comprising larger (ethane-sized) monomers (M) (with σmm = 0.44 nm, and M
m bond angle of 111.0°) and a larger lengthscale polymer (denoted CG25) comprising larger (ethane-sized) monomers (M) (with σmm = 0.44 nm, and M M bond length of 0.25 nm). The M
M bond length of 0.25 nm). The M M
M M bond angle potential was not used in simulations of the larger lengthscale CG25 polymer, and dihedral potentials were not used for either polymers. We note that we are not interested in detailed studies of specific alkanes in water but in conformational transitions of flexible hydrophobic chains driven by hydrophobic interactions and their dependence on lengthscales, attractions, and temperature. The inclusion of additional intrapolymer interactions (e.g., a dihedral potential) will change the conformational preferences but not affect the qualitative hydration contributions or their temperature dependence that are of primary interest here. Furthermore, the larger lengthscale polymer, CG25, can be thought of approximately as a coarse-grained equivalent of a 50-mer of methane-sized monomers. The M
M bond angle potential was not used in simulations of the larger lengthscale CG25 polymer, and dihedral potentials were not used for either polymers. We note that we are not interested in detailed studies of specific alkanes in water but in conformational transitions of flexible hydrophobic chains driven by hydrophobic interactions and their dependence on lengthscales, attractions, and temperature. The inclusion of additional intrapolymer interactions (e.g., a dihedral potential) will change the conformational preferences but not affect the qualitative hydration contributions or their temperature dependence that are of primary interest here. Furthermore, the larger lengthscale polymer, CG25, can be thought of approximately as a coarse-grained equivalent of a 50-mer of methane-sized monomers. The M M
M M angle potential was therefore turned off to capture the higher conformational flexibility of that polymer.
M angle potential was therefore turned off to capture the higher conformational flexibility of that polymer.
To quantify effects of polymer–water interactions on collapse transitions, we used three different interactions for C25 and CG25 polymers. (i) Full Lennard–Jones (LJ), where monomers interact with other nonbonded monomers and water via LJ interactions [σmw = (σmm + σww)/2 and ]. For m–m and M–M interactions we used εmm = 0.5856 kJ/mol, εmm = 0.85 kJ/mol, and εww = 0.6502 kJ/mol, respectively. The strength of attractions used here is similar to that of typical alkane–water attractive interactions (34). (ii) Half LJ, where monomer–monomer attraction was reduced to half its value in i, and monomer–water εmw correspondingly by a factor of . (iii) WCA, where Weeks–Chandler–Andersen (35) repulsive potential was used to describe monomer–monomer and monomer–water interactions. We denote the full LJ and WCA versions of the two polymers by C25-LJ, C25-WCA, CG25-LJ, and CG25-WCA, respectively.
Fig. 1 shows the PMF, W(Rg), for conformational sampling of polymers along the radius of gyration reaction coordinate obtained from umbrella sampling simulations (see Methods). Several interesting features are apparent. For the C25-WCA polymer, tightly wound helical configurations (with Rg ∼ 0.42 nm) are stabilized by ≈20 kJ/mol relative to extended configurations in the unfolded basin. Although the PMF decreases monotonically as the polymer folds, the slope is not uniform: PMF is relatively flat beyond Rg of 0.65 nm and rapidly decreases below that value.
Fig. 1.
Lengthscale- and attraction-dependent hydrophobic collapse. Shown is the PMF, W(Rg), at 298 K, for the smaller C25 and larger CG25 polymers interacting with water via attractive (LJ) and repulsive (WCA) interactions. Conformations with Rg < Rgcut are defined as compact states: Rgcut = 0.6 nm for C25, and 0.73 nm for CG25 polymers, respectively, as shown by arrows. PMF for C25 and CG25 polymers are zeroed at Rg values of 0.83 and 1.4 nm, respectively. To make the comparison clear, the horizontal axis is Rg − Rgmin, with Rgmin = 0.385 nm for C25 and 0.474 nm for CG25 polymers, respectively.
For the CG25-WCA polymer comprising larger monomer units, the driving force for folding is significantly higher. The folded states are stabilized by almost 100 kJ/mol relative to extended states. Indeed, in absence of the umbrella potential, the extended states of this polymer collapse rapidly into compact states over a time scale of ≈100 ps. Once folded, no significant partial or complete unfolding events are observed indicating the strength of hydrophobic driving force for the polymer of this size.
To what extent do attractive interactions change the folding-unfolding of hydrophobic polymers in water? Fig. 1 also shows PMF profiles for C25-LJ and CG25-LJ polymers interacting with full LJ potential having magnitude similar to typical alkane–water attractions. Here, the attractive interactions directly affect two opposing enthalpic terms: the monomer–monomer attractions favor folding of the polymer into compact states in absence of other effects (Fig. 2c), whereas monomer–water attractions favor unfolding (Fig. 2b) to increase the favorable energetic interactions with water.
Fig. 2.
