Abstract
A cell's ability to regulate gene transcription depends in large part on the energy with which transcription factors (TFs) bind their DNA regulatory sites. Obtaining accurate models of this binding energy is therefore an important goal for quantitative biology. In this article, we present a principled likelihood-based approach for inferring physical models of TF–DNA binding energy from the data produced by modern high-throughput binding assays. Central to our analysis is the ability to assess the relative likelihood of different model parameters given experimental observations. We take a unique approach to this problem and show how to compute likelihood without any explicit assumptions about the noise that inevitably corrupts such measurements. Sampling possible choices for model parameters according to this likelihood function, we can then make probabilistic predictions for the identities of binding sites and their physical binding energies. Applying this procedure to previously published data on the Saccharomyces cerevisiae TF Abf1p, we find models of TF binding whose parameters are determined with remarkable precision. Evidence for the accuracy of these models is provided by an astonishing level of phylogenetic conservation in the predicted energies of putative binding sites. Results from in vivo and in vitro experiments also provide highly consistent characterizations of Abf1p, a result that contrasts with a previous analysis of the same data.
Keywords: binding energy, likelihood, transcription factor, mutual information
Transcription factors (TFs) are central to the cell's ability to regulate gene expression (1). Any quantitative understanding of gene regulation will therefore require an accurate characterization of the specificity with which these proteins recognize their DNA target sites. This problem is usually phrased in one of two ways: can we find a statistical pattern (or “motif”) that distinguishes TF binding sites from the genomic background or, alternatively, can we find a faithful representation of the TF's sequence-dependent binding energy (SDBE) to DNA? For a variety of reasons, the development of motif-finding algorithms addressing the statistical question has received much more attention than attempts to directly model TF binding energy. Although the statistical and the energetic pictures are related, they are not equivalent. Indeed, knowledge of binding energy is essential for an understanding of gene regulatory dynamics: a TF binds, not in response to a statistical P value, but to the physical affinity of a site. In this article we describe a method for inferring physical models of binding energy from genome-scale TF binding assays, such as ChIP–chip (2, 3) or protein binding microarrays (PBMs) (4). Instead of seeking a “best” model, our method finds ensembles of models with a high likelihood of being responsible for the data. This probabilistic approach allows direct comparison of results from different experiments and provides additional ways of making biologically relevant predictions.
To set a context for our proposal, we briefly compare and contrast the statistical and energetic approaches to TF specificity. Because high-throughput binding assays generally localize TFs to within only ∼500 bp, one often faces the problem of predicting where, precisely, a TF binds within experimentally bound regions of DNA. The statistics-based algorithms that have been proposed to do this locate binding sites (see ref. 5 for a concise summary) typically proceed by positing a model for “background” DNA and then searching for statistical motifs describing short DNA sequences that appear more often in TF-bound regions than otherwise expected.
In a seminal work, Berg and von Hippel (6) [see also the work of Stormo (7) and Stormo and Fields (8)] proposed a method for estimating the SDBE of a TF from the sequence statistics of known binding sites, and this method is often used to estimate TF–DNA binding energies from the output of motif-finding programs. However, Berg and von Hippel's derivation assumes (i) that all of the binding sites of a given TF experience the same selection pressure on their energy and (ii) that such sites evolve out of random background DNA (see refs. 9 and 10 for a more general discussion of how binding site sequence, energy, and fitness relate to one another). In reality, different sites may need to have different binding energies for functional reasons, and the noncoding DNA of many organisms is clearly not random [Plasmodium falciparum is an instructive example: see supporting information (SI) Table 1]. Although the Berg–von Hippel formula provides a plausible connection between a TF's SDBE and the sequences of its target sites, it is unlikely to be exact or universally valid.
In fact, it should not be necessary to invoke such a connection at all in analyzing PBM and ChIP–chip data, because both assays probe the SDBE of the TF rather directly. This is clear for PBM experiments, where the TF is directly bound in vitro to dsDNA-spotted microarrays. Things are more subtle in ChIP–chip experiments where in vivo effects can modify TF binding (1): in yeast, chromatin can obscure binding sites and other transfactors can alter TF–DNA binding energy. Although such effects are biologically important, they generally vary from site to site, whereas the SDBE of the TF acts in the same way throughout the genome. Thus, for some purposes, it is possible to regard these in vivo effects as a type of “noise” that obscures, but does not negate, the influence of the TF's SDBE.
