Abstract
VP40, the putative matrix protein of both Ebola and Marburg viruses, possesses a conserved proline-rich motif (PY motif) at its N terminus. We demonstrate that the VP40 protein can mediate its own release from mammalian cells, and that the PY motif is important for this self-exocytosis (budding) function. In addition, we used Western-ligand blotting to demonstrate that the PY motif of VP40 can mediate interactions with specific cellular proteins that have type I WW-domains, including the mammalian ubiquitin ligase, Nedd4. Single point mutations that disrupted the PY motif of VP40 abolished the PY/WW-domain interactions. Significantly, the full-length VP40 protein was shown to interact both physically and functionally with full-length Rsp5, a ubiquitin ligase of yeast and homolog of Nedd4. The VP40 protein was multiubiquitinated by Rsp5 in a PY-dependent manner in an in vitro ubiquitination assay. These data demonstrate that the VP40 protein of Ebola virus possesses a PY motif that is functionally similar to those described previously for Gag and M proteins of specific retroviruses and rhabdoviruses, respectively. Last, these studies imply that VP40 likely plays an important role in filovirus budding, and that budding of retroviruses, rhabdoviruses, and filoviruses may proceed via analogous mechanisms.
Ebola and Marburg viruses, members of the Filoviridae family, have been the cause of deadly outbreaks of severe hemorrhagic disease in humans (1–4). Filoviruses possess a nonsegmented, negative-sense RNA genome encoding seven viral proteins: NP (nucleoprotein), VP35 (phosphoprotein), VP40 (putative matrix protein), GP (glycoprotein), VP30 (nucleocapsid-associated protein), VP24 (putative secondary matrix protein), and L (RNA-dependent RNA polymerase; refs. 5–13). Aside from recent insights into the structure and function of the GP of filoviruses, and the recent finding that NP, VP35, and L proteins of Marburg virus are required for RNA replication and transcription, little is known regarding the functional properties of VP24, VP30, and VP40 (8, 9, 11–20). Because the filoviruses are closely related to members of the Rhabdoviridae family (e.g., vesicular stomatitis virus; VSV), the roles of specific filoviral proteins have been postulated based upon known activities of homologous proteins of negative-sense RNA viruses such as VSV. For example, the VP40 protein of Ebola virus is thought to be the functional homolog of the matrix (M) protein of VSV based largely on its position in the genome, its hydrophobicity, and its abundance within the virion (5, 6).
While the amino acid sequences of Ebola VP40 and VSV M proteins are highly divergent, a short, proline-rich region is conserved at the amino termini of both proteins (5, 21). Wills et al. were the first to demonstrate that a similar proline-rich motif (PPxY or PY motif, where P = proline, x = any amino acid, and Y = tyrosine) within the p2b region of the Gag polyprotein of Rous sarcoma virus (RSV) plays a critical role during a late stage of virus budding (22–25). As a result, the PY motif of RSV Gag was termed the L-domain for late budding domain (24, 25). More recently, a functional L-domain having the PPxY core consensus sequence was identified in the M protein of VSV, and this core sequence is highly conserved in the M proteins of other rhabdoviruses (26, 27). In addition to the PPxY core consensus sequence, L-domains that have the core consensus sequences of PTAPP and YxxL have been identified in the Gag polyproteins of HIV-1 and equine infectious anemia virus, respectively (23, 28–33). When expressed alone in mammalian cells, both Gag and M proteins are capable of being released (bud) into the culture media in the form of “virus-like” particles, a phenomenon dependent on having a functional L-domain (24, 26, 27, 34, 35). Thus, retroviral Gag and rhabdoviral M proteins play a major role in the budding process of their respective viruses; however, the mechanism by which their L-domains function during virus-cell separation remains unknown.
In addition to their role in virus budding, L-domains have been shown to mediate interactions with cellular proteins that may facilitate or promote virus budding (22, 27, 33, 36). For example, the PY motifs within the RSV Gag, VSV M, and rabies virus M proteins were shown to interact with WW-domains of specific cellular proteins (22, 27). WW-domains are modular, hydrophobic domains found in a wide variety of cellular proteins, and these domains are classified as types I–IV, depending on the core sequence of the ligand (e.g., PPxY) with which they interact (36–42). The main function of WW-domains is to facilitate protein–protein interactions in the cell (36). It should be noted that the viral L-domains are not limited to binding WW-domain-containing proteins. For example, a YxxL-based L-domain of the equine infectious anemia virus Gag protein was shown to mediate interactions with the cellular AP2 protein, a component of the endocytosis pathway in mammalian cells (33).
In this report we present evidence that the VP40 protein of Ebola virus possesses a potential L-domain having the core consensus sequence of PPxY. VP40 was capable of mediating its own release from mammalian cells and of mediating interactions with specific WW-domains of cellular proteins. Mutations that disrupted the PY motif led to a significant decrease in the release of VP40 from mammalian cells and a complete abolishment of WW-domain binding. Importantly, the full-length VP40 protein was able to interact both physically and functionally with the full-length Rsp5 protein, a ubiquitin ligase having multiple WW-domains (43, 44). As a result of this protein–protein interaction, VP40 was covalently modified with multiple copies of ubiquitin. Thus, our data are the first to suggest that filoviruses, like the retro- and rhabdoviruses, encode a structural protein that has L-domain activity. Furthermore, these results imply that the budding machinery used by members of these three RNA virus families may be comparable. Last, the presence of an L-domain within an Ebola virus protein may have important ramifications for the development of novel antivirals designed to block L-domain activity and thus inhibit virus budding.