The hydration and other contributions to PMF for polymer folding at 298 K. (a) Whyd(Rg) contribution for C25 and CG25 polymers interacting via full LJ (filled circles), half LJ (inverted triangles), and repulsive WCA (open circles) interactions. (b) Polymer–water attractive contribution, 〈Upw(Rg)〉. Lines are guide to the eye. (c) Intrapolymer Wvac(Rg) contribution for LJ and WCA versions of C25 and CG25 polymers.
For the smaller, C25-LJ polymer, balance of these interactions leads to two distinct minima corresponding to folded and unfolded ensembles separated by a barrier. Overall, the polymer–water attractive interactions dominate over other contributions, leading to a slight stabilization of the unfolded states compared with the folded ones. In contrast, for the larger CG25-LJ polymer, despite the significant polymer–water attraction contribution favoring the unfolded states (Fig. 2b), the qualitative nature of the PMF is similar to that of CG25-WCA polymer: folded states are favored significantly over the unfolded states.
The total PMF, W(Rg), governs the overall conformational behavior of polymers in water and can be separated into three contributions (36):
where Wvac is the PMF profile in vacuum, 〈UPW〉 is the ensemble averaged polymer–water interaction energy (for given Rg), and we define Whyd as the hydration contribution. We note that [W(Rg) − Wvac(Rg)] is traditionally referred to as the “solvent contribution.” However, its separation in Eq. 1 into 〈UPW(Rg)〉 and Whyd(Rg) is similar to that used in perturbation theory approaches applied previously to solvation phenomena (10, 37). The rationale for writing Eq. 1 is that each individual term may be separately predicted by using theoretical arguments or simplified models (36), as we illustrate below for the Whyd(Rg) term. For nonpolar solutes in water, the solute–water attractive energy is generally smaller in magnitude than the hydration contribution, whereas for polar or ionic solutes, it will dominate the solvation free energy. In either case, a similar separation may provide a useful predictive framework.
Fig. 2 shows Whyd(Rg), 〈UPW(Rg)〉, and Wvac(Rg) contributions to W(Rg) for LJ and WCA versions of the C25 and CG25 polymers. For both the polymers, the hydration contribution is large, ≈40 kJ/mol and 140 kJ/mol, respectively, and monotonically favors folding of polymers into compact states. Despite the significant attractive interactions for LJ polymers, Whyd makes the dominating contribution to the overall PMF. More importantly, the Whyd(Rg) contribution appears to be similar for all C25 or CG25 polymers (full LJ, half LJ, or WCA), relatively insensitive to the strength of attraction. This supports the separation of the PMF in Eq. 1 and indicates that the Whyd term captures the essential physics of hydrophobic hydration that may be described by using either cavity formation (for small lengthscales) or interface formation process (for larger lengthscales).
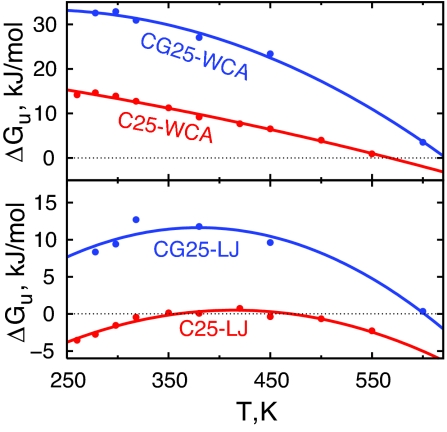
Once the overall W(Rg) profile is available from simulations, the total free energy of unfolding a polymer in water, ΔGu, can be obtained by integration of that PMF (38). Fig. 3 shows the temperature dependence of ΔGu, calculated in this manner. Effects of both lengthscale and attractive interactions on the free energy of unfolding are apparent. For the smaller and attractive C25-LJ polymer, the ΔGu profile is parabolic with compact states being only marginally stable (by ≈1 kJ/mol) for 350 < T < 450 K. Thus, this polymer shows signatures of both warm and cold denaturation processes. Increasing the lengthscale increases the hydrophobic driving force for collapse. Indeed, folded states of CG25-LJ polymer are stabilized by over 10 kJ/mol at the temperature of maximum stability. Reducing the polymer–water attractions or removing them completely also increases the driving force for folding. For example, at 300 K, the ΔGu values for C25-WCA and CG25-WCA polymers are ≈10 and 30 kJ/mol, respectively.
Fig. 3.
Free energy of polymer unfolding. The free energy ΔGu obtained by integrating W(Rg) curve as exp(−ΔGu/kBT) = ∫RgcutRgmax exp(−W(Rg)/kBT)dRg/∫RgminRgcut exp(−W(Rg)/kBT)dRg is shown by circles. The curves are obtained by fitting the data to form ΔGu(T) = ΔHu(Tref) − TΔSu(Tref) + ΔCp−u[(T − Tref) − Tln(T/Tref)] shown by solid lines, where Tref = 298 K was used.