One should therefore be able to model this energy directly from ChIP–chip and PBM data without calling on any assumptions about binding site evolution or the statistics of background DNA. Foat et al. (11) have recently developed such an approach: they postulate a simple parametrized model of the TF's SDBE and then find the parameters that best fit experimental data. Although it is a big step in the right direction, the Foat et al. analysis leaves some things to be desired: it makes the implicit assumption that measurement noise is Gaussian, an assumption that is not always supported by the data (see SI Fig. 5); more importantly, it returns only the best set of parameters, rather than giving the full range of parameters consistent with given observations. In this article, we address these problems and in the process develop a simple but general method for analyzing TF–DNA binding data.
Our basic method is to use experimental data to specify a probability distribution on the parameters of a model of the TF's SDBE. By model we mean a function, involving parameters denoted collectively by θ, that takes a potential binding site sequence as input and gives an energy as output. The distribution on models is then used to make predictions (e.g., for binding site energies) that have mean values and variances. We think of the data as a set of values {zi}i=1N (such as observed fluorescence intensities) obtained for the N regions of DNA probed by the experiment. Because of experimental noise, particular model parameters θ will produce the observed data with a probability p({zi}|θ) (also referred to as the “likelihood”), which can be computed if the statistics of the noise are known. Given a prior distribution p(θ) on allowable model parameters, we can use Bayes' theorem to turn this into a posterior distribution on model parameters, given the observed data:
Unfortunately, traditional methods of computing p({zi}|θ) require a quantitative model of the experimental noise (or “error model”), something that is not usually available; moreover, using the wrong error model will generally lead to incorrect inferences. To deal with this problem, we take the further Bayesian step of averaging p({zi}|θ) over the space of all possible error models to obtain an “error-model-averaged” (EMA) likelihood that can be explicitly evaluated and used in Eq. 1. Our major result is that this relaxed version of likelihood still allows real data to constrain energy models: in effect the data are used to determine both the energy model and the error model.
To make predictions by using p(θ|{zi}) we use a Markov chain Monte Carlo (MCMC) algorithm to generate a large ensemble of models Θ = {θ1, θ2, …,θT}, sampled according to this distribution. Θ is then used to give concrete probabilistic answers to questions about TF binding behavior conditioned on the experimental data. We demonstrate this approach on ChIP–chip (3) and PBM (4) studies of the yeast TF Abf1p and find that the data determine the parameters of simple binding models with remarkably low statistical uncertainty. This finding contrasts with the commonly held view that high-throughput experiments can give only rough characterizations of TF specificity. Although the microarray data are noisy, the sheer number of regions probed, along with the large amount of DNA sequence in each region, allows for a precise characterization of SDBE. We have found the same to be true for some other broad-acting yeast TFs (data not shown).
Results
We first present the results of a likelihood analysis of the PBM data of Mukherjee et al. (4) for the yeast TF Abf1p. In this assay, epitope-tagged TFs were bound directly to dsDNA spotted on a glass microarray and then visualized with fluorescent antibodies. The fluorescent intensity observed for each of the ∼6,000 microarray spots (representing virtually all of the intergenic regions of Saccharomyces cerevisiae) was then normalized by the amount of DNA in each spot. After averaging the data over replicates and further processing, Mukherjee et al. reported log intensity ratios (LIRs) for N = 5,812 of these intergenic sequences.
Our goal was to find models of Abf1p–DNA binding that were likely to have produced these LIRs. In Methods, we derive an expression (Eq. 3) for the EMA likelihood of different energy models for a given set of binding assay data. Because this equation applies to discretized data, we began our analysis by binning the N intergenic sequences {si}i=1N according to their LIRs into “z-bins.” We chose 20 sequences per bin, for a total of m = 292 bins. The bin size is, of course, arbitrary, but the results of our analysis were found to depend only weakly on this choice (see SI Text and SI Fig. 6). Each sequence si was thus assigned an integer zi identifying the bin into which it was placed. This set of intergenic sequences {si} and their corresponding bin numbers {zi} constituted the sole input to the rest of our analysis.
The parametrized energy model we chose for Abf1p was a 4 × 20 “energy matrix” where each base in a site of length 20 contributes additively to the overall binding energy. We use this energy matrix to classify sites as “bound” (having a substantial TF occupancy in the experiment) if their energies lie below some threshold μ; otherwise they are classified as “unbound.” The energy baseline was fixed by setting the lowest element in each column to zero, and the overall scale was fixed by setting μ = 1. The elements of this matrix are the model parameters θ. See SI Text for more details.
