Abstract
The massive increase in information on the human DNA sequence and the development of new technologies will have a profound impact on the diagnosis and treatment of cardiovascular diseases. The microarray is a micro-hybridisation based assay. The filter, called microchip or chip, is a special kind of membrane in which are spotted several thousands of oligonucleotides of cDNA fragments coding for known genes or expressed sequence tags. The resulting hybridisation signal on the chip is analysed by a fluorescent scanner and processed with a software package utilising the information on the oligonucleotide or cDNA map of the chip to generate a list of relative gene expression. Microarray technology can be used for many different purposes, most prominently to measure differential gene expression, variations in gene sequence (by analysing the genome of mutant phenotypes), or more recently, the entire binding site for transcription factors. Measurements of gene expression have the advantage of providing all available sequence information for any given experimental design and data interpretation in pursuit of biological understanding. This research tool will contribute to radically changing our understanding of cardiovascular diseases.
Keywords: DNA, microarray, genome, cardiovascular disease
The collection of genes that are expressed or transcribed from genomic DNA is sometimes referred to as the expression profile or “transcriptome”. In terms of understanding the functions of genes, knowing when, where, and to what extent a gene is expressed is central to investigating the activity and biological roles of its encoded protein. In addition, changes in the multigene patterns of expression can provide clues about regulatory mechanisms and broader bioactivity functions. This knowledge may increase the basic understanding of the cause and consequences of diseases, how drugs and drug candidates work in cells and organisms, and what gene products might have therapeutic uses or may be appropriate targets for therapeutic intervention. Furthermore, profiling genomic transcripts may also assist in the identification of diagnostic indicators and prognostic markers that might direct individualised clinical management. Nevertheless, it is important to emphasise that no new approach replaces conventional methods. Indeed, standard methods such as northern blots, western blots, and reverse transcriptase polymerase chain reaction (RT-PCR) are simply used in more targeted fashion to complement the broader measurements and to follow up on the genes, pathways, and mechanisms identified by new techniques such as microarrays. Here we will illustrate the scientific rationale of the microarray technique and its implications for cardiovascular medicine on the basis of previously published work using this investigational approach.
MICROARRAY ANALYSIS
Microarray technique
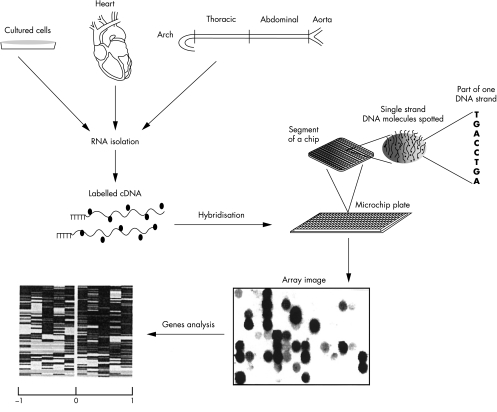
The microarray is a new microhybridisation based assay which encodes as many as 20 000 genes, offers the opportunity to make a parallel hybridisation of thousands different genes simultaneously, and thus provides unprecedented resolution for relatively unbiased genetic profiling. In analogy to hybridisation assays, it involves a microfilter or “chip” made of a porous membrane or materials such as glass, plastic, silicon, gold, or gel,1,2 in which either oligonucleotides or cDNA fragments are spotted or synthesised at high density (for example, 10 000 per cm2). Probes for the microarray can be complementary DNA (cDNA), RNA, genomic DNA, or plasmid libraries, which are appropriately labelled and hybridised to the chip.3 To measure the resulting hybridisation signals, radioactive and fluorescence detection strategies are used. The result is an image obtained by fluorescence scanner or phosphorimager and that can be processed with computer software to generate a spreadsheet of gene expression values (fig 1). The application of multiple types of statistical analysis to microarray data allows classification and clustering of genes according to their upregulation or downregulation. In principle, data can be linked to expression values to define a list of genes.4,5 Furthermore, microarrays also can be used to analyse genomic DNA rather than mRNA, to characterise the interactions of proteins,6,7 and potentially to characterise other types of molecule with double stranded DNA.8
Figure 1.
The stages of a typical microarray experiment, divided into different steps: (1) preparation of probe from different sources; (2) labelling the probe; (3) hybridisation to microchip filter; (4) hybridisation of positive signals that appear as variation of signal intensity and/or colour; (5) transformation of these signals into a list of up- or downregulated genes.
Preparation of chip microarray
One of the most important applications for arrays so far is the monitoring of the gene expression (mRNA abundance). The most significant obstacle for extraction of reproducible data from experiments is the quality of chip filter. One difficulty is spotting oligonucleotides or cDNAs at high density and high efficiency. Several strategies have been developed for this purpose. The two groundbreaking technologies were the photolithography masks for light directed synthesis of oligonucleotides (pioneered at Kirkland, Washington, USA)9 and the robotic deposition of cDNAs introduced by the Brown laboratory (address).2 Rosetta Inpharmatics (Affymetrix, Inc, Santa Clara, California, USA) developed an approach for generating flexible oligonucleotide arrays based on inkjet technology that allows synthesis of more than 25 000 different sequences on a single 2.5 × 7.5 cm glass slide.10 New techniques are also emerging that will give users the flexibility to create custom arrays. Indeed, Singh-Gasson and colleagues described the use of digitally controlled micromirrors instead of photolithographic masks for light directed synthesis of oligonucleotides in an array to increase the density of spotting.11
Data analysis: many options
Once data are collected, correct analyses have to be applied. Outputs can be displayed as lists, but are more typically visualised by some variation of the red/green light, originally introduced by Eisen and colleagues.12 Many computational analysis tools and packages are then available for data analysis of microarray derived data (some examples are listed in table 1) (also reviewed by Stoeckert and colleagues13). The statistical analysis is important to detect whether there are reliable biologically relevant differences in the expression level. Packages typically include spot quantification, data storage and retrieval, and higher level analysis. The statistical analysis is usually done using either clustering algorithms or conventional statistical tests. A selection of published clustering techniques for expression profile data analysis is listed in table 27,12,14–23 (also reviewed by Bittnerr and colleagues24). The processing of data includes background subtraction, normalisation, and detection of outliers.