The hydration contribution, ΔGuhyd, to the total free energy of unfolding estimated by using Whyd(Rg) is shown in Fig. 4a. Several features of these curves are noteworthy. The ΔGuhyd is positive at all temperatures, is larger in magnitude for the CG25 polymers, and decreases monotonically with increasing temperature for all polymers. Because the polymer–water attractions are not included in this term, the ΔGuhyd(T) profile is similar and favors folding of both WCA and LJ polymers. Figs. 3 and 4 together show that for the purely repulsive WCA polymers, ΔGuhyd determines the temperature dependence of the overall ΔGu, whereas for the attractive LJ versions of the polymer, the additional polymer–water energy contribution (which favors unfolding) makes the ΔGu(T) profile parabolic.
Fig. 4.
Hydration contribution to the free energy and entropy of polymer unfolding. (a) ΔGuhyd(T) estimated from simulations (symbols) by subtracting appropriately integrated polymer–water and intrapolymer energy and entropy contributions: LJ polymers (filled circles), WCA polymers (open circles), with red and blue indicating C25 and CG25 polymers. Intrapolymer contributions were obtained from vacuum runs of the polymers at the same temperature. Lines in a are predictions of the area model, ΔGuhyd = γ(T)ΔAu, where ΔAu for C25 and CG25 are 0.73 and 1.66 nm2, respectively, obtained by taking differences of ensemble average area of folded and unfolded states, and γ(T) = [∂Whyd/∂A]T. (b) Comparison of ΔGuhyd predicted by using the area–volume model (lines) for C25 and CG25 polymers with simulation data (symbols). c and d show the relative conformational hydration free energy, ΔWhyd, plotted as Whyd(Rg) − Whyd(Rgcut), as a function of solvent-excluded volume (SEV) and SASA, respectively. SEV and SASA are plotted relative to their average values for Rgcut conformers, indicated by ΔV and ΔA. Dashed lines in c and d with respective slopes c(298 K) and γ(298 K) (values indicated) bracket the observed behavior for different lengthscales. (e) Hydration contribution to entropy of unfolding. Also shown is entropy of hydration for WCA-methane obtained by using test particle insertion calculations (37).
The hydration contribution quantifies the difference in hydration free energy of folded and unfolded state ensembles, ΔGuhyd(T) = Guhyd(T) − Gfhyd(T). To predict ΔGuhyd(T), we can (i) explore the applicability of simpler models that predict free energies of hydration of representative folded and unfolded states using lengthscale-dependent physics of hydration or (ii) attempt to model the entire conformation-dependent PMF Whyd(Rg) and obtain ΔGuhyd by appropriate integration. Below we explore path i first.
The folded globular states of C25 and CG25 have equivalent radius [(3V/4π)1/3 or (A/4π)1/2] of ≈0.65 and 0.95 nm, respectively, where V is the solvent-excluded volume (SEV) and A is the solvent-accessible surface area (SASA) of the polymer. These radii values are in the “crossover” region (9, 4), and we expect the hydration free energy of the folded state to be adequately described by the surface tension model, Gfhyd = γA (36). In contrast, the lengthscale of unfolded polymers is more difficult to quantify. If we assume the relevant lengthscale to be small, equal to that of a monomer, then the free energy of hydration of unfolded polymers will be roughly proportional to the solvent excluded volume (4, 7), approximated as n × Gmonhyd(T) (where n is the number of monomers and the subscript “mon” indicates the value for a monomer) or more quantitatively as, V × c(T), where the volume coefficient c(T) = Gmonhyd/Vmon (or alternatively, = ∂Gmonhyd/∂Vmon).
Fig. 4b shows the hydration contribution to the free energy of unfolding predicted using the “area–volume” model, ΔGuhyd(T) = V × c(T) − γ(T)A. We obtained V and A values using the molecular surface package of Connolly (39). Values of c(T) (≈150–170 kJ/mol/nm3) obtained independently from test particle insertions of C25 and CG25 monomers into pure water systems are in excellent agreement with that used by ten Wolde (36) recently. The surface tension, γ(T), for folded states can be approximated by that of an equivalent sphere. These data are difficult to obtain by test particle insertions (9) but can be estimated by using the data in figure 2 of Huang and Chandler (7) (as Δμ/A), by using Tolman corrected macroscopic surface tension, γ(T) = γ∞(T) (1–2δ(T)/R) [where the Tolman length, δ(T), is taken from ref. 40, and γ∞(T) is the surface tension of water (41)], or by using γ(T) = [∂Whyd/∂A]T. Fig. 4b shows the prediction using the area–volume model, where γ(T) = [∂Whyd/∂A]T was used. The prediction not only differs numerically from simulation values of ΔGuhyd but shows opposite temperature dependence. We find that the other two methods of estimating γ(T) give similar results.