Materials and Methods
Cells and Plasmids.
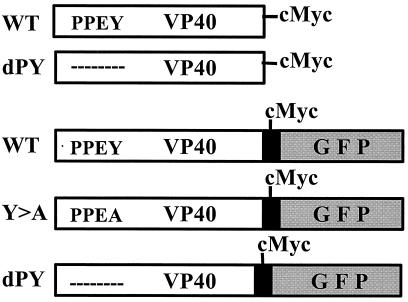
Stocks of CV-1 and COS-7 cells were maintained in DMEM (Life Technologies, Grand Island, NY) supplemented with 10% FCS (vol/vol) (HyClone). The VP40 gene from Ebola (Zaire) in vector pGem3 was kindly provided by H.-D. Klenk (Institut for Virology, Marburg, Germany). Nucleotide sequences encoding amino acids 2–21 of VP40 were joined in-frame with bacterial alkaline phosphatase in vector pMY101 (kindly provided by B. Kay, University of Wisconsin, Madison) to generate plasmid VP40-bacterial alkaline phosphatase (BAP; wild type, WT). Site-directed mutagenesis was used to change tyrosine (Y) at amino acid position 13 to alanine (A) to generate plasmid VP40-BAP (Y>A). The c-myc epitope tag (EQKLISEEDL) was joined in-frame to the C terminus of VP40 in vector pcDNA3.1 (Invitrogen) to generate plasmid VP40-W.T. In addition, the PY motif of VP40 (amino acids 10–13) was deleted to generate plasmid VP40-dPY. Three plasmids encoding VP40-GFP fusion proteins were generated using pEGFP-N3 (CLONTECH). Wild-type VP40 and the Y>A and dPY mutant forms of full-length VP40 were joined in-frame to the N terminus of GFP.
Glutathione S-Transferase (GST)-WW Fusion Proteins and BAP Binding Assay.
Purification of GST-WW fusion proteins (kindly provided by M. Sudol, Mount Sinai Medical Center New York, NY, and C. Ross, Johns Hopkins Medical Center, Baltimore, MD) and Western-ligand blotting assays were performed as described previously (27).
Ubiquitination Assay.
In vitro ubiquitination assays were performed as described previously (45). Briefly, wild-type and mutant forms of VP40 were first translated in vitro, using a coupled transcription/translation system (TNT, Promega) in the presence of [35S]methionine (NEN Dupont), and then incubated in a reaction mixture containing 25 mM Tris (pH 8.0), 125 mM NaCl, 2 mM MgCl2, 50 μM DTT, 2 mM ATP, 3.0 μg ubiquitin (Sigma), E1 enzyme, and E2 protein (UBC8 from Arabidopsis thaliana), in the absence or presence of Rsp5 (E3 ubiquitin ligase). In the indicated reactions, Rsp5 was replaced with the Rsp5 C-A mutant, which contains a Cys-to-Ala change in the active site of the enzyme, rendering the protein inactive (45).
VP40 Budding Assay.
Budding assays were performed basically as described previously (27, 34). Briefly, 35-mm dishes of either CV-1 or COS-7 cells were first infected with VvT7 for 30 min and then transfected with the appropriate VP40 expression plasmid, using N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (Roche Molecular Biochemicals). Cells were metabolically labeled 2 h after transfection with 100 μCi of [35S]Met-Cys (NEN Dupont), and both the cells and culture media were harvested 8–10 h after transfection. Cells were lysed in RIPA buffer (50 mM Tris, pH 8.0/150 mM NaCl/1.0% Nonidet P-40/0.5% deoxycholate/0.1% SDS), and 900 μl of culture medium was mixed with 100 μl of 10× NTE buffer (0.5 M Tris⋅HCl, pH 7.5/1.5 M NaCl/1.0% Nonidet P-40/10 mM EDTA/2.5% gelatin/0.2 M sodium azide). Immunoprecipitated proteins were analyzed by SDS/PAGE, visualized by autoradiography, and quantitated using National Institutes of Health image Version 2 software.
Confocal Microscopy.
Subcellular localization of VP40-GFP fusion proteins was determined by observing GFP fluorescence in COS-7 cells transfected with the indicated plasmid. At various times after transfection, cells grown on coverslips were fixed in acetone:methanol (1:1) for 15 min on ice. Cells were rinsed several times with 1× PBS, and the coverslips were inverted onto a drop of Flourmount (Fisher) on glass slides. Images of fluorescing cells were observed and documented using a Leica TCS4D confocal microscope.
Results
PY Motif of VP40 Interacts with Cellular WW Domains.