Table 1.
Software packages available for gene expression analysis, categorised by package format as either enterprise-wide, software for desktop use, or useful websites
| Company | Software | URL |
| Enterprise-wide | ||
| GeneData | Expressionist | www.genedata.com |
| Informax | Genomax | www.informaxinc.com |
| LION Bioscience | ArrayScout | www.lionbioscience.com |
| NetGenics | Synergy | www.netgenics.com |
| Rosetta Inpharmatics | Rosetta resolver | www.rii.com |
| Desktop | ||
| Biodiscovery | Genesight | www.biodiscovery.com |
| Silicon Genetics | Genespring | www.sigenetics.com |
| Web based | ||
| Genome Biology | Bioarray software environment | Genomebiology.com/2002/3 |
| AMADA | Analysing microarray data | Web.hku.hk/xxia/software |
| Albert Einstein College of Medicine | GNU and GPL scatterplots | biotech.aecon.yu.edu |
| Ontario Cancer Institute | Microarray database | www.uhnres.utoronto.ca/microarray |
| European Bioinformatics Institute | ArrayExpress | www.ebi.ac.uk/arrayexpress |
| National Center Biotechnology Information NIH | Gene expression omnibus (GEO) | ncbi.nlm.nih.gov/geo |
| Microarray Gene Expression Data Society | MGED | www.mged.org |
| MIT Department of Biology | Microarray scanning | Web.mit.edu/biology/AbCam |
| Spotfire | Array explorer | www.spotfire.com |
A growing appreciation of the microarray results has led to the creation of public repositories for microarray data. More information on microarray databases is given in reference 13.
Table 2.
Analysis techniques for microarray expression profiling
| Method | Type of analysis |
| Hierarchical agglomerative | Forms clusters starting at lowest level12 |
| Hierarchical divisive | Forms clusters starting at the highest level14,15 |
| Self organising maps | Partitions genes/experiments into prespecific number of groups by mapping onto nodes16,17 |
| K-means | Partitions genes/experiments into prespecific number of groups by finding centroids7 |
| Quality cluster algorithm | Forms groups based on diameter quality measure and jackknife correlations18 |
| Super vector machines | Machine learning process that incorporates non-array information19 |
| Singular value decomposition | Groups genes/experiments by reducing data matrix to eigen vectors (similar to principal component analysis)20,21 |
| Gene shaving | Seeks groups of genes to maximise variation among experiments22 |
| Class prediction | Identifies genes whose behaviour predicts defined classes23 |
The difficulties of data analysis derive from the myriad of potential sources of random and systematic measurement error in the microarray process and from the small number of samples relative to the large number of variables (probes). This issue complicates data analysis and interpretation and jeopardises the validity of many of the microarray findings reported to date.25 For example, statistically significant differences in gene expression may reflect the biasing effects of extraneous factors rather than the biology. Lack of statistical significance can indicate low experimental sensitivity rather than absence of biological effect, while low sensitivity can be caused by an inadequate number of replicates or failure to control extraneous factors that contribute to random error. When analysing gene expression data it is also necessary to evaluate the error derived from measurements. Random error is minimised by repeating the measurements (replicates). Systematic error (bias) is controlled experimentally as far as possible, although additional statistical correction is invariably necessary with current microarray technology. Extraneous factors that contribute to random error or bias of microarray expression values include target accessibility, which is affected by variation in absorbency of discrete nylon membrane, target fixation on membrane, and variation in washing procedures.
Microarray analysis has already provided valuable insights into cancer biology and has assisted in the identification and classification of gene expression patterns in several types of cancer. Although the magnitude of changes in gene expression and the availability of tissue for testing purposes are often more limited in cardiovascular medicine, rapidly accumulating data support the considerable potential of this technique in exploring the mechanisms of disease and the effect of treatment.
Quality control for hybridisation
Multiple spotting of target DNA on a slide provides a means of assessing the quality of data for a gene on that slide. The coefficient of variation (CV) is defined as the standard deviation divided by the mean of the spot intensity for each gene.26,27 The quality of data on the expression level is inversely correlated with its CV. When a CV for one gene is greater than 10% among genes used as internal controls, the data for that gene are considered unreliable. In calibration experiments the same sample is labelled with two different dyes and hybridised to the same slide. The calibration experiment provides a control to investigate possible systemic errors, such as slide effect, dye effect, and gene label effect.26,27 The slide effect is caused by variation in imaging between slides hybridised with the same probe. Factors affecting hybridisation may include the amount of probe DNA immobilised on the slide during array fabrication and the amount of labelled cDNA added to the slide and the local environment in each hybridisation chamber. Fluorescent dyes have different efficiency incorporation during the labelling and they are detected by the scanner with different efficiencies.26,27 The effect of these factors is accounted for by the normalisation curve in normalisation experiments, in which two differentially expressed mRNA pool are separately labelled with two dyes and are co-hybridised to the same slide defining the curve calibration.26 Comparison of expression levels across different cDNA microarray experiments is easier when a common reference is co-hybridised to every microarray. Usually this reference consists of one experimental control sample, a pool of cell lines, or a mix of all samples to be analysed. A mix of the products that are printed and their subsequent amplification toward either sense and antisense cRNA provides an excellent alternative common reference.27
Bootstrapping cluster analysis
A general technique for making statistical inferences from clustering tools applied to gene expression microarray data uses an analysis of variance model to achieve normalisation and estimate differential expression of genes across multiple conditions. Statistical inferences are based on the application of a complicated randomisation technique called bootstrapping (reviewed by Kerr and Churchill14). The bootstrapping procedure, based on the reliability of clustering algorithm analysis (various simulated datasets based on the statistical models are created), relies on experimental replication and good design in microarray experiments. When accuracy is high, the bootstrap estimates of relative expression will be more like the original estimates and the bootstrap clustering will be more like the original clustering. Without an assessment of the clusters one cannot make valid inferences about genes that show similar behaviour. Whatever algorithm is chosen, it is mandatory to assess whether the results are statistically reliable relative to the noise in the data.