A specific energy matrix classifies each region as bound if it contains at least one bound site, and otherwise classifies it as unbound. The numbers of bound and unbound regions in each z-bin suffice to calculate the posterior probability of the matrix p(θ|{zi}) using Eq. 3 of Methods. We performed multiple MCMC runs on the Abf1p data of Mukherjee et al. (4) to obtain an ensemble of 4 × 104 matrices (which we denote by ΘPBM) sampled according to this distribution. Appropriate tests were used to verify MCMC convergence (see SI Text and Fig. 7).
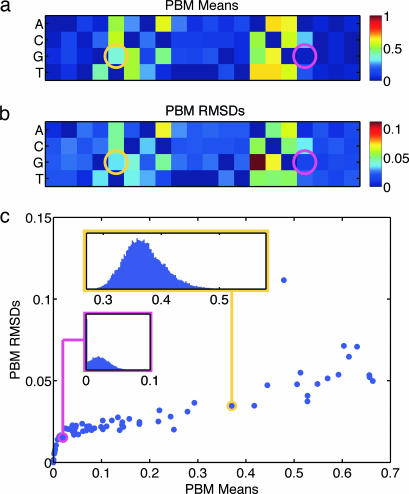
Our results are generally in line with the qualitative motif RTCRYNNNNNACG known for Abf1p (12). However, inspection of ΘPBM shows that the parameters of the Abf1p energy matrix are determined with remarkable precision. The mean and rmsd of each matrix element across all models in ΘPBM are shown in Fig. 1a and b. A scatter plot of the same data is given in Fig. 1c along with the marginal ΘPBM distribution for two representative matrix elements. Matrix elements that differ significantly from zero generally have uncertainties much smaller than their means, and their distributions have a roughly Gaussian shape (Fig. 1c Upper Inset). Other elements are consistently assigned the lowest energy in their respective columns and are thus set by our normalization to be precisely zero in much of the ensemble (e.g. Fig. 1c Lower Inset). The matrix elements are determined with a degree of precision that makes it possible to see meaningful structure even in the center of the binding site, in positions that contribute little to overall TF specificity. That this precision is not an artifact of overfitting was verified by showing that parameter distributions derived from disjoint halves of the data provide consistent predictions for the energy matrix. We also verified that different choices for the matrix width led to consistent parameter distributions (see SI Text and SI Figs. 8 and 9).
Fig. 1.
PBM data determines Abf1p energy model parameters with surprising precision. (a and b) Mean and rmsd of energy matrix elements over the ΘPBM ensemble of Abf1p models. All-blue columns contribute little to TF binding specificity. Overall, these matrices match the known Abf1p motif RTCRYNNNNNACG well. (c) Scatter plot of matrix element means versus rmsds: all rmsds are small compared with the binding cutoff of 1. (Insets) Marginal distributions of two representative matrix elements, circled in corresponding color in a and b.
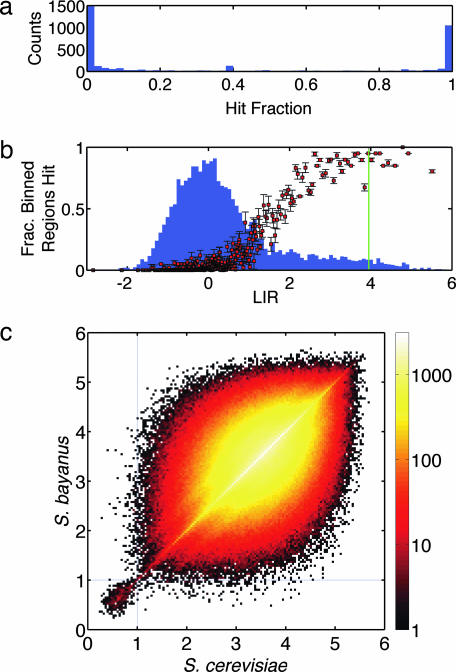
The model ensemble ΘPBM provides a direct way of predicting putative binding sites. For any 20-bp sequence of DNA, we can determine the fraction of models θ in ΘPBM that “hit” that site (i.e., assign it an energy < 1). Fig. 2a histograms this “hit fraction” (HF) for every possible 20-bp site in the intergenic DNA of S. cerevisiae [as defined in Kellis et al. (12)]. The plot shows that the distribution of HFs is strongly bimodal (1,469 sites have HF > 50%, whereas 1,182 of these also have HF > 90%). We adopt HF > 50% as a plausible criterion for predicting a site to be bound.
Fig. 2.