Why does the physically motivated area–volume model not provide a good description of ΔGuhyd obtained from simulations? This model attempts to predict the temperature-dependent excess chemical potentials of hydration (or the vacuum-to-water transfer free energies) of folded and unfolded states independently, a truly challenging task. We note that only the difference ΔGuhyd(T) and not these independent values is currently available from simulations. Therefore, we can only make qualitative arguments here regarding the applicability of the area–volume model to the present systems. For example, the unfolded states of hydrophobic polymers comprise monomers joined together by covalent bonds, as distinct from fully separated and well dispersed monomers in solution. In addition, there is conformational diversity in the unfolded states that includes fully as well as partially extended conformations with different local curvatures. A single small-monomer-like lengthscale therefore does not appear to describe the hydration free energy of unfolded states and, especially, its temperature dependence. In contrast, many other processes do involve assembly of small lengthscale moieties into a larger lengthscale aggregate [e.g., in micelle formation (42)] where the area–volume model may provide a good description of hydrophobic contributions to the thermodynamics of assembly.
We also explored the suitability of alternate models (i) ΔGuhyd(T) = c(T)ΔVu and (ii) ΔGuhyd(T) = γ(T)ΔAu, where ΔVu and ΔAu are the volume and areas of unfolding, respectively, determined from our simulations. Here, volume- or area-based description alone is assumed to apply to both folded and unfolded state ensembles. We find that the volume-based calculation (model i) that uses c(T) obtained from thermodynamics of small solute transfer significantly underpredicts the ΔGuhyd at room temperature, as well as its temperature dependence (data not shown). In contrast, the area-based description (model ii) provides a quantitative prediction of ΔGuhyd(T) as shown in Fig. 4a. γ(T) constitutes a critical input to this model and was obtained by using γ(T) = [∂Whyd/∂A]T for both polymers. The numerical value of γ is larger for CG25 polymer compared with that for the smaller lengthscale C25 polymer. Temperature dependence, γ(T), for both polymers is qualitatively similar to that of vapor–liquid interfacial tension measured in experiments or in simulations of the SPC/E water (41).
In contrast to the area–volume model, the models i or ii used above based on volume or area alone do not attempt to predict the excess chemical potentials of folded or unfolded states independently but implicitly assume that the PMF Whyd varies linearly with volume or area over the entire Rg range. How good is the assumption of linearity? Fig. 4 c and d shows Whyd plotted as a function of V and A, respectively. All values are referenced to their respective values at Rgcut. For the CG25-WCA polymer, variation of Whyd is not linear but roughly sigmoidal with respect to V. The slope, c = ∂Whyd/∂V, is more than twice that of cmon obtained from small solute studies (≈171 kJ/mol/nm3) and approaches cmon only at the ends. For the CG25-LJ polymer, the behavior is similar if somewhat less nonlinear. As the effective lengthscale of the polymer is reduced, the slope decreases and for C25-LJ polymer approaches the cmon(298 K) value. Inspection of these curves over a range of temperatures shows a similar behavior. For C25 polymers, even though Whyd is approximately linear with V at all temperatures, the slope c(T) is different from cmon(T), explaining the failure of volume-based ΔGuhyd(T) = cmon(T)ΔVu model i above.
Fig. 4d shows that Whyd varies roughly linearly with area. The slope, γ = ∂Whyd/∂A, is higher for CG25 polymers and decreases (from 50 dynes/cm to 25 dynes/cm) as the effective lengthscale is reduced from CG25-WCA to CG25-LJ to C25-WCA to C25-LJ. The temperature dependence of γ for C25 and CG25 polymers is apparent from Fig. 4a (as ΔAu values are constant). γ decreases monotonically over the temperature range of interest qualitatively similar to the vapor–liquid surface tension of water. In the absence of detailed simulation data, how one may predict a priori the exact numerical value of γ(T) for a polymer of a given lengthscale is not entirely clear. Prediction using the Tolman equation (typically used for spherical solutes) provides a qualitatively correct picture but is not quantitative (data not shown). Application of Lum, Chandler, and Weeks theory (6, 27) to explore lengthscale and temperature dependence of γ for nonspherical solutes may be one computationally efficient alternative. Nevertheless, the approximate linearity with area and the corresponding temperature dependence of the slope explain the success of the ΔGuhyd = γ(T)ΔAu model.
The free energies ΔGu(T) and ΔGuhyd(T) obtained here for hydrophobic polymers have important connections with the thermodynamics of protein folding. The ΔGuhyd(T) is positive, favors folding, and decreases in magnitude with increasing temperature, a trend similar to that of ΔGu(T) for WCA polymers. The polymer–water attractions in contrast favor unfolding and more so at lower temperatures. Inclusion of attractions therefore reduces the stability of folded states and makes the ΔGu(T) curve roughly parabolic in T, similar to that for proteins. To summarize, increasing the lengthscale increases the magnitude of ΔGu(T) and decreases the temperature, T*, of maximum ΔGu, whereas addition of attractive interactions has the opposite effect.
These observations have two important consequences. First, the increased stability of folded polymers with reduction of polymer–water attractive interactions as well as the shift of T* to lower values with increasing lengthscale will make it difficult to observe cold denaturation over temperature range accessible to experiments without assistance from other denaturing influences (43, 44). Second, because the magnitude and temperature dependence of ΔGuhyd (and therefore, of ΔGu) is sensitive to lengthscales, the corresponding temperature-dependent entropy contributions will also vary with polymer (or protein) size. Sufficient variation in protein lengthscales will lead to blurring of the “entropy convergence” observed in thermal unfolding of globular proteins (7, 45–48).