CARDIOVASCULAR DISEASES AND THE MICROARRAY TECHNIQUE
Endothelium and shear stress
The microarray technique has been important in investigating cellular mechanisms of vascular physiology and pathophysiology. The DNA microarray approach was used to investigate the gene expression profile in cultured endothelial cells exposed for 24 hours to shear stress.28,29 Similarly, it was shown that mechanical stimuli induced changes in transcription profile of smooth muscle cells.30 Endothelial cells exposed to shear stress for seven days increase the expression of several genes, but in particular a unique endothelial transcription factor, Kruppel-like factor (KLF-2).31 KLF-2 is a target for angiotensin II signalling and is an essential regulator of cardiovascular remodelling.32 The biological response of human endothelial cells to Gram negative lipopolysaccharide has also been assessed by microarray.33,34 These studies led through genomic investigation to the identification of molecules downstream of transforming growth factor β and tumour necrosis factor α signalling, and clarified the chemotactic pathways that participate in infection related vascular dysfunction.35,36
Vascular dysfunction, atherogenesis, and pulmonary circulation
Atherosclerosis is a multifactorial and temporally dynamic disease37,38 that can be modulated by a large number of environmental and genetic factors. The microarray technique is therefore particularly suitable for sifting through the spectrum of candidate genes in an attempt to focus on specific culprits and identify novel pathways associated with atherogenesis. For example, it was demonstrated that homocysteine can activate 3-hydroxy 2-methylglutayl coenzyme reductase in vascular endothelial cells, thereby promoting atherogenesis.39 In smooth muscle cells, a new gene pathway participating in vascular inflammation was identified that involved the eotaxin gene and its receptor CCR3.40 Chip experiments on high density lipoprotein deficient mice revealed several novel genes involved in oxidative processes, vascular dysfunction, and sterol metabolism.41 Recently, we have described a large list of early genes involved in vascular dysfunction and atherogenesis in hypercholesterolaemic mice.42 The microarray has also been used more recently to study vascular function in a murine model of hypertension. It has been used in rats which had quantitative trait loci (QTL) for blood pressure to characterise their related phenotypes on every chromosome.43 New information on gene expression profiles in spontaneously hypertensive rats and Wistar-Kyoto rats is now also available.44
The study of the pulmonary circulation has generated much interest owing to unsolved therapeutic issues such as pulmonary hypertension. Recent studies which attempted to correlate gene expression with pulmonary hypertension and tissue remodelling have also used microarrays.45–47
A very recent study identified an upregulation of collagens in a model of flow induced pulmonary vascular remodelling.48 Thus microarray technology will help us to find causal pathogenic mechanisms involved in pulmonary hypertension and vascular remodelling.
Transplantation
Chronic rejection of allografts is a major clinical and therapeutic problem during heart transplantation, and microarrays can identify early genes involved in this phenomenon.49 Microarrays undertaken on Norway rats after cardiac grafts secondary to ischaemia and reperfusion identified a large number of genes involved in this pathology.50 Although the largest numbers of downregulated genes were seen early, the ischaemia and reperfusion effects may be continuing in many transcripts that remain downregulated up to seven days after transplantation.50 In a rat model subjected to cardiac allografts, interferon (IFN) γ inducible genes were specifically upregulated, suggesting that signalling mediated by IFNγ may play an important role in the late phase of acute rejection in vivo.51 However, the fact that IFNγ deficient mice can rapidly reject cardiac allografts suggests an alternative pathway. DNA microarray analysis also revealed a new pattern of mRNA expression in allografts in IFNγ deficient mice that involved a group of specific genes including macrophage inflammatory protein, C-10-like chemokine, and platelet factor 4.52
Finally, using the Brown Norway to Lewis heterotopic heart transplant model, highly purified RNA was isolated from cardiac tissue on postoperative days 3, 5, and 7 and hybridised onto microarrays by using log average ratio (LAR) analysis.53 Of the 8800 transcripts studied, 2864 were increased on day 3, 1418 on day 5, and 2745 on day 7. Downregulated transcripts included many novel molecules such as SC1 and decorin. Thus LAR provides a useful approach to the analysis of microarray data and the results were well correlated with RT-PCR.