Predictions derived from the ensemble of Abf1p energy models. (a) Histogram showing the ΘPBM HF of all 20-bp sites in the intergenic DNA of S. cerevisiae. The distribution is strongly bimodal, efficiently separating bound sites (HF near 1) from unbound sites (HF near 0). The left-most bin contains the vast majority of sites and has been truncated for readability. (b) Mean fraction of regions in each z-bin declared bound by models in ΘPBM, plotted against the mean LIR of those regions. Error bars show the rmsd variation in this fraction from model to model. The distribution of experimentally determined LIRs is shown in the background for reference. The green line is the threshold used by Mukherjee et al. (4) to identify bound regions. (c) 2D histogram of mean energies assigned by ΘPBM to ungapped orthologous intergenic site pairs in S. cerevisiae and S. bayanus. Sites lying below the E = 1 binding cutoff (dashed lines) in one species are highly likely to fall below this cutoff in the other species.
Fig. 2b plots the mean fraction of sequences in each z-bin declared to be bound by models in ΘPBM against the mean LIR of those sequences. Error bars show the variation in these predictions across different models in ΘPBM. A histogram of the actual measured LIRs is shown in the background. The most striking feature of this plot is the sigmoidal relationship between model predictions and measured LIRs, showing a rapid transition from mostly not bound to mostly bound sequences at the beginning of the heavy tail of the LIR distribution. Whereas its general shape is exactly as expected, this outcome is in no way predetermined: Eq. 3 places no a priori weight on models that make similar predictions for sequences in neighboring z-bins or that declare sequences with large LIR to be bound. The consistency between the shape of this scatter plot and our physical expectation is an independent confirmation of the validity of EMA likelihood.
The Abf1p models in ΘPBM appear to account for the data to within Mukherjee et al.'s (4) estimate of the experimental error. The green line in Fig. 2b indicates the LIR cut used by Mukherjee et al. to define bound regions. Of the 186 regions passing this cut, the experimenters estimated that 7–9% were false positives. We find that 167 (89.8%) of these regions have HF > 90%, 18 (9.7%) have HF < 10%, and only 1 region has an intermediate HF. So, although energy matrix models are simplistic, they appear to account for this Abf1p PBM data about as well as one could hope for any model, regardless of sophistication.
Mukherjee et al. (4) were obliged to adopt a stringent threshold to minimize false positives, in the process rejecting an unknown number of bound sequences. Our model-based approach, by contrast, can tease out regions that are likely to be bound, regardless of where they lie in the raw data distribution. In this case, we find 840 regions with HF > 50% (and therefore bound by our criterion) and with LIR below the cut chosen by Mukherjee et al. (4). In short, we find many more bound regions lying below the experimenters' threshold than lying above it.
This is a strong statement and one for which one would like independent confirmation. The best evidence would come from the direct in vitro measurement of large numbers of putative site binding energies. Energy measurements for Abf1p have been carried out for a small number of sites (13), but the results agree neither with our predictions nor with the analysis of Mukherjee et al. (4) or Lee et al. (3). It is possible that these measurements are in error, and we believe further in vitro studies are necessary to resolve this issue. Recently developed high-throughput techniques for direct measurement of binding affinities give promise of providing data of the quality and scope needed to test these models in quantitative detail (S. Quake, personal communication).
Although direct energy data are lacking, phylogenetic analysis provides alternative evidence in support of our binding energy predictions. Using the intergenic alignments of Kellis et al. (12), we identified all pairs of ungapped orthologous intergenic 20-bp sequences in S. cerevisiae and Saccharomyces bayanus. We then computed the mean predicted binding energy (using ΘPBM) of each site in S. cerevisiae, as well as that of its ortholog. A scatter plot of the resulting energies (Fig. 2c) reveals a large and well separated population of sites whose putative energies lie below 1 (i.e., are predicted to be bound) in both genomes: of the 676 S. cerevisiae sites with energy < 1, a total of 501 (74%) have an S. bayanus ortholog whose energy also lies < 1. By contrast, sites with energy > 1 in either genome tend to have orthologs with highly randomized energy. In short, the large majority of alignable sites predicted to be bound by our models have strongly conserved putative energies. The fact that our models are found directly from in vitro binding data provides a compelling case that they also describe the free energy of Abf1p–DNA binding in vivo, and that this binding energy plays a major role in determining which sites have biological function.
Next, we performed a similar analysis of Lee et al.'s ChIP–chip data (3) to determine whether it gives a description of Abf1p consistent with that obtained from the PBM data. In these ChIP–chip experiments, TFs were cross-linked in vivo to their binding sites, after which TF-bound fragments of DNA were isolated, amplified, labeled, and hybridized in competition with reference DNA to a ssDNA microarray of yeast intergenic regions. The enrichment observed in each microarray spot was characterized by an “X-statistic” based on the single array error model of Hughes et al. (14). Assuming a Gaussian distribution for these X-statistics in the absence of TF binding, Lee et al. reported an enrichment P value for each region.