Fig. 4e shows that for a methane-sized solute, the hydration entropy, ΔSuhyd = −(∂ΔGuhyd/∂T), is large and negative at room temperature, characteristic of hydrophobic hydration at small lengthscales (49). For C25 and CG25 polymers, the ΔSuhyd is positive over the entire range of T, with its magnitude being larger for the CG25 polymer. These observations are qualitatively consistent with predictions by Huang and Chandler (7), who argued that such lengthscale dependence of entropy explains the blurring of the “entropy convergence” observed in unfolding of globular proteins. Also, the hydration contribution to the heat capacity of unfolding, ΔCp−uhyd = T(∂ΔSuhyd/∂T), is positive, similar to that for proteins.
Is the folding or collapse of hydrophobic polymers observed here induced by the formation of a vapor–liquid-like interface? In their study, ten Wolde and Chandler (27) found that as a hydrophobic polymer folds to more compact states, it goes through dry or dewetted transition states with low vicinal water densities. In Fig. 5, we quantify the vicinal water density by calculating number, Nw, of water molecules in the first hydration shell divided by the polymer SASA. These local density values are plotted as a function of the polymer SASA and are normalized to 1 in the extended states of polymers.
Fig. 5.
Water density fluctuations and weak dewetting of polymers. (a and b) Local water density in the vicinity of C25 and CG25 polymers, respectively, calculated as average number of hydration waters divided by average SASA, 〈Nw〉/SASA. These values are normalized to 1 in the wet extended states of the polymers. (c and d) Measure of hydration shell compressibility or fluctuations obtained as σ2Nw/〈Nw〉2 SASA, where σ2Nw is the variance of water number fluctuations in the hydration shell of the polymer. A water molecule is considered to be in the hydration shell if it is less than a certain cut off distance (0.6 nm for C25 and 0.7 nm for CG25) from any monomer of the polymer. The top x axis indicates 〈Rg − Rgmin〉 values corresponding to SASA on the bottom x axis.
The normalization factor, equal to the vicinal density in the extended state, is 14.1 for C25-LJ, 13.2 for C25-WCA, 13.3 for CG25-LJ, and 12.8 for CG25-WCA polymers (all in units of molecules per nm2). For isolated C25 and CG25 monomers, the local surface densities obtained from independent simulations are essentially identical for LJ and WCA versions, equal to ≈16 and 15 molecules per nm2, respectively. Thus, by this criterion, the extended states of polymers themselves are already fairly dewetted compared with isolated monomers, highlighting the differences between fully separated monomers and unfolded states of polymers.
Folding of these polymers leads to further dewetting. The extent of that dewetting is rather small, ≈3% for the C25-LJ polymer. When the attractive interactions are removed (e.g., for the C25-WCA case), the local density decreases and goes through a minimum (≈10% reduction) before reaching the folded states, for which the reduction is ≈6% (Fig. 5a). This depletion is consistent with a small increase in the lengthscale upon folding of the polymer. These effects are enhanced for the larger CG25 polymers, which show density depletion even for attractive polymer–water interactions (Fig. 5b).
The fluctuations of water density in the hydration shell are also noteworthy. Fig. 5 c and d shows normalized water number density fluctuations in the first hydration shell, σ2Nw × SASA/〈Nw〉2, which quantify approximately the compressibility of the first hydration shell. (We note that σ2Nw also shows similar features.) It is clear that for the small attractive C25-LJ polymer, the hydration shell compressibility changes little as it folds. In contrast, the larger and repulsive CG25-WCA polymer shows increased absolute fluctuations as well as a peak in the magnitude of fluctuations as the polymer folds to compact states. Interestingly, although the nature of W(Rg) and the dewetting is qualitatively similar for CG25-WCA and CG25-LJ polymers, an explicit peak in fluctuations is not observed during folding of the CG25-LJ polymer. This is expected because the polymer–water attractions will dampen the longitudinal density fluctuations. In addition, solute–water attractions increase the crossover lengthscale or, alternately, reduce the effective lengthscale of the solute. In this sense, CG25-LJ lengthscale is smaller than that of CG25-WCA polymer.
Collectively, for the polymers of the nanometer lengthscale studied here, we find that the unfolded states are dewetted relative to separated monomers, and the folding process is accompanied by further dewetting of the polymer surface. More importantly, the increased density fluctuations for repulsive polymers indicate that the polymer–water interface is soft, qualitatively similar to water-hydrophobic liquid–liquid (50) or liquid–vapor interfaces. In addition, the positive hydration entropy indicates that the unfavorable free energy is dominated by the enthalpic contribution similar to that for interface formation (4, 9), supporting the above assertion. Extension of the above results would predict increased dewetting, higher density fluctuations, and larger driving forces for the folding of longer polymers.