Myocardial infarction, hypertrophy, and heart failure
Cardiac hypertrophy, remodelling, and the progression to heart failure represent prime areas for genomic discovery. Little is known about the differentiation of the cardiomyocytes that are directly involved in cardiac disease. Microarray analysis has identified multiple novel genes and expressed sequence tags involved in the cardiovascular system54 and the genetic programme of cardiomyocyte differentiation.55,56
The programmed cell death of cardiomyocytes is a key event in the development of heart failure and is associated with chronic Akt activation in the heart.57,58 Microarray analysis helped to identify the Nix protein upregulated in myocardial hypertrophy which is necessary for apoptosis of cardiomyocytes and results in decompensation in the murine model.59 Experimental myocardial infarction in animals has many features in common with human myocardial infarction. Indeed, Stanton and colleagues found that more than 200 of 4000 genes were differentially expressed in microarray experiments after myocardial infarction in rats,60 and statistical analysis showed patterns of genes expressed in wound healing, cell signalling, and energetics. Sehl and associates also used microarrays to analyse rat myocardial infarction tissue to identify 14 novel mRNAs that changed their expression pattern significantly after infarction.61 The first examination of a human mRNA population from left ventricular tissues of patients with idiopathic dilated cardiomyopathy and from control tissue identified several genes upregulated in affected individuals.62 The gene coding for ventricular myosin light chain type 2 is a potential clinical target.63 Barrans and colleagues constructed the first human cardiovascular cDNA microarray containing 10 368 genes and selected 38 genes from failing hearts.64 Using a human cardiovascular based cDNA microarray, a molecular profile of dilated cardiomyopathy was also obtained.65 A more detailed analysis of failing versus non-failing human hearts revealed 19 of 7000 genes that were differentially expressed, including genes involved in cytoskeleton, protein turnover, and energetics.66 There is also a decreased SLIM1 expression and increased gelsolin expression in failing human hearts.67 These examples of myocardial expression profiling show that myocardial remodelling draws on multiple interactive pathways that are largely undefined.68
Heart failure resulting from dilated cardiomyopathy or hypertrophic cardiomyopathy appears to develop through different remodelling and molecular pathways.69 Microarrays of RNA samples of left ventricular tissue from patients with dilated cardiomyopathy and hypertrophic cardiomyopathy were hybridised against normal adult heart. The results showed that more than 100 genes were highly expressed in both dilated cardiomyopathy and hypertrophic cardiomyopathy, and several genes were differentially expressed.69 Thus microarray technology provides us with a genomic approach to explore the genetic markers and molecular mechanisms leading to heart failure. Moreover, there is now evidence of a divergent transcriptional responses to independent genetic causes of cardiac hypertrophy.70
Similar techniques show great potential for identifying therapeutic targets. For instance, Jin and colleagues evaluated the effects of captopril on myocardial remodelling for one day after infarction in the rat.71 The results of these experiments showed 37 differentially expressed genes. This strategy has the potential to reveal novel targets in remodelling that are not currently affected by angiotensin converting enzyme inhibitors.
Cardiovascular inflammation and cardiac metabolism
In a rat model of abdominal aortic aneurysm, microarray analysis revealed that pro-oxidant/antioxidant and inflammatory genes were increased more than twofold.72 Indeed, an intervention study with an antioxidant (AEOL 10150) showed an attenuated response of inflammatory genes in response to ischaemia and reperfusion in a mouse model.73 Similarly, a novel pathway of vascular inflammation was identified in human atherosclerosis that involved the overexpression of eotaxin and its receptor CCR3.42 Messenger RNA from rodent abdominal aortic aneurysm tissue was harvested for cDNA labelling and hybridisation to microarray.74 Twenty nine genes were differentially expressed in aneurysm tissue (relative intensity/relative intensity of control ratio > 1.5 and < 0.67), including haem oxygenase 1. Interestingly, α tocopherol was found to be as effective as flow loading in limiting aneurysm enlargement. Thus flow loading may attenuate aneurysm enlargement through wall shear or reduction in oxidative stress.
Microarrays can also be useful to explore pathways involved in cardiac metabolism. As insulin can affect a multitude of pathways in cardiovascular disease, microarrays have been used to study the effects of insulin-like growth factor 1 (IGF-1) on the gene expression profile in cardiomyocytes.75 IGF-1 modulated the expression of several functional categories, such as cell cycle, cellular respiration, and mitochondrial function. The results also showed that the majority of IGF-1 regulated genes required the activation of both ERK and PI 3 kinase. Thus PI 3 kinase and ERK coordinately mediate the transcriptional regulatory effects of IGF-1 in cardiac muscle cells. These findings provide a novel insight into how IGF-1 signalling modulates the programming of cardiac muscle gene expression.75 Nicotine is able to change the expression pattern of genes involved in energy metabolism and the signal transduction pathway. Indeed, using microarray technology demonstrated that rats fed with nicotine for three months have altered expression of genes involved in energy metabolism and signal transduction, such as the mitochondrial ATP synthase β subunit, liver mitochondrial aldehyde dehydrogenase 2, metabotropic glutamate receptor 2, calcium calmodulin dependent protein kinase II β subunit, and so on, when compared with rats fed normally.76