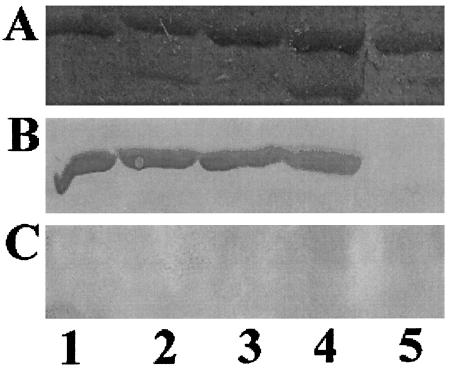
We noted the presence of a proline-rich motif (PY motif) at the N terminus of the VP40 protein that was reminiscent of those described for the Gag and M proteins of retroviruses and rhabdoviruses, respectively. One characteristic of these PY motifs was their ability to mediate interactions with select WW-domains of cellular proteins (22, 27). To determine whether the VP40 PY motif could mediate interactions with WW domains of cellular proteins, Western-ligand blotting assays were used. To this end, amino acids 2–21 of VP40 containing the PY motif were joined in-frame to BAP to generate a VP40-BAP (WT) fusion protein (Fig. 1). As a negative control, a plasmid encoding a Y13A mutant VP40-BAP fusion protein was also constructed (Fig. 1). Equivalent amounts of the enzymatically active fusion proteins were isolated from Escherichia coli and used to probe a panel of GST-WW domain fusion proteins immobilized on nitrocellulose filters (Fig. 2). Each lane contained an equivalent amount (1.0 μg) of the various GST-WW-domain fusion proteins, as illustrated by staining with Coomassie brilliant blue (Fig. 2A). The VP40-BAP (WT) probe interacted with WW-domains 2 and 3 from the Nedd4 protein (Fig. 2B, lanes 1 and 2) and with WW-domains 1 and 2 from the YAP protein (Fig. 2B, lanes 3 and 4). Both Nedd4, a membrane-associated ubiquitin ligase, and YAP, a Yes kinase-associated protein, contain multiple type I WW-domains (37, 40). The VP40-BAP (WT) probe did not interact with a type II WW domain derived from the FE65 (36) protein (Fig. 2B, lane 5). The VP40-BAP (Y>A) mutant did not interact with any of the cellular WW-domains tested (Fig. 2C, lanes 1–5). It should be noted that in addition to the GST-WW-domain fusion proteins shown (Fig. 2), the VP40-BAP (WT) probe also interacted with type I GST-WW-domain proteins AIP1, AIP3/WWP3, and Rsp5 (yeast homolog of Nedd4), but failed to interact with WW-domains from dystrophin or ESS1 (refs. 36 and 44; data not shown).
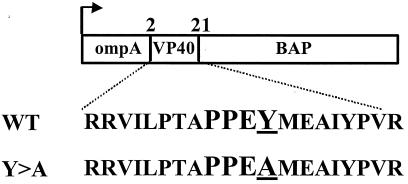
Figure 1.
Diagram of VP40-BAP fusion proteins. Amino acids 2–21 of VP40 were inserted in-frame between the secretory signal sequence of ompA and the full-length BAP ORF in vector pMY101. The amino acid sequence of the VP40 inserts are shown to illustrate the wild-type (PPEY) and mutant (PPEA) PY motifs.
Figure 2.
Far-Western binding assay. One microgram each of GST-Nedd4WW2 (lane 1), GST-Nedd4WW3 (lane 2), GST-YapWW1 (lane 3), GST-YapWW2 (lane 4), and GST-FE65 (lane 5) was subjected to SDS/PAGE. The GST fusion proteins were either stained with Coomassie brilliant blue (A) or immobilized on nitrocellulose filters and probed with equivalent amounts of VP40-BAP (WT) (B) or VP40-BAP (Y>A) (C).
Because amino acids 2–21 of VP40 are rich in proline residues and contain a potential ligand sequence (PxxP) necessary for binding to SH3-domains (38), we determined whether VP40-BAP (WT) could mediate interactions with select SH3-domains from cellular proteins. In contrast to its ability to bind to type I WW-domains, the VP40-BAP (WT) probe was unable to interact with SH3-domains derived from the cellular proteins Src, Yes, Lyn, and Fyn (data not shown); however, the possibility that VP40-BAP (WT) may interact with other SH3-domains not tested cannot be ruled out. These results are the first to demonstrate that (i) amino acids 2–21 of VP40 are sufficient to mediate interactions with type I WW-domains, and (ii) the PY motif was critical for this interaction.
Physical and Functional Interaction Between Full-Length VP40 and Full-Length Rsp5.
Because of the potential involvement of endocytosis-related cellular proteins in virus budding (33) and the potential role of ubiquitin in the process of virus budding (31), we sought to determine whether the full-length VP40 protein could interact with the full-length Rsp5 ubiquitin ligase. Our reasoning was as follows: (i) a functional ubiquitination assay employing a stable, enzymatically active form of Rsp5 exists, whereas a similar, reliable assay employing the mammalian Nedd4 ubiquitin ligase does not, and (ii) both Rsp5 and Nedd4 are localized at the plasma membrane, contain multiple WW-domains, and participate in endocytosis. Furthermore, the Nedd4 protein was identified from a mouse cDNA library screen, using the L-domain from the VSV M protein as the bait (R.N.H., J. Paragas, and P. Palese, unpublished data).