Conclusions
We have presented results from extensive molecular dynamics simulations of folding–unfolding of hydrophobic polymers in water with focus on understanding effects of polymer lengthscale and polymer–water attractive interactions on thermodynamic and structural aspects of polymer folding. The folding of model polymers provides a route to characterizing the many-body hydrophobic effects in the context of realistic biological self-assembly process, such as protein folding. We studied conformational transitions of 25-mers comprising monomers of methane-like and larger particles. To study the effects of polymer–water attractions, we used attractive LJ as well as purely repulsive WCA interactions between monomers and water. We found that the smaller attractive polymer displays a parabolic protein-like unfolding free energy, ΔGu(T), profile as a function of temperature. The folded states of this polymer are only marginally stable at room temperature and can be unfolded by heating or cooling the solution. Increasing the lengthscale or reducing the polymer–water attractions not only increases the stability of folded states but also shifts the temperature of maximum stability to lower values. In addition, the dependence of hydration entropy on polymer lengthscale is consistent with predictions of Huang and Chandler (7) on the lengthscale dependence of hydrophobic hydration that provide a basis for understanding of the blurring of entropy convergence observed in protein unfolding thermodynamics.
The hydration contribution to folding of hydrophobic 25-mers studied here is large and strongly favors compact folded states. We explored the applicability of physically motivated models based on area and/or volume descriptions of hydration of polymer conformations to predict the hydration contribution, ΔGuhyd. Our analysis indicates that predictions using ΔGuhyd = γ(T)ΔAu are in excellent agreement with simulation data. The surface tension γ is larger for CG25 polymers and decreases monotonically with temperature for all polymers. Structural details indicate that the polymer–water interface, especially in the vicinity of the folded state ensemble, is soft, weakly dewetted, and characterized by enhanced density fluctuations. Correspondingly, ΔGuhyd is enthalpy-dominated, consistent with interface-driven folding process.
The polymer models presented here or the ones used by ten Wolde and Chandler, present promising model systems of potential importance in understanding the role of hydration in thermodynamics of protein folding and aggregation (51). The ability to change intrapolymer and polymer–water interactions and polymer size allows for systematic studies of the free energy landscape in the extended parameter space. Furthermore, investigation of the folding–unfolding of these polymers over a range of solution conditions, such as high pressures, in the presence of cosolutes and solvents (31, 32), will provide important insights into protein stability over the broader thermodynamic space.
Methods
Intrapolymer Interactions.
The intrapolymer potentials for bond lengths in C25 and CG25 polymers and bond angles in C25 polymers were harmonic in nature with the following parameters. For m m bond length in C25 polymers, Vb = 0.5kb(r − r0)2, where kb = 334720.0 kJ/mol/nm2 and r0 = 0.153 nm, and for m
m bond length in C25 polymers, Vb = 0.5kb(r − r0)2, where kb = 334720.0 kJ/mol/nm2 and r0 = 0.153 nm, and for m m
m m bond angle, Vθ = 0.5kθ (θ − θ0)2, where kθ = 462.0 kJ/mol/deg2 and θ0 = 111.0 deg. Similarly, for M
m bond angle, Vθ = 0.5kθ (θ − θ0)2, where kθ = 462.0 kJ/mol/deg2 and θ0 = 111.0 deg. Similarly, for M M bond length in CG25 polymers, we used kb = 60,702.0 kJ/mol/nm2 and r0 = 0.25 nm.
M bond length in CG25 polymers, we used kb = 60,702.0 kJ/mol/nm2 and r0 = 0.25 nm.
Free Energy Calculations.
We used the umbrella sampling technique (52) to characterize the conformational free energy of hydrophobic polymers in water as a function of polymer radius of gyration, Rg. A harmonic potential, (1/2)ku(Rg − Rg0)2, was applied to efficiently sample polymer conformations in a window near Rg0. For C25 polymers, we used 13 equally spaced windows spanning a range of Rg between the most compact (≈0.37 nm) and most extended (≈0.92 nm) states, whereas for CG25 polymers, we used 20 equally spaced windows with Rg ranging from ≈0.45 nm to ≈1.50 nm. Simulations of C25 polymers included 1,700 water molecules [SPC/E model (53)]. For CG25 polymers, depending on Rg0 value, the number of water molecules ranged from 4,000 to 12,000. Additional simulations in which 12,000 water molecules were used uniformly in all windows confirmed that the results presented here are insensitive to system size.
The values of ku ranged between 4,000 and 12,000 kJ/mol/nm2, based on the local gradient of the free energy, dW(Rg)/dRg, estimated from preliminary simulations. Data from simulation trajectories [26 ns for the C25 polymer, and 40 ns for the CG25 polymer (2 ns per window)] were combined by using the WHAM formalism (54–56) to generate a PMF profile along the Rg coordinate for each polymer. Similar calculations were performed in vacuum to obtain the PMF in the absence of solvent.
Simulation Details.