LIMITATIONS OF THE MICROARRAY ANALYSIS
One of the most important problems in microarray experiments is the reproducibility in interpreting gene expression results. To identify the complexity of the subcellular changes that take place and to make more rapid progress in identifying causes and cures of cardiovascular disease, we must consider the emerging high throughput gene profiling technologies. Several reports have already appeared identifying changes in classes and clusters of genes whose expression changes during cardiac remodelling hypertrophy or heart failure.60,62,77 Collectively, these studies represent a major leap forward in sorting out the different pathways in the heart or isolated cardiac myocytes, where changes in gene expression and very probably in protein expression have occurred. However, it is necessary to consider several important issues pertinent to data analysis and interpretation. To have good quality data, it is necessary at the outset to produce high quality labelled RNA or cDNA. Recently, the SMART system for synthesis of a cDNA probe was shown to produce highly reproducible results and to yield gene expression profiles that represent the majority of transcripts detected.78
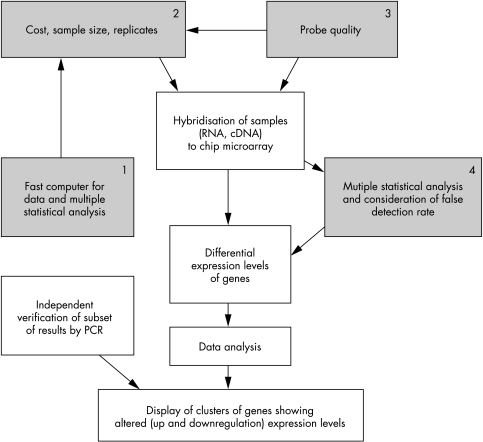
For the data analysis, it is necessary to consider several issues, the first being the volume of data. For storage and manipulation of data, gigabytes of computer storage and a fast network connection are required. Second, we need to consider the magnitude of changes in gene expression that should be considered significant. Liu and colleagues showed how increasing the number of replicates reduces false detection rates of gene expression change during cardiac remodelling.75 Another approach is to conduct multiple statistical tests and bootstrapping on the same data (see the section on statistical analysis above). In this case, the usual error rates applied to each test are no longer valid. Whereas some theoretical work along these lines has been done, it has not yet been extended to the microarray problem to any great extent.79 However, the numbers of original studies employing microarray expression profiling and the availability of computational tools for analysis of large datasets increase every year. The power of these studies is to produce vast quantities of data that represent the changes in transcription profile of cells or biopsy tissues from selected patients. However, this power is also a limitation of the arrays technique,30,80,81 as one of the most challenging aspects of gene expression analysis is the selection among the vast quantities of data those genes that could have a real causal role. Indeed, the change in the transcriptional level of a gene is not correlated with the causal role of that gene. Sometimes small changes in a key gene can produce a large biological effect. Moreover, when considering microarray data few studies address the combinatorial nature of transcription, a well established phenomenon in eukaryotes.82 In addition, changes in gene expression are not invariably associated with changes in protein synthesis, in which case the significance of altered gene expression may be questionable. Furthermore, rather than altered expression, subcellular translocalisation or post-translational modifications of proteins are also considerably meaningful for their biological effects, but would not be disclosed by microarray techniques. Thus the microarray techniques may be viewed as guiding tools to point the investigator in the right direction, with more conclusive functional information collected in subsequent complementary studies (fig 2).
Figure 2.
Flow chart showing issues that need to be addressed during the design of microarray experiments. Topics are divided in four groups: (1) availability of computers; (2) costs, samples size, and replicates; (3) verification of the probe quality; and (4) statistical analysis of results.
CONCLUSIONS
Much progress has been made in the technologies available to assess global alterations in mRNA levels in clinical research samples. Although such transcript profiling can provide a powerful research tool, the broad range of options can be bewildering for the inexperienced investigator, and more often than not the limitations and pitfalls of this approach are not fully appreciated. It is important to recognise that the major goals of transcript profiling experiments can be to identify novel therapeutic targets or delineate complex patterns of gene expression that provide a potentially pathognomonic phenotype. Furthermore, similar technologies are being used to develop antibody based protein microarray assays,83 which can detect and quantify the expression of many proteins simultaneously, and require minimal amounts of reagents and samples.84 Such assays are therefore potentially powerful tools to address the need for high throughput analysis techniques, but a careful postanalysis follow up and validation of microarray experiments will be needed soon.85 We have presented a review of the clinical and scientific applications of DNA microarray technology with an emphasis on important limitations and implications for cardiovascular medicine, and have discussed possible standards for contextual validation. A projected decrease in cost and complexity may facilitate increased availability of this technology in the physician’s office, and future applications may then allow tailoring of clinical diagnosis and treatment strategies based on individual need.
Acknowledgments
We thank Dr Wulf Palinski for his critical review of the manuscript. Our work (references 38 and 42) was supported by grant HL-56989 from the National Institutes of Health.