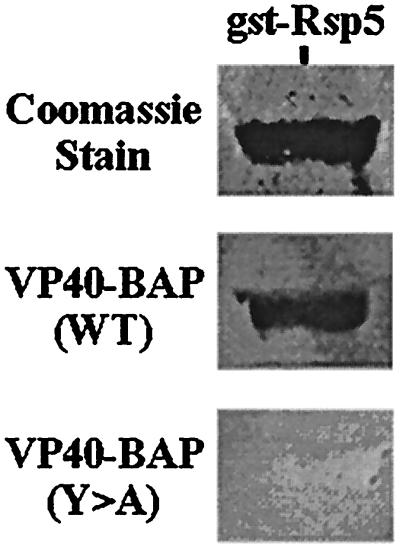
Western-ligand blotting was first used to determine whether the VP40-BAP fusion proteins could interact with a full-length GST-Rsp5 fusion protein (Fig. 3). Indeed, the VP40-BAP WT probe interacted with full-length Rsp5, whereas the VP40-BAP Y>A mutant did not (Fig. 3).
Figure 3.
Far-Western binding assay. Two micrograms of a GST-Rsp5 (full-length Rsp5) fusion protein was stained with Coomassie brilliant blue (Top) or probed with VP40-BAP WT (Middle) or with VP40-BAP Y>A (Bottom).
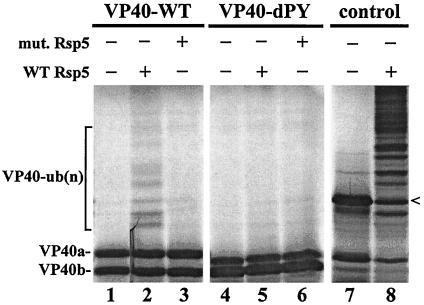
We next took advantage of a functional ubiquitination assay employing Rsp5 as the terminal E3 ubiquitin ligase (43, 45). The ability of a target protein to be ubiquitinated by Rsp5 in this assay is indicative of both a physical and functional interaction between Rsp5 and the target protein, VP40. Full-length VP40-WT and full-length VP40-dPY were translated in vitro, and the radiolabeled proteins were incorporated into the ubiquitination assay (Fig. 4). In the absence of full-length Rsp5, both VP40-WT and VP40-dPY remained nonubiquitinated, as expected (Fig. 4, lanes 1 and 4). However, in the presence of WT Rsp5, multiubiquitinated forms of VP40 were detected, as evidenced by a series of higher molecular weight protein species (Fig. 4, lane 2). In contrast, multiubiquitinated forms of VP40-dPY were not observed in the presence of WT Rsp5 (Fig. 4, lane 5). As further proof of a physical and functional interaction between VP40 and Rsp5, an enzymatically inactive mutant of Rsp5 (Rsp5 C-A mutant) failed to ubiquitinate both VP40-WT and VP40-dPY (Fig. 4, lanes 3 and 6). A Saccharomyces cerevisiae protein known to be ubiquitinated by Rsp5 served as a positive control (Fig. 4, lanes 7 and 8). These results demonstrated that full-length VP40 and full-length Rsp5 interacted in a PY-dependent manner, leading to the covalent modification of VP40 with ubiquitin [VP40-Ub(n)].
Figure 4.
In vitro ubiquitination of VP40. VP40-WT and VP40-dPY were transcribed and translated in vitro (lanes 1 and 4, respectively). In vitro translated proteins were incubated with recombinant E1 and E2 (UBC8) proteins, ATP, ubiquitin, and either WT Rsp5 (lanes 2 and 5) or the C-A mutant form of Rsp5, as indicated (lanes 3 and 6). The position of the VP40 doublet (lanes 1–6) and multiubiquitinated forms of VP40 [VP40-ub(n); lane 2] are indicated. A positive control (52-kDa yeast protein encoded by the YHL002w gene; G.W. and J.H., unpublished data) for ubiquitination by Rsp5 is shown (lanes 7 and 8). The arrowhead depicts the location of the nonubiquitinated form of the 52-kDa protein. The presence or absence of WT Rsp5 or mut. Rsp5 is indicated by a + or −.
Expression and Detection of Full-Length VP40 in Mammalian Cells.
Because the retroviral and rhabdoviral PY motifs have been shown to function as late budding domains, we used a functional assay and plasmids expressing full-length VP40 (Fig. 5) to determine whether the VP40 PY motif could promote the release (budding) of VP40 from transfected cells. Western blot analysis demonstrated that the c-myc epitope tag was functional and that the full-length VP40 proteins were stably expressed in transfected cells (data not shown).
Figure 5.
Diagram of full-length VP40 and VP40-GFP fusion proteins. The position of the c-myc epitope tag is indicated. The dashed line indicates that the PPxY sequence has been deleted. Three forms of c-myc-tagged VP40 were joined in-frame to the N terminus of GFP (shaded box) in vector pEGFP-N3 to generate the indicated VP40-GFP fusion proteins.
Release of VP40 from Mammalian Cells.