MD simulations were performed by using GROMACS (57, 58), with suitable modifications to implement umbrella sampling as well as the WCA interaction scheme. Periodic boundary conditions were applied and the particle mesh Ewald method (59) was used to calculate the electrostatic interactions with a grid spacing of 0.1 nm. NPT simulations were performed at 1 atm and 300 K maintained using Berendsen algorithm (60). The SETTLE algorithm (61) was used to constrain OH and HH distances in water with a geometric tolerance of 0.0001 Å. Simulations were also carried out over a range of temperatures to calculate temperature dependence of the free energy of folding along the saturation curve of water (45).
Acknowledgments
We acknowledge Prof. David Chandler and Prof. Hank Ashbaugh for fruitful discussions. S.G. acknowledges financial support through a National Science Foundation CAREER grant, a Nanoscale Science and Engineering Center grant, and through the National Institutes of Health/Rensselaer Exploratory Center for Cheminformatics Research grant. T.M.T. acknowledges financial support through the David and Lucile Packard Foundation, the Alfred P. Sloan Foundation, and National Science Foundation CAREER Award CTS-0448721.
Abbreviations
- LJ
Lennard–Jones
- PMF
potential of mean force
- SASA
solvent-accessible surface area
- WCA
Weeks–Chandler–Andersen.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Kauzmann W. Adv Protein Chem. 1959;14:1–63. doi: 10.1016/s0065-3233(08)60608-7. [DOI] [PubMed] [Google Scholar]
- 2.Tanford C. The Hydrophobic Effect: Formation of Micelles and Biological Membranes. New York: Wiley; 1973. [Google Scholar]
- 3.Dill KA. Biochemistry. 1990;29:7133–7155. doi: 10.1021/bi00483a001. [DOI] [PubMed] [Google Scholar]
- 4.Chandler D. Nature. 2005;437:640–647. doi: 10.1038/nature04162. [DOI] [PubMed] [Google Scholar]
- 5.Pratt LR. Annu Rev Phys Chem. 2002;53:409–436. doi: 10.1146/annurev.physchem.53.090401.093500. [DOI] [PubMed] [Google Scholar]
- 6.Lum KA, Chandler D, Weeks JD. J Phys Chem B. 1999;103:4570–4577. [Google Scholar]
- 7.Huang DM, Chandler D. Proc Natl Acad Sci USA. 2000;97:8324–8327. doi: 10.1073/pnas.120176397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandler D. Nature. 2002;417:491. doi: 10.1038/417491a. [DOI] [PubMed] [Google Scholar]
- 9.Rajamani S, Truskett TM, Garde S. Proc Natl Acad Sci USA. 2005;102:9475–9480. doi: 10.1073/pnas.0504089102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pratt LR, Chandler D. J Chem Phys. 1977;67:3683–3704. [Google Scholar]
- 11.Hummer G, Garde S, Garcia AE, Pohorille A, Pratt LR. Proc Natl Acad Sci USA. 1996;93:8951–8955. doi: 10.1073/pnas.93.17.8951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stillinger FH. J Solution Chem. 1973;2:141–158. [Google Scholar]
- 13.Hummer G, Garde S. Phys Rev Lett. 1998;80:4193–4196. [Google Scholar]
- 14.Wallqvist A, Berne BJ. J Phys Chem. 1995;99:2885–2892. [Google Scholar]
- 15.Wallqvist A, Berne BJ. J Phys Chem. 1995;99:2893–2899. [Google Scholar]
- 16.Huang X, Margulis CJ, Berne BJ. Proc Natl Acad Sci USA. 2003;100:11953–11958. doi: 10.1073/pnas.1934837100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choudhury N, Pettitt BM. J Am Chem Soc. 2005;127:3556–3567. doi: 10.1021/ja0441817. [DOI] [PubMed] [Google Scholar]
- 18.Li L, Bedrov D, Smith GD. Phys Rev E. 2005;71 doi: 10.1103/PhysRevE.71.011502. 011502. [DOI] [PubMed] [Google Scholar]
- 19.Smith DE, Haymet ADJ. J Chem Phys. 1993;98:6445–6454. [Google Scholar]
- 20.Ghosh T, Garcia AE, Garde S. J Chem Phys. 2002;116:2480–2486. [Google Scholar]
- 21.Tsai J, Gerstein M, Levitt M. Protein Sci. 1997;6:2606–2616. doi: 10.1002/pro.5560061212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raschke TM, Tsai J, Levitt M. Proc Natl Acad Sci USA. 2001;98:5965–5969. doi: 10.1073/pnas.111158498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosh T, Garcia AE, Garde S. J Phys Chem B. 2003;107:612–617. [Google Scholar]
- 24.Shimizu S, Chan HS. Proteins. 2002;48:15–30. doi: 10.1002/prot.10108. [DOI] [PubMed] [Google Scholar]