Abbreviations
CV, coefficient of variation
IFNγ, interferon γ
IGF-1, insulin-like growth factor 1
KLF-2, Kruppel-like factor
LAR, log average ratio
RT-PCR, reverse transcriptase polymerase chain reaction
REFERENCES
- 1.Chese M, Yang R, Hubbell E, et al. Accessing genetic information with high-density DNA array. Science 1996;224:610–14. [DOI] [PubMed] [Google Scholar]
- 2.Schena M, Shalson D, Davis RW, et al. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995;270:467–70. [DOI] [PubMed] [Google Scholar]
- 3.Gray DE, Case-Green SC, Fell TS, et al. Ellipsometric and interferometric characterization of DNA probe immobilized on a combinatorial array. Langmuir 1997;13:2833–42. [Google Scholar]
- 4.Barlund M, Monni O, Kononen J, et al. Multiple genes at 17q23 undergo amplification and overexpression in breast cancer. Cancer Res 2000;60:5340–4. [PubMed] [Google Scholar]
- 5.Virtaneva K, Wright FA, Tanner SM, et al. Expression profiling reveals fundamental biological differences in acute myeloid leukemia with isolation trisomy 8 and normal cytogenetics. Proc Natl Acad Sci USA 2001;98:1124–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roth FP, Hughes JD, Estep PW, et al. Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantification. Nat Biotechnol 1998;16:939–45. [DOI] [PubMed] [Google Scholar]
- 7.Tavazoie S, Hughers JD, Campbell MJ, et al. Systematic determination of genetic network architecture. Nat Genet 1999;22:281–5. [DOI] [PubMed] [Google Scholar]
- 8.Bulyk ML, Gentalem E, Lockhart DJ, et al. Quantifying DNA–protein interactions by double stranded DNA arrays. Nat Biotechnol 1999;17:573–7. [DOI] [PubMed] [Google Scholar]
- 9.Fodor SP, Read R, Pirrung MC, et al. Light-directed spatially addressable parallel chemical synthesis. Science 1991;251:767–73. [DOI] [PubMed] [Google Scholar]
- 10.Blanchard AP, Kaiser RJ, Hood LE. High-density oligonucleotide arrays. Biosensor Bioelectronics 1996;6/7:687–90. [Google Scholar]
- 11.Singh-Gasson S, Green RD, Yue Y, et al. Maskless fabrication of light-directed oligonucleotide microarrays using a digital micromirror array. Nat Biotechnol 1999;17:974–8. [DOI] [PubMed] [Google Scholar]
- 12.Eisen MB, Spellman PT, Brown PO, et al. Cluster analysis and display of genome wide expression patterns. Proc Natl Acad Sci USA 1998;95:14863–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoeckert CJ, Causton HC, Ball CA. Microarray databases: standards and ontologies. Nat Genet 2002;32:469–73. [DOI] [PubMed] [Google Scholar]
- 14.Kerr MK, Churchill GA. Bootstrapping cluster analysis: assessing the reliability of conclusions from microarray experiments. Proc Natl Acad Sci USA 2001;98:8961–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alon U, Barkai N, Notterman DA, et al. Broad patterns of gene expression revealed by clustering analysis of tumor and normal colon tissue probed by oligonucleotide arrays. Proc Natl Acad Sci USA 1999;96:6745–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Toronen P, Kolehmainen M, Wong G, et al. Analysis of gene expression data using self-organizing maps. FEBS Lett 1999;451:142–6. [DOI] [PubMed] [Google Scholar]
- 17.Tamayo P, Slonim D, Mesirov J, et al. Interpreting patterns of gene expression with self-organizing maps: methods and application to hematopoietic differentiation. Proc Natl Acad Sci USA 1999;96:2907–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heyer LJ, Kruglyak S, Yooseph S. Exploring expression data: identification and analysis of coexpressing genes. Genome Res 1999;9:1106–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown MP, Grundy WN, Lin D, et al. Knowledge-based analysis of microarray gene expression data by using support vector machines. Proc Natl Acad Sci USA 2000;97:262–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holter NS, Mitra M, Maritan A, et al. Fundamental patterns underlying gene expression profiles: simplicity from complexity. Proc Natl Acad Sci USA 2000;97:8409–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alter D, Brown PO, Botstein D. Singular value decomposition for genome wide expression data processing and modeling. Proc Natl Acad Sci USA 2000;97:10101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hastie T, Tibshirani R, Eisen MB, et al. Gene shaving as a method for identifying distinct sets of gene with similar expression patterns. Genome Biol 2000;1:research 00031.1–0003.21 (www). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Golub TR, Slonim DK, Tamayo P, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science 1999;286:531–7. [DOI] [PubMed] [Google Scholar]
- 24.Bittnerr M, Meltzer P, Trent J. Data analysis and integration: of steps and arrows. Nat Genet 1999;22:213–15. [DOI] [PubMed] [Google Scholar]
- 25.Miller RA, Galecki A, Shmookler-Reis RJ. Interpretation, design, and analysis of gene array expression experiments. J Gerontol Ser A–Biol Sci Med Sci 2001;56:B52–7. [DOI] [PubMed] [Google Scholar]
- 26.Tseng GC, Oh MK, Rohlin L, et al. Issues in cDNA microarray analysis: quality filtering, channel normalization, models of variations and assessment of gene effects. Nucleic Acid Res 2001;29:2549–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sterrenburg E, Turk R, Boer JM, et al. A common reference for cDNA microarray hybridization. Nucleic Acid Res 2002;30:e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen BP, Lii YS, Zhao Y, et al. DNA microarray analysis of gene expression in endothelial cells in response to 24-h shear stress. Physiol Genomics 2001;7:55–63. [DOI] [PubMed] [Google Scholar]
- 29.Garcia-Gardena G, Comander J, Anderson KR, et al. Biochemical activation of vascular endothelium as determinant of its function and phenotype. Proc Natl Acad Sci USA 2001;98:4478–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng Y, Yang JH, Huang H, et al. transcriptional profile of mechanically induced genes in human vascular smooth muscle cells. Circ Res 1999;5:1118–23. [DOI] [PubMed] [Google Scholar]