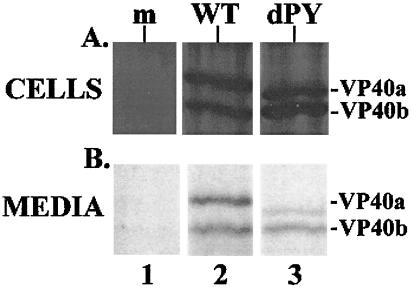
To determine whether VP40 could be released from mammalian cells, plasmids expressing c-myc-tagged VP40-WT and VP40-dPY were used. COS-7 cells were first infected with VvT7 and then either mock-transfected or transfected with the appropriate VP40 expression plasmid. At various times after transfection, both the radiolabeled cell extract and culture media were harvested and subjected to immunoprecipitation with the c-myc monoclonal antibody (Fig. 6). As observed in Western blot analysis (data not shown), two species of VP40 were observed in this assay (VP40a and VP40b). Although the origin of this VP40 doublet remains to be determined, the presence of these two species was highly reproducible, and VP40b likely results from the initiation of translation at an internal AUG codon. Identical amounts of VP40-WT and VP40-dPY were detected in the cell extract (Fig. 6A, lanes 2 and 3). As expected, VP40 was not detected in mock-transfected cells (Fig. 6A, lane 1). More importantly, the VP40-WT protein was detected in the media, indicating that VP40-WT was indeed released from transfected cells (Fig. 6B, lane 2). The ability of VP40-WT to be released into the culture media was highly reproducible in multiple experiments using various cell types (data not shown). Although the VP40a-dPY protein was also detected in the media, the level of VP40a-dPY was reduced significantly by 75% (4-fold) as compared with that of VP40a-WT (Fig. 6B, lane 3). This approximately 4-fold decrease is reminiscent of that observed for PY mutants of VSV M and RSV Gag proteins in similar functional assays (25–27).
Figure 6.
VP40 budding assay. (A) Radiolabeled cell extracts from COS-7 cells that received no DNA (mock, lane 1), pVP40-WT (lane 2), and pVP40-dPY (lane 3) were immunoprecipitated and analyzed by SDS/PAGE. (B) Radiolabeled proteins released into the media covering cells transfected with no DNA (mock, lane 1), pVP40-WT (lane 2), and pVP40-dPY (lane 3) were immunoprecipitated as above. Immunoprecipitated proteins were quantitated using National Institutes of Health image Version 2 software.
To ensure that the cytopathic nature of VvT7 was not promoting the release of VP40, we assessed the ability of VP40 to be released from cells, using a Vaccinia-free assay system. VP40-WT was inserted downstream of an internal ribosome entry site and expressed from the pCite4a vector in BSR-T7 cells, a cell line that constitutively expresses T7 polymerase (kindly provided by K-Klaus Conzelmann, Max V. Pettenkofer-Institut für Virologie, Munich, Germany). Indeed, VP40-WT was readily detected in both the cell lysate and media when expressed in BSR-T7 cells (R.N.H. and M. Brown, unpublished data).
Last, VP40-Gag (Rous sarcoma virus) fusion proteins were constructed in which the PY motif of RSV Gag was replaced with that of VP40 (amino acids 1–47). The VP40-Gag fusion proteins were used in a functional retroviral budding assay (26). Indeed, the VP40 PY motif was able to functionally substitute for that of RSV Gag, as determined by the release of virus-like particles into the media (A. Patnaik, R. Craven, J. Wills, and R.N.H., unpublished data). Taken together, these results strongly indicate that the PY motif of VP40 functions as a late budding domain.
Cellular Localization of VP40.
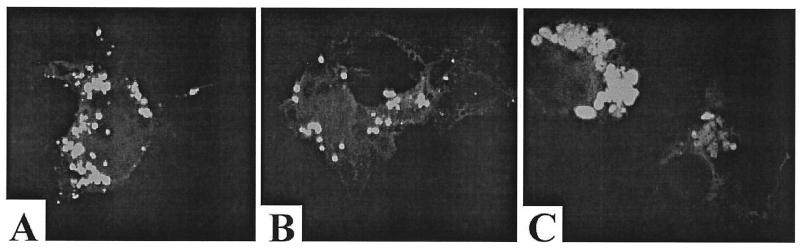
One possible explanation for why the VP40-dPY mutant was not released from cells as efficiently as VP40-WT is that the VP40-dPY protein is mislocalized in transfected cells. To rule out this possibility, we examined the intracellular localization patterns of VP40-WT and PY mutants of VP40, using confocal microscopy. COS-7 cells were transfected with the indicated plasmid (Fig. 5), and expression of the VP40/GFP fusion proteins was observed in positively fluorescing cells by confocal microscopy (Fig. 7). VP40/GFP-WT, VP40/GFP-Y>A, and VP40/GFP-dPY (Fig. 7 A, B, and C, respectively) exhibited virtually identical localization patterns within the cells. Interestingly, intensely fluorescing globules, which may represent VP40 aggregation, as well as more diffuse cytoplasmic staining, were visible for all of the VP40/GFP fusion proteins (Fig. 7). No nuclear fluorescence was observed for any of the VP40/GFP fusion proteins. In a small percentage of the cells, a potential enrichment of the VP40/GFP fusion proteins at the periphery of the cell was observed (e.g., Fig. 7C, bottom right). Although the nature of these globules remains undefined, a similar globular pattern of fluorescence was observed for several Marburg virus proteins expressed from alphavirus vectors in Vero cells (46). These results indicate that no gross differences in the cellular localization patterns were evident for the various VP40/GFP fusion proteins, and thus the budding defect exhibited by VP40a-dPY (Fig. 6B) was likely not due to mislocalization of the PY mutant within the cell.
Figure 7.