- 25.Czaplewski C, Liwo A, Ripoll DR, Scheraga HA. J Phys Chem B. 2005;109:8108–8119. doi: 10.1021/jp040691b. [DOI] [PubMed] [Google Scholar]
- 26.Rank JA, Baker D. Protein Sci. 1997;6:347–354. doi: 10.1002/pro.5560060210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.ten Wolde PR, Chandler D. Proc Natl Acad Sci USA. 2002;99:6539–6543. doi: 10.1073/pnas.052153299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wallqvist A, Covell DG. Biophys J. 1996;71:600–608. doi: 10.1016/S0006-3495(96)79260-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ashbaugh HS, Garde S, Hummer G, Kaler EW, Paulaitis ME. Biophys J. 1999;77:645–654. doi: 10.1016/S0006-3495(99)76920-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.LunaBarcenas G, Bennett GE, Sanchez IC, Johnston KP. J Chem Phys. 1996;104:9971–9973. [Google Scholar]
- 31.Ghosh T, Kalra A, Garde S. J Phys Chem B. 2005;109:642–651. doi: 10.1021/jp0475638. [DOI] [PubMed] [Google Scholar]
- 32.Paschek D, Nonn S, Geiger A. Phys Chem Chem Phys. 2005;7:2780–2786. doi: 10.1039/b506207a. [DOI] [PubMed] [Google Scholar]
- 33.Sun L, Siepmann JI, Schure MR. J Phys Chem B. 2006;110:10519–10525. doi: 10.1021/jp0602631. [DOI] [PubMed] [Google Scholar]
- 34.Jorgensen WL, Tirado-Rives J. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 35.Weeks JD, Chandler D, Andersen HC. J Chem Phys. 1971;54:5237–5247. [Google Scholar]
- 36.ten Wolde PR. J Phys Condens Matter. 2002;14:9445–9460. [Google Scholar]
- 37.Garde S, Garcia AE, Pratt LR, Hummer G. Biophys Chem. 1999;78:21–32. doi: 10.1016/s0301-4622(99)00018-6. [DOI] [PubMed] [Google Scholar]
- 38.Hill TL. Statistical Mechanics. New York: McGraw–Hill; 1956. pp. 195–196. [Google Scholar]
- 39.Connolly ML. J Mol Graphics. 1993;11:139–141. doi: 10.1016/0263-7855(93)87010-3. [DOI] [PubMed] [Google Scholar]
- 40.Ashbaugh HS, Pratt LR. Rev Mod Phys. 2006;78:159–178. [Google Scholar]
- 41.Alejandre J, Tildesley D, Chapela GA. J Chem Phys. 1995;102:4574–4583. [Google Scholar]
- 42.Maibaum L, Dinner AR, Chandler D. J Phys Chem B. 2004;108:6778–6781. [Google Scholar]
- 43.Griko YV, Privalov PL. Biochemistry. 1992;31:8810–8815. doi: 10.1021/bi00152a017. [DOI] [PubMed] [Google Scholar]
- 44.Kunugi S, Tanaka N. Biochim Biophys Acta. 2002;1595:329–344. doi: 10.1016/s0167-4838(01)00354-5. [DOI] [PubMed] [Google Scholar]
- 45.Garde S, Hummer G, García AE, Paulaitis ME, Pratt LR. Phys Rev Lett. 1996;77:4966–4968. doi: 10.1103/PhysRevLett.77.4966. [DOI] [PubMed] [Google Scholar]
- 46.Baldwin RL. Proc Natl Acad Sci USA. 1992;89:7110–7113. doi: 10.1073/pnas.89.15.7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Privalov PL. Adv Prot Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- 48.Ashbaugh HS, Truskett TM, Debenedetti PG. J Chem Phys. 2002;116:2907–2921. [Google Scholar]
- 49.Lazaridis T, Paulaitis ME. J Phys Chem. 1992;96:3847–3855. [Google Scholar]
- 50.Patel HA, Nauman EB, Garde S. J Chem Phys. 2003;119:9199–9206. [Google Scholar]
- 51.Liu P, Huang XH, Zhou RH, Berne BJ. Nature. 2005;437:159–162. doi: 10.1038/nature03926. [DOI] [PubMed] [Google Scholar]
- 52.Frenkel D, Smit B. Understanding Molecular Simulation. New York: Academic; 2001. pp. 167–200. [Google Scholar]
- 53.Berendsen HJC, Grigera JR, Straatsma TP. J Phys Chem. 1987;91:6269–6271. [Google Scholar]
- 54.Ferrenberg AM, Swendsen RH. Phys Rev Lett. 1989;63:1195–1198. doi: 10.1103/PhysRevLett.63.1195. [DOI] [PubMed] [Google Scholar]
- 55.Kumar S, Bouzida D, Swendsen RH, Kollman PA, Rosenberg JM. J Comput Chem. 1992;13:1011–1021. [Google Scholar]
- 56.Roux B. Comput Phys Commun. 1995;91:275–282. [Google Scholar]
- 57.Berendsen HJC, van der Spoel D, van Drunen R. Comput Phys Commun. 1995;91:43–56. [Google Scholar]
- 58.Lindahl E, Hess B, van der Spoel D. J Mol Model. 2001;7:306–317. [Google Scholar]
- 59.Darden T, York D, Pedersen L. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 60.Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 61.Miyamoto S, Kollman PA. J Comput Chem. 1992;13:952–962. [Google Scholar]