- 31.Dekker RJ, van Soest S, Fontijn RD, et al. Prolonged fluid shear stress induces a distinct set of endothelial cell genes, most specifically lung Kruppel-like factor (KLF2). Blood 2002;100:1689–98. [DOI] [PubMed] [Google Scholar]
- 32.Shindo T, Manabe I, Fukushima Y, et al. Kruppel-like zinc-finger transcription factor KLF5/BTEB2 is a target for angiotensin II signaling and an essential regulator of cardiovascular remodeling. Nat Med 2002;8:856–63. [DOI] [PubMed] [Google Scholar]
- 33.Neilsen PO, Zimmerman GA, McIntyre TM. Escherichia coli Braun lipoprotein induces a lipopolysaccharide-like endotoxic response from primary human endothelial cells. J Immunol 2000;167: 5231–9. [DOI] [PubMed] [Google Scholar]
- 34.Zhao B, Bowden RA, Stavchansky SA, et al. Human endothelial cell response to Gram-negative lipopolysaccharide assessed with cDNA microarrays. Am J Physiol Cell Physiol 2001;281:C587–95. [DOI] [PubMed] [Google Scholar]
- 35.Haley KJ, Lilly CM, Yang JH, et al. Overexpression of eotaxin and CCR3 receptor in human atherosclerosis: using genomic technology to identify a potential novel pathway of vascular inflammation. Circulation 2000;102:2185–9. [DOI] [PubMed] [Google Scholar]
- 36.Murakami T, Mataki C, Nagao C, et al. The gene expression profile of human umbilical vein endothelial cells stimulated by tumor necrosis factor alpha using DNA microarray analysis. J Atheroscler Thromb 2000;7:39–44. [DOI] [PubMed] [Google Scholar]
- 37.Ross R. Atherosclerosis – an inflammatory disease. N Engl J Med 1999;340:115–26. [DOI] [PubMed] [Google Scholar]
- 38.Palinski W, Napoli C. The fetal origins of atherosclerosis: maternal hypercholesterolemia, and cholesterol-lowering or antioxidant treatment during pregnancy influence in utero programming and postnatal susceptibility to atherogenesis. FASEB J 2002;16:1348–60. [DOI] [PubMed] [Google Scholar]
- 39.Li H, Lewis A, Brodsky S, et al. Homocysteine induces 3-hydroxy 3-methylglutaryl coenzyme a reductase in vascular endothelial cells: a mechanism for development of atherosclerosis? Circulation 2002;105:1037–43. [DOI] [PubMed] [Google Scholar]
- 40.Haley KJ, Lilly CM, Yang JH, et al. Overexpression of eotaxin and the CCR3 receptor in human atherosclerosis: using genomic technology to identify a potential novel pathway of vascular inflammation. Circulation 2000;102:2185–9. [DOI] [PubMed] [Google Scholar]
- 41.Callow MJ, Dadoit S, Gong EL, et al. Microarray expression profiling identifies genes with altered expression in HDL-deficient mice. Genome Res 2000;110:2022–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Napoli C, de Nigris F, Welch JS, et al. Maternal hypercholesterolemia during pregnancy promotes early atherogenesis in LDL receptor-deficient mice and alter aortic gene expression determined by microarray. Circulation 2002;105:1360–7. [DOI] [PubMed] [Google Scholar]
- 43.Kwitek-Black AE, Jacob HJ. The use of designer rats in the genetic dissection of hypertension. Curr Hypertens Rep 2001;3:12–18. [DOI] [PubMed] [Google Scholar]
- 44.Okuda T, Sumiya T, Iwai N, et al. Difference of gene expression profiles in spontaneous hypertensive rats and Wistar-Kyoto rats from two sources. Biochem Biophys Res Commun 2002;296:537–43. [DOI] [PubMed] [Google Scholar]
- 45.Huang W, Sher YP, Peck K, et al. Correlation of gene expression with physiological functions: Examples of pulmonary blood vessel rheology, hypoxic hypertension, and tissue remodeling. Biorheology 2001;38:75–87. [PubMed] [Google Scholar]
- 46.Leikauf GD, McDowell SA, Bachurski CJ, et al. Functional genomics of oxidant-induced lung injury. Adv Exp Med Biol 2001;500:479–87. [DOI] [PubMed] [Google Scholar]
- 47.Sheppard D: Roger S. Mitchell lecture. Uses of expression microarrays in studies of pulmonary fibrosis, asthma, acute lung injury, and emphysema. Chest 2002;121:21–5S. [DOI] [PubMed] [Google Scholar]
- 48.Medhora M, Bousamra M, Zhu D, et al. Upregulation of collagens detected by gene array in a model of flow-induced pulmonary vascular remodeling. Am J Physiol Heart Circ Physiol 2002;282:H414–22. [DOI] [PubMed] [Google Scholar]
- 49.Xu B, Sakkas LI, Slachta CA, et al. Apoptosis in chronic rejection of human cardiac allografts. Transplantation 2001;71:1137–46. [DOI] [PubMed] [Google Scholar]
- 50.Stegall MD, Park WD, Kim DY, et al. Changes in intragraft gene expression secondary to ischemia reperfusion after cardiac transplantation. Transplantation 2002;74:924–30. [DOI] [PubMed] [Google Scholar]
- 51.Saiura A, Mataki C, Murakami T, et al. A comparison of gene expression in murine cardiac allografts and isografts by means DNA microarray analysis. Transplantation 2001;72:320–9. [DOI] [PubMed] [Google Scholar]
- 52.Saiura A, Kohro T, Ymamoto T, et al. Detection of an up regulation of a group of chemokine genes in murine cardiac allograft in the absence of interferon-gamma by mean of DNA microarray. Transplantation 2002;73:1480–9. [DOI] [PubMed] [Google Scholar]
- 53.Stegall M, Park W, Kim D, Kremers W. Gene expression during acute allograft rejection: novel statistical analysis of microarray data. Am J Transplant 2002;2:913–25. [DOI] [PubMed] [Google Scholar]
- 54.Dempsey AA, Dzau VJ, Liew CC. Cardiovascular genomics: estimating the total number of genes expressed in the human cardiovascular system. J Mol Cell Cardiol 2001;33:1879–86. [DOI] [PubMed] [Google Scholar]
- 55.Peng CF, Wei Y, Levsky JM, et al. Microarray analysis of global changes in gene expression during cardiac myocyte differentiation. Physiol Genomics 2002;9:145–55. [DOI] [PubMed] [Google Scholar]
- 56.Zhao XS, Gallardo TD, Lin L, et al. Transcriptional mapping and genomic analysis of the cardiac atria and ventricles. Physiol Genomics 2002;12:53–60. [DOI] [PubMed] [Google Scholar]