Intracellular localization of VP40 using GFP fusion proteins and confocal microscopy. COS-7 cells were transfected with VP40/GFP-WT (A), VP40/GFP-Y>A (B), and VP40/GFP-dPY (C). Sections of fluorescing cells were observed using a Leica TCS4D confocal microscope.
Discussion
In this report, we present the following findings regarding the VP40 protein of Ebola virus: (i) a conserved PPxY motif within the VP40 protein is able to mediate interactions with specific cellular WW-domains, (ii) a PY/WW-domain interaction between full-length VP40 and a full-length ubiquitin ligase resulted in the generation of multiubiquitinated forms of VP40, (iii) efficient release of VP40 from mammalian cells was facilitated by the PY motif, and (iv) VP40 exhibited both a diffuse and globular cytoplasmic distribution in transfected cells. Taken together, these data strongly suggest that VP40 possesses a late budding domain with characteristics similar to those described for L-domains identified in Gag and M proteins of retroviruses and rhabdoviruses, respectively (23, 24, 26–30, 32, 33, 47, 48).
The L-domain was first identified and characterized in the Gag polyprotein of RSV, and, as its name implies, L-domains function at a late stage of virus–cell separation (24). Although all L-domains possess the same budding function, the core consensus sequence of these L-domains varies from PPxY to PTAPP to YxxL. Viral proteins containing functional L-domains have the unique property of being released from mammalian cells in the absence of any other viral protein (24, 27). Indeed, in repeated experiments the VP40a-WT protein was clearly able to be released efficiently from transfected cells in a PY-dependent manner. The amount of VP40a-dPY protein detected in the medium (approximately 4-fold less than WT VP40) was similar to the amounts of PY mutant forms of Gag and M proteins reported previously (25, 27). The fact that the VP40a-dPY mutant was capable of being released at a low level suggests that additional sequences within VP40 may be important for its release. Interestingly, the PY motif of VP40 is actually overlapped by a PTAPP sequence (PTAPPxY), which matches the core sequence of the L-domain present within the HIV-1 Gag protein (28–30). The contribution of the PTAPP core sequence to the release of VP40 from mammalian cells has yet to be determined.
Analysis of the intracellular localization of WT and mutant forms of VP40 suggests that the decreased ability of the dPY mutant to be released from cells is not due to mislocalization in the cells. All of the VP40/GFP fusion proteins exhibited both a diffuse cytoplasmic and globular pattern of fluorescence with some enrichment at the plasma membrane. The intensely fluorescing globules may be indicative of VP40 aggregation.
In addition to their biological activity in budding, an intriguing characteristic of the L-domains is their ability to mediate interactions with cellular proteins (22, 27, 33). It has been hypothesized that L-domain activity may be dependent on a virus–host interaction(s), perhaps at the budding site on the plasma membrane (22, 26, 27, 33). For example, the functional L-domain (YxxL) of the equine infectious anemia virus Gag protein was shown to interact with the cellular AP2 protein, a critical component of the endocytic pathway (33). This observation led to the proposal that budding of equine infectious anemia virus may occur at sites on the plasma membrane that are actively undergoing endocytosis/exocytosis, and perhaps the L-domain functions to recruit the appropriate cellular protein(s) to the budding site.
Although viral PY motifs have not been shown to interact with AP2, they have been shown to interact with specific cellular proteins that have WW-domains (22, 27). Of these cellular proteins, Nedd4/Rsp5 are of particular interest because (i) both proteins contain multiple type I WW-domains; (ii) both proteins interacted strongly with the viral PY motifs; (iii) both proteins possess ubiquitin ligase activity and are known to function in endocytosis in mammalian cells and in Saccharomyces cerevisiae, respectively; and (iv) full-length Rsp5 interacted with and ubiquitinated full-length VP40 in vitro (43, 44, 49, 50). Although we cannot yet conclude that Nedd4 plays a role in the budding process of any of these RNA viruses, these recent findings designate Nedd4 as an attractive candidate for further study. The ability of full-length, biologically active Rsp5 to interact both physically and functionally with full-length VP40 in a PY-dependent manner suggests that a similar interaction may occur in vivo between VP40 and Nedd4.
At first glance, the role of ubiquitin in virus budding is not apparent; however, evidence supporting a functional link between L-domains and ubiquitin is mounting (31). Indeed, ubiquitin was found to be covalently attached to the p6 and p12 regions of HIV-1 Gag and Moloney murine leukemia virus Gag proteins, respectively, and ubiquitin has been detected in purified HIV-1 virions (31). In addition, the M protein of VSV is modified by ubiquitin in a PY-dependent manner (R.N.H., G. Wang, and J. Huibregsle, unpublished data). Thus, ubiquitinated forms of VP40 may be important in promoting the late stage of virus–cell separation. Ubiquitin and ubiquitinated proteins are known to be involved in cellular processes including endocytosis/exocytosis (51). Thus, one possibility is that ubiquitinated forms of VP40 may function to recruit components of the endocytic pathway to the site of budding. The potential recruitment of proteins involved in endocytosis by viral budding proteins has been proposed previously (33); however, recruitment of proteins involved in exocytosis has yet to be demonstrated. Alternatively, ubiquitinated forms of VP40 may be specifically targeted to areas on the plasma membrane that are active in endocytosis. Indeed, much more work needs to be performed before the precise molecular aspects of budding are truly understood.