We assigned these probed sequences to z-bins according to their P values (equivalently, according to their X-statistics). MCMC analysis then gave an ensemble ΘChIP of 4 × 104 matrix models. Results of a single-ensemble analysis of ΘChIP were similar to those of ΘPBM (see SI Text and SI Figs. 10–14). Note that, by integrating over all possible error models, we avoided having to model the noise contributions from each individual step in the ChIP–chip protocol. We also avoided having to estimate in vivo contributions to the experimental noise, such as the fraction of valid binding sites likely to be obscured by chromatin.
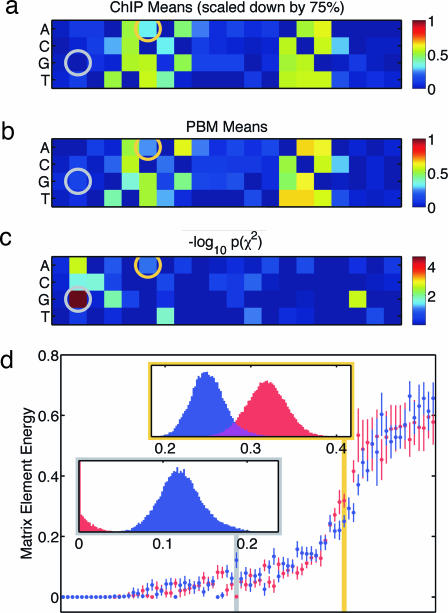
Although the ChIP–chip and PBM results are quite similar, the energy matrix elements derived from ΘChIP are systematically larger than those of ΘPBM. Our procedure, though, produces energy matrices artificially scaled so that the energy cutoff is equal to 1. Thus, when comparing ensembles, we are free to rescale the matrices in one ensemble so as to bring them into accord with those of the other. The resulting difference between the rescaled cutoffs has a natural interpretation: binding site occupancy, which we approximate by a step function at the energy cutoff, should vary with TF concentration and may well differ between experiments; the energy matrix itself, on the other hand, reflects an intrinsic property of the TF molecule that should, in principle, not vary between experiments (if other factors, such as ion concentration and pH, are kept at similar levels). In the case at hand, we found that rescaling the ΘChIP energy cutoff to 0.75, while keeping the ΘPBM cutoff at 1, brought the ΘChIP energy matrix elements into close agreement with those of ΘPBM. Fig. 3a and b illustrates this close agreement between the mean matrix elements in the two ensembles. Fig. 3d provides a direct comparison of the ΘChIP and ΘPBM distributions for each matrix element. In most cases, values that could plausibly have been drawn from either the ΘChIP or ΘPBM distribution can be identified (illustrated in Fig. 3d Upper Inset, which shows the raw ΘChIP and ΘPBM histograms for the orange-circled matrix element in Fig. 3 a and b). Although the two histograms are not identical, they overlap enough that a matrix element value consistent with both distributions can be found.
Fig. 3.
Comparison of ΘPBM and ΘChIP parameter distributions. (a) Mean values of rescaled matrix elements in ΘChIP. (b) Mean matrix elements in ΘPBM (same as Fig. 1a). (c) χ2 P values quantifying the element-by-element consistency of the ΘChIP and ΘPBM distributions. (d) Mean and rmsd uncertainty of each matrix element according to the ΘPBM (blue) and rescaled ΘChIP (red) distributions. Elements are arranged from left to right in order of increasing mean. (Insets) Raw MCMC histograms show the values obtained for the matrix elements circled in a–c and highlighted below each Inset in d. The ΘChIP matrix element distribution (Lower Inset) has most of its weight at precisely 0; the corresponding histogram has been truncated at this bin.