- 57.Cook SA, Matsui T, Li L, et al. Transcriptional effects of chronic Akt activation in the heart. J Biol Chem 2002;277:22528–33. [DOI] [PubMed] [Google Scholar]
- 58.Steenbergen C, Afshari CA, Petranka JG, et al. Alterations in apoptotic signaling in human idiopathic cardiomyopathic hearts in failure. Am J Physiol Heart Circ Physiol 2003;284:H268–76. [DOI] [PubMed] [Google Scholar]
- 59.Yussman MG, Toyokawa T, Odley A, et al. Mitochondrial death protein Nix induced in cardiac hypertrophy and triggers apoptotic cardiomyopathy. Nat Med 2002;8:725–30. [DOI] [PubMed] [Google Scholar]
- 60.Stanton LW, Garrard LJ, Damm D, et al. Altered patterns of gene expression in response to myocardial infarction. Circ Res 2000;86:939–45. [DOI] [PubMed] [Google Scholar]
- 61.Sehl PD, Tai JT, Hillan KJ, et al. Application of cDNA microarrays in determining molecular phenotype in cardiac growth development, and response in injury. Circulation 2000;101:1990–9. [DOI] [PubMed] [Google Scholar]
- 62.Friddle CJ, Koga T, Rubin EM, et al. Expression profiling reveals distinct sets of genes altered during induction and regression of cardiac hypertrophy. Proc Natl Acad Sci USA 2000;97:6745–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haase D, Lehmann MH, Korner MM, et al. Identification and validation of selective upregulation of ventricular myosin light chain type 2 mRNA in idiopathic dilated cardiomyopathy. Eur J Heart Fail 2002;4:23–31. [DOI] [PubMed] [Google Scholar]
- 64.Barrans JD, Stamatiou D, Liew C. Construction of a human cardiovascular cDNA microarray: portrait of the failing heart. Biochem Biophys Res Commun 2001;280:964–9. [DOI] [PubMed] [Google Scholar]
- 65.Barrans JD, Allen PD, Stamatiou D, et al. Global gene expression profiling of end-stage dilated cardiomyopathy using a human cardiovascular-based cDNA microarray. Am J Pathol 2002;160:2035–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yang J, Moravec CS, Sussman MA, et al. Expression profiling reveals distinct sets of genes altered during induction and regression of cardiac hypertrophy. Proc Natl Acad Sci USA 2000;97:6745–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang J, Moravec CS, Sussman MA, et al. Decreased SLIM1 expression and increased gelsolin expression in failing human hearts measured by high-density oligonucleotide arrays. Circulation 2000;102:3046–52. [DOI] [PubMed] [Google Scholar]
- 68.Chien KR. Genomic circuits and the integrative biology of cardiac diseases. Nature 2000;407:227–32. [DOI] [PubMed] [Google Scholar]
- 69.Hwang JJ, Allen PD, Tsseng GC, et al. Microarray gene expression profiles in dilated and hypertrophic cardiomyopathic end-stage heart failure. Physiol Genomics 2002;10:31–44. [DOI] [PubMed] [Google Scholar]
- 70.Aronow BJ, Toyokawa T, Canning A, et al. Divergent transcriptional responses to independent genetic causes of cardiac hypertrophy. Physiol Genomics 2001;6:19–28. [DOI] [PubMed] [Google Scholar]
- 71.Jin H, Yang R, Awal TA, et al. Effects of early angiotensin-converting enzyme inhibition on cardiac gene expression after acute myocardial infarction. Circulation 2001;103:736–42. [DOI] [PubMed] [Google Scholar]
- 72.Yajima N, Masuda M, Miyazaki M, et al. Oxidative stress is involved in the development of experimental abdominal aortic aneurysm: a study of the transcription profile with complementary DNA microarray. J Vasc Surg 2002;36:379–85. [DOI] [PubMed] [Google Scholar]
- 73.Bowler R, Sheng H, Enghild J, et al. A catalytic antioxidant (AEOL 10150) attenuates expression of inflammatory genes in stroke. Free Radic Biol Med 2002;33:1141. [DOI] [PubMed] [Google Scholar]
- 74.Nakahashi TK, Hoshina K, Tsao PS, et al. Flow loading induces macrophage antioxidative gene expression in experimental aneurysms. Arterioscler Thromb Vasc Biol 2002;22:2017–22. [DOI] [PubMed] [Google Scholar]
- 75.Liu TJ, Lai Hc, Wu W, et al. Developing a strategy to define the effects of insulin-like growth factor-1 on gene expression profile in cardiomyocytes. Circ Res 2001;88:1231–8. [DOI] [PubMed] [Google Scholar]
- 76.Hu D, Cao K, Peterson-Wakeman R, et al. Altered profile of gene expression in rat hearts induced by chronic nicotine consumption. Biochem Biophys Res Commun 2002;297:729. [DOI] [PubMed] [Google Scholar]
- 77.Taylor LA, Carthy CM, Yang D, et al. Host gene regulation during coxsackievirus B3 infection in mice/assessment by microarray. Circ Res 2000;87:328–34. [DOI] [PubMed] [Google Scholar]
- 78.Vernon SD, Unger ER, Rajeevan M, et al. Reproducibility of alternative probe synthesis approaches for gene expression profiling with arrays. J Mol Diagn 2000;3:124–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rao JS, Bond M. Microarrays managing the data deluge. Circ Res 2001;88:1226–7. [DOI] [PubMed] [Google Scholar]
- 80.McCormick SM, Eskin SG, McIntire LV, et al. DNA microarray reveals changes in gene expression of shear stressed human umbilical vein endothelial cells. Proc Natl Acad Sci USA 2001;98:8955–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Butte A. The use and analysis of microarray data. Nat Rev Drug Discov 2002;1:951–60. [DOI] [PubMed] [Google Scholar]
- 82.Pilpel Y, Sudarsanam P, Church GM. Identifying regulatory networks by combinatorial analysis of promoter elements. Nat Genet 2001;29:153–9. [DOI] [PubMed] [Google Scholar]
- 83.Huang RP. Detection of multiple proteins in an antibody-based protein microarray system. J Immunol Methods 2001;255:1–13. [DOI] [PubMed] [Google Scholar]
- 84.Haab BB. Advances in protein microarray technology for protein expression and interaction profiling. Curr Opin Drug Discov Devel 2001;4:116–23. [PubMed] [Google Scholar]
- 85.Chuaqui RF, Bonner RF, Best CJM, et al. Post-analysis follow-up and validation of microarray experiments. Nat Genet 2002;32:509–14. [DOI] [PubMed] [Google Scholar]