We are currently mapping the regions of VP40 required for modification by ubiquitin. In addition, we are attempting to determine whether ubiquitinated forms of VP40 are present in mammalian cells, whether the Nedd4 protein plays a role in VP40 release, and whether specific proteasome inhibitors have an effect on the release of VP40 from mammalian cells. The conserved nature of these L-domains across different RNA virus families hints at their broad-based importance to virus budding. Thus, it is likely that a better understanding of the role of these L-domains will be crucial for elucidating the molecular mechanisms of retroviral, rhabdoviral, and filoviral budding. Importantly, in addition to enhancing our understanding of the interplay between virus and host proteins during the budding process, future studies may also lead to the development of novel antiviral agents designed to inhibit this potential shared step in the budding pathways of these important pathogens.
Acknowledgments
We thank H.-D. Klenk, J. Paragas, A. Patnaik, R. Craven, J. Wills, and M. Sudol. Funding was provided in part to R.N.H. from the University of Pennsylvania Research Foundation.
Abbreviations
- M
matrix protein
- GST
glutathione S-transferase
- VSV
vesicular stomatitis virus
- L-domain
late budding domain
- BAP
bacterial alkaline phosphatase
- GFP
green fluorescent protein
- WT
wild type
- RSV
Rous sarcoma virus
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.250277297.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.250277297
References
- 1.Feldmann H, Klenk H D. Adv Virus Res. 1996;47:1–52. doi: 10.1016/s0065-3527(08)60733-2. [DOI] [PubMed] [Google Scholar]
- 2.Feldmann H, Klenk H D, Sanchez A. Arch Virol Suppl. 1993;7:81–100. doi: 10.1007/978-3-7091-9300-6_8. [DOI] [PubMed] [Google Scholar]
- 3.Takada A, Kawaoka Y. Trends Microbiol. 1998;6:258–259. doi: 10.1016/s0966-842x(98)01309-2. [DOI] [PubMed] [Google Scholar]
- 4.Volchkov V, Volchkova V, Eckel C, Klenk H D, Bouloy M, LeGuenno B, Feldmann H. Virology. 1997;232:139–144. doi: 10.1006/viro.1997.8529. [DOI] [PubMed] [Google Scholar]
- 5.Bukreyev A A, Volchkov V E, Blinov V M, Netesov S V. FEBS Lett. 1993;322:41–46. doi: 10.1016/0014-5793(93)81107-b. [DOI] [PubMed] [Google Scholar]
- 6.Elliott L H, Kiley M P, McCormick J B. Virology. 1985;147:169–176. doi: 10.1016/0042-6822(85)90236-3. [DOI] [PubMed] [Google Scholar]
- 7.Feldmann H, Muhlberger E, Randolf A, Will C, Kiley M P, Sanchez A, Klenk H D. Virus Res. 1992;24:1–19. doi: 10.1016/0168-1702(92)90027-7. [DOI] [PubMed] [Google Scholar]
- 8.Feldmann H, Volchkov V E, Volchkova V A, Klenk H D. Arch Virol Suppl. 1999;15:159–69. doi: 10.1007/978-3-7091-6425-9_11. [DOI] [PubMed] [Google Scholar]
- 9.Muhlberger E, Lotfering B, Klenk H D, Becker S. J Virol. 1998;72:8756–8764. doi: 10.1128/jvi.72.11.8756-8764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez A, Kiley M P, Klenk H D, Feldmann H. J Gen Virol. 1992;73:347–357. doi: 10.1099/0022-1317-73-2-347. [DOI] [PubMed] [Google Scholar]
- 11.Volchkov V E, Volchkova V A, Stroher U, Becker S, Dolnik O, Cieplik M, Garten W, Klenk H D, Feldmann H. Virology. 2000;268:1–6. doi: 10.1006/viro.1999.0110. [DOI] [PubMed] [Google Scholar]
- 12.Wool-Lewis R J, Bates P. J Virol. 1999;73:1419–1426. doi: 10.1128/jvi.73.2.1419-1426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Volchkova V A, Feldmann H, Klenk H D, Volchkov V E. Virology. 1998;250:408–414. doi: 10.1006/viro.1998.9389. [DOI] [PubMed] [Google Scholar]
- 14.Ito H, Watanabe S, Sanchez A, Whitt M A, Kawaoka Y. J Virol. 1999;73:8907–8912. doi: 10.1128/jvi.73.10.8907-8912.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muhlberger E, Weik M, Volchkov V E, Klenk H D, Becker S. J Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takada A, Robison C, Goto H, Sanchez A, Murti K G, Whitt M A, Kawaoka Y. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volchkov V E, Volchkova V A, Slenczka W, Klenk H D, Feldmann H. Virology. 1998;245:110–119. doi: 10.1006/viro.1998.9143. [DOI] [PubMed] [Google Scholar]
- 18.Volchkov V E, Feldmann H, Volchkova V A, Klenk H D. Proc Natl Acad Sci USA. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Volchkova V A, Klenk H D, Volchkov V E. Virology. 1999;265:164–171. doi: 10.1006/viro.1999.0034. [DOI] [PubMed] [Google Scholar]
- 20.Wool-Lewis R J, Bates P. J Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gill D, Banerjee A. Virology. 1986;150:308–312. doi: 10.1016/0042-6822(86)90293-x. [DOI] [PubMed] [Google Scholar]