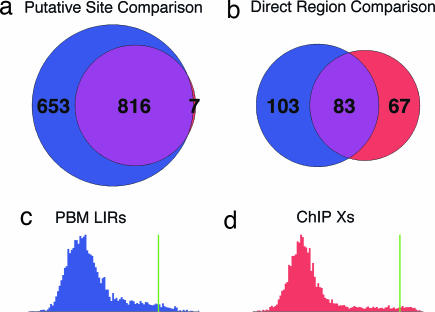
In SI Text we argue that a simple χ2 test provides a valid way of quantifying such consistency. The resulting χ2 P values for each matrix element are shown in Fig. 3c, with lower P values corresponding to poorer consistency between ensemble distributions. There are a few matrix elements for which ΘChIP and ΘPBM give inconsistent distributions by this test (the red and yellow squares in Fig. 3c). The raw histograms for the most inconsistent matrix element (the red square in Fig. 3c), plotted in Fig. 3d Lower Inset, display the poor overlap between the two distributions. Although the inconsistency is significant (even accounting for multiple hypotheses), it occurs outside of the binding site proper and may not have much practical impact. One way to assess the impact of such discrepancies is to compare the binding site predictions of ΘChIP and ΘPBM. If the matrices in both ensembles were identical, the sites selected by the smaller (ChIP–chip) cutoff would be a subset of the sites selected by the larger (PBM) cutoff. Fig. 4a shows that this is in fact nearly the case: of the 823 ΘChIP-predicted sites, all but 7 are predicted by ΘPBM. We stress that these two sets of predictions were made by using data from different experimental platforms and performing separate analyses involving no free parameters.
Fig. 4.
EMA likelihood analysis leads to compatible binding site predictions from different experimental data sets, whereas the more standard method of thresholding the experimental signal leads to substantial disagreement. (a) The 20-bp intergenic sites in S. cerevisiae having ΘChIP HF > 50% (red) are a nearly perfect subset of those with ΘPBM HF > 50% (blue). (b) In contrast, the intergenic regions selected by Mukherjee et al.'s (4) LIR threshold on PBM data (blue) overlap poorly with those selected by Lee et al.'s (3) (P value threshold on ChIP–chip data (red). (c and d) The thresholds chosen by the experimenters are indicated by the green lines on the experimental LIR histogram of PBM data in c and on the X-statistic histogram of ChIP–chip data in d.
Our consistency analysis contrasts with that of Mukherjee et al. (4), who compared their putatively bound regions (identified by a stringent LIR threshold; see Fig. 4c) with those of Lee et al. (3) (identified by a stringent P value threshold; see Fig. 4d) and obtained the Venn diagram reproduced in Fig. 4b. Although the overlap between the two sets of regions is certainly significant, neither is even approximately a subset of the other. This problem cannot be avoided by choosing lower thresholds, as that would result in more false-positive predictions. Even though both data sets are of high quality, this discrepancy is an inevitable result of experimental noise, in vivo effects on binding, etc. However, by “bottlenecking” PBM and ChIP–chip data through a simple parametric model, one obtains almost entirely consistent pictures of Abf1p specificity.
Discussion
A quantitative understanding of biological systems will require the ability to deduce quantitative models from data in a transparent and principled way. Not only must optimal model parameters be found, but the confidence one has in the values of these parameters must be characterized and parameters determined from different data sets must be compared for consistency. For these and other reasons, we believe likelihood inference using MCMC, an approach that has had enormous success in physics, should become an important tool in 21st-century biology.
Modern biology, however, faces challenges very different from those of physics. Although physicists often take great care to understand and minimize experimental noise, high-throughput assays have sources of noise that may vary from system to system or even from laboratory to laboratory and may be impractical to characterize quantitatively. The central point of this article is that, given enough data, one can precisely characterize interesting biological phenomena without having to model uninteresting effects, such as the experimental errors. We believe this is of particular importance for the study of TFs, although it may find application in other areas of quantitative biology as well.
Furthermore, the high-throughput platforms currently available for probing TF binding appear, at least for broad-acting TFs, to provide sufficient data for such analyses. In our study of Mukherjee et al.'s (4) PBM data for Abf1p, we were essentially able to infer the values of 352 independent parameters describing both the TF and the experimental error model (although the error model parameters were integrated out analytically). Nonetheless, most of the 80 energy matrix elements describing the TF were determined to a precision of ≤5% of the functional range. This precision is not caused by overfitting; it simply reflects the large amount of information contained in the data.
Our analysis is meant only as a proof-of-principle demonstration and could be extended in many ways. Perhaps our most unrealistic assumption is that TF binding sites are either bound or not bound. This was done primarily to keep the analysis simple and speed up the MCMC algorithm, but a more physically realistic computation of region occupancy, such as that used by the program GOMER (15), could be used instead. The assumption that each nucleotide contributes independently to the TF's SDBE (see ref. 16 for a critique) could also be relaxed by allowing couplings between positions. Indeed, a great advantage of likelihood inference is the way it accommodates such refinements without changing the conceptual basis of the analysis. The only obstacle one faces when implementing such changes is the need for computational power.
Another important aspect of likelihood inference is the possibility of refining quantitative models by using data from multiple experiments. For example, if two data sets {zi} and {z′j} are available for some TF, one can perform a likelihood analysis by using the product of each data set's likelihood as the combined likelihood of both data sets, i.e., p({zi},{z′j}|θ) = p({zi}|θ)p({z′j}|θ). Extending this to more than two data sets is straightforward, and data from different experimental platforms may be combined in this way. For example, ChIP–chip data might be combined with low-throughput EMSA measurements and high-throughput in vitro data by using synthetic DNA probes (17–19). Also, any knowledge one has about the experimental errors in any of these experiments may be used, either by assuming an explicit error model or biasing the error model prior (see SI Text). The combination of data from different experiments is a powerful analysis technique, and our method should help facilitate its application to high-throughput biological data.
Methods
Software and Results.
The ΘPBM and ΘChIP matrix ensembles are available on request. The software used in this analysis, including our MCMC algorithm, was written in Matlab and C++ and is available on request.
Likelihood With Unknown Error Models.
Suppose an experiment assigns a value zi to each DNA region si (i = 1, …, N) and let xi = θ(si) be the corresponding prediction made by the TF binding model for a particular choice of parameters θ. The zi and xi can be quite different quantities: a model might predict whether or not a TF is likely to be bound to a particular DNA sequence in thermal equilibrium, while an experiment might observe the fluorescence intensity of the corresponding spot on a microarray. Because of experimental noise, the two quantities are related by an error model E(z|x), giving the probability of observing z when the state predicted by the model is really x. We make the simplifying assumption that the error model is the same for all regions probed in any given experiment, although different experiments may be subject to different error models.
We want to find specific parameters θ such that the N model predictions {xi}i=1N account well for the measurements {zi}, i.e., give a large value to the likelihood p({zi}|θ). Our analysis relies on three further assumptions: (i) There is a “correct” set of parameters θ that accurately describes the TF's behavior in the experiment. (ii) The likelihood of the data {zi} depends on the model parameters θ only through the model predictions {xi}. Thus, p({zi}|θ) = p({zi}|{xi}). (iii) The experimental results for each sequence are independent, so that p({zi}|{xi}) = ∏i p(zi|xi).
Our method is most simply implemented if both the observations and the predictions are discrete. Accordingly, we group the N ∼ 6,000 DNA sequences in a binding assay into ∼100–300 equipopulated z-bins on the basis of observed fluorescences; at the same time the model predictions assign each sequence to an x-bin (bound or not bound). If we know the error model, we can then write an explicit expression for the likelihood:
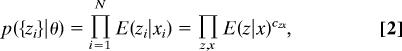 |
where czx is the number of regions assigned simultaneously to bin z and bin x. This expression depends on the parameters θ only through the way regions are assigned to x-bins. Because error models applicable to high-throughput biological experiments are usually unknown, in practice we cannot evaluate Eq. 2. We propose to deal with this problem by averaging over the space of all error models with some reasonable prior p(E). The explicit expression we obtain for the EMA likelihood under a uniform prior on the error models is (see SI Text for more details):
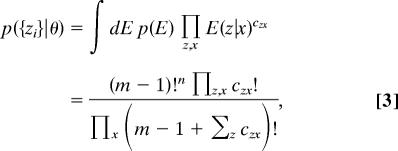 |
where m and n, respectively, denote the number of possible values that z and x can take on. We stress that this equation for EMA likelihood is completely general and may be applied to any situation in which both the data and the model predictions are quantized. Note that the specific numerical values of the experimental data and model predictions serve only to cluster sequences together and are otherwise unused in this analysis. Thus, monotonic reparametrizations of these values do not affect our results.
It is interesting to note that Eq. 3 may be rewritten (see SI Text) as:
where I(z; x) is the empirical mutual information (20) between {zi} and {xi} (and thus depends on θ), H(z) is the empirical entropy of {zi} (and does not depend on θ), and Δ is a nonnegative correction, accounting for finite data and the choice of error model prior p(E). For a large class of error model priors, including the uniform prior used to compute Eq. 3, Δ vanishes as N becomes large. In this limit, the per-datum log likelihood becomes, up to an additive constant, the mutual information between model predictions and data. The emergence of mutual information helps explain how one can meaningfully evaluate likelihood without prior knowledge of which model predictions should correspond to which data: the best model will make predictions that provide the most information about the experimental results, regardless of how this correspondence is realized.
Sampling Model Space.
The primary goal of our analysis is to characterize the distribution of model parameters θ specified by the data {zi} by using the posterior distribution in Eq. 1 with likelihood specified by Eq. 3. To do this we used MCMC, a powerful computational method for sampling from such distributions (21). An essential feature of MCMC is that it does not require that one know how to normalize the distribution being sampled, which frees us from having to estimate the proportionality constant in Eq. 1.