- 22.Garnier L, Wills J W, Verderame M F, Sudol M. Nature (London) 1996;381:744–745. doi: 10.1038/381744a0. [DOI] [PubMed] [Google Scholar]
- 23.Parent L J, Bennett R P, Craven R C, Nelle T D, Krishna N K, Bowzard J B, Wilson C B, Puffer B A, Montelaro R C, Wills J W. J Virol. 1995;69:5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wills J W, Cameron C E, Wilson C B, Xiang Y, Bennett R P, Leis J. J Virol. 1994;68:6605–6618. doi: 10.1128/jvi.68.10.6605-6618.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xiang Y, Cameron C E, Wills J W, Leis J. J Virol. 1996;70:5695–5700. doi: 10.1128/jvi.70.8.5695-5700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Craven R C, Harty R N, Paragas J, Palese P, Wills J W. J Virol. 1999;73:3359–3365. doi: 10.1128/jvi.73.4.3359-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harty R N, Paragas J, Sudol M, Palese P. J Virol. 1999;73:2921–2929. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Accola M A, Strack B, Gottlinger H G. J Virol. 2000;74:5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gottlinger H G, Dorfman T, Sodroski J G, Haseltine W A. Proc Natl Acad Sci USA. 1991;88:3195–3199. doi: 10.1073/pnas.88.8.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang M, Orenstein J M, Martin M A, Freed E O. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ott D E, Coren L V, Copeland T D, Kane B P, Johnson D G, Sowder R C, II, Yoshinaka Y, Oroszlan S, Arthur L O, Henderson L E. J Virol. 1998;72:2962–2968. doi: 10.1128/jvi.72.4.2962-2968.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puffer B A, Parent L J, Wills J W, Montelaro R C. J Virol. 1997;71:6541–6546. doi: 10.1128/jvi.71.9.6541-6546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Puffer B A, Watkins S C, Montelaro R C. J Virol. 1998;72:10218–10221. doi: 10.1128/jvi.72.12.10218-10221.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Justice P A, Sun W, Li Y, Ye Z, Grigera P R, Wagner R R. J Virol. 1995;69:3156–3160. doi: 10.1128/jvi.69.5.3156-3160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Y, Luo L, Schubert M, Wagner R R, Kang C Y. J Virol. 1993;67:4415–4420. doi: 10.1128/jvi.67.7.4415-4420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sudol M. Prog Biophys Mol Biol. 1996;65:113–132. doi: 10.1016/s0079-6107(96)00008-9. [DOI] [PubMed] [Google Scholar]
- 37.Chen H I, Einbond A, Kwak S J, Linn H, Koepf E, Peterson S, Kelly J W, Sudol M. J Biol Chem. 1997;272:17070–17077. doi: 10.1074/jbc.272.27.17070. [DOI] [PubMed] [Google Scholar]
- 38.Kay B K, Williamson M P, Sudol M. FASEB J. 2000;14:231–241. [PubMed] [Google Scholar]
- 39.Macias M J, Hyvonen M, Baraldi E, Schultz J, Sudol M, Saraste M, Oschkinat H. Nature (London) 1996;382:646–649. doi: 10.1038/382646a0. [DOI] [PubMed] [Google Scholar]
- 40.Staub O, Rotin D. Structure (London) 1996;4:495–499. doi: 10.1016/s0969-2126(96)00054-8. [DOI] [PubMed] [Google Scholar]
- 41.Sudol M, Chen H I, Bougeret C, Einbond A, Bork P. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 42.Sudol M. Trends Biochem Sci. 1996;21:161–163. [PubMed] [Google Scholar]
- 43.Huibregtse J M, Scheffner M, Beaudenon S, Howley P M. Proc Natl Acad Sci USA. 1995;92:2563–2567. doi: 10.1073/pnas.92.7.2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G, Yang J, Huibregtse J M. Mol Cell Biol. 1999;19:342–352. doi: 10.1128/mcb.19.1.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huibregtse J M, Yang J C, Beaudenon S L. Proc Natl Acad Sci USA. 1997;94:3656–3661. doi: 10.1073/pnas.94.8.3656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hevey M, Negley D, Pushko P, Smith J, Schmaljohn A. Virology. 1998;251:28–37. doi: 10.1006/viro.1998.9367. [DOI] [PubMed] [Google Scholar]
- 47.Yasuda J, Hunter E. J Virol. 1998;72:4095–4103. doi: 10.1128/jvi.72.5.4095-4103.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yuan B, Li X, Goff S P. EMBO J. 1999;18:4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plant P J, Yeger H, Staub O, Howard P, Rotin D. J Biol Chem. 1997;272:32329–32336. doi: 10.1074/jbc.272.51.32329. [DOI] [PubMed] [Google Scholar]
- 50.Staub O, Dho S, Henry P, Correa J, Ishikawa T, McGlade J, Rotin D. EMBO J. 1996;15:2371–2380. [PMC free article] [PubMed] [Google Scholar]
- 51.Bonifacino J S, Weissman A M. Annu Rev Cell Dev Biol. 1998;14:19–57. doi: 10.1146/annurev.cellbio.14.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]