MCMC starts from a seed model θ0 and stochastically wanders from model to model in such a way that the ensemble Θ = {θ1, θ2, …, θT} of models visited is eventually distributed according to a desired distribution (in our case Eq. 1). The expected value of any θ-dependent quantity q(θ), given the observations {zi}, is then estimated by the ensemble average:
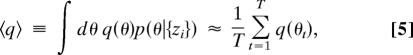 |
where q(θ) might be the value of a particular matrix element, the square deviation of that element from its mean, or a binary variable describing whether or not a particular site is bound. See SI Text for a detailed discussion of the particular MCMC algorithm used in this analysis.
Supplementary Material
Acknowledgments
We thank Manuel Llinás and William Bialek for generous advice and constructive criticism; William Press, Steven Quake, and Jason Lieb for valuable criticism of this manuscript; Michael Buck, Casey Bergman, Neil Clarke, Thomas Gregor, Michael Lässig, Saeed Tavazoie, Martin Vingron, and the participants of the 2006 Otto Warburg Summer School for helpful discussions; and Martin Jones for helping produce Fig. 2c. C.G.C. and J.B.K. were supported by National Institutes of Health Grant P50GM071508. C.G.C. was also supported by Department of Energy Grant DE-FG02-91ER40671. G.T. was supported by the Burroughs-Wellcome Fund.
Abbreviations
- TF
transcription factor
- SDBE
sequence-dependent binding energy
- PBM
protein binding microarray
- EMA
error-model-averaged
- MCMC
Markov chain Monte Carlo
- LIR
log intensity ratio
- HF
hit fraction.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0609908104/DC1.
References
- 1.Ptashne M, Gann A. Genes and Signals. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2002. [Google Scholar]
- 2.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 3.Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I, et al. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- 4.Mukherjee S, Berger MF, Jona G, Wang XS, Muzzey D, Snyder M, Young RA, Bulyk ML. Nat Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tompa M, Li N, Bailey TL, Church GM, De Moor B, Eskin E, Favorov AV, Frith MC, Fu Y, Kent WJ, et al. Nat Biotechnol. 2005;23:137–144. doi: 10.1038/nbt1053. [DOI] [PubMed] [Google Scholar]
- 6.Berg OG, von Hippel PH. J Mol Biol. 1987;193:723–750. doi: 10.1016/0022-2836(87)90354-8. [DOI] [PubMed] [Google Scholar]
- 7.Stormo GD, Fields DS. Trends Biochem Sci. 1998;23:109–113. doi: 10.1016/s0968-0004(98)01187-6. [DOI] [PubMed] [Google Scholar]
- 8.Stormo GD. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 9.Mustonen V, Lässig M. Proc Natl Acad Sci USA. 2005;102:15936–15941. doi: 10.1073/pnas.0505537102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg J, Willmann S, Lässig M. BMC Evol Biol. 2004;4:42. doi: 10.1186/1471-2148-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foat BC, Morozov AV, Bussemaker HJ. Bioinformatics. 2006;22:141–149. doi: 10.1093/bioinformatics/btl223. [DOI] [PubMed] [Google Scholar]
- 12.Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 13.Beinoravčiūtė-Kellner R, Lipps G, Krauss G. FEBS Lett. 2005;579:4535–4540. doi: 10.1016/j.febslet.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 14.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, et al. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 15.Granek JA, Clarke ND. Genome Res. 2005;6:R87. doi: 10.1186/gb-2005-6-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benos PV, Bulyk ML, Stormo GD. Nucleic Acids Res. 2002;30:4442–4451. doi: 10.1093/nar/gkf578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bulyk ML, Gentalen E, Lockhart DJ, Church GM. Nat Biotechnol. 1999;17:573–577. doi: 10.1038/9878. [DOI] [PubMed] [Google Scholar]
- 18.Bulyk ML, Huang X, Choo Y, Church GM. Proc Natl Acad Sci USA. 2001;98:7158–7163. doi: 10.1073/pnas.111163698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren CL, Kratochvil NC, Hauschild KE, Foister S, Brezinski ML, Dervan PB, Phillips GN, Jr, Ansari AZ. Proc Natl Acad Sci USA. 2006;103:867–872. doi: 10.1073/pnas.0509843102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cover TM, Thomas JA. Elements of Information Theory. New York: Wiley; 1991. pp. 12–49. [Google Scholar]
- 21.Gilks WR, Richardson S, Spiegelhalter DJ. Markov Chain Monte Carlo in Practice. New York: Chapman & Hall; 1996. pp. 1–16. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.