Abstract
One of the ways a cell can rapidly and tightly regulate gene expression is to target specific mRNAs for rapid decay. A number of mRNA instability sequences that mediate rapid mRNA decay have been identified, particularly from multicellular eukaryotes, but pinpointing the cellular components that play critical roles in sequence-specific decay in vivo has been more difficult. In contrast, general pathways of mRNA degradation in yeast have been well established through the analysis of mutants affecting the general mRNA decay machinery. Strategies to isolate mutants in sequence-specific mRNA decay pathways, although extremely limited so far, have the potential to be just as powerful. In the study reported here, a selection in transgenic plants allowed the isolation of rare mutants of Arabidopsis thaliana that elevate the abundance of mRNAs that contain the plant mRNA instability sequence called DST (downstream element). This instability sequence is highly conserved in unstable small auxin up RNA (SAUR) transcripts. Genetic analysis of two dst mutants isolated via this selection showed that they are incompletely dominant and represent two independent loci. In addition to affecting DST-containing transgene mRNAs, mutations at both loci increased the abundance of the endogenous DST-containing SAUR-AC1 mRNA, but not controls lacking DST sequences. That these phenotypes are caused by deficiencies in DST-mediated mRNA decay was supported by mRNA stability measurements in transgenic plants. Isolation of the dst mutants provides a means to study sequence-specific mRNA degradation in vivo and establishes a method to isolate similar mutants from other organisms.
The wide range of mRNA decay rates observed in eukaryotic cells allows enhanced control over the expression of specific genes. For example, some genes with very specific temporal and spatial expression patterns have been found to encode unstable mRNAs, which facilitates this tight control. In cases where these mRNAs have been examined in detail, specific cis-acting sequence elements have been identified that target transcripts for rapid degradation (1).
Most studies of mRNA degradation in multicellular eukaryotes have focused on genes encoding unstable transcripts that must be expressed transiently. The early response genes of mammalian cells are prominent examples. AU-rich elements (AREs), found in the 3′ untranslated regions (UTRs) of many of the corresponding transcripts, have been studied in great detail and represent the archetypal mRNA instability determinant (2). Although AREs can be classified into different types based on sequence, many contain multiple overlapping AUUUA motifs. Plants also recognize multiple overlapping AUUUA sequences as instability determinants (3).
In contrast to AREs, the plant instability sequence DST (downstream element) is not AU-rich, lacks AUUUA sequences, and has only been reported to function in plants thus far. The DST element is found in the 3′ UTR of the small auxin up RNA (SAUR) transcripts, which are among the most unstable mRNAs in plants, having half-lives of 10–50 min (4–6). On the basis of their expression properties, the SAUR genes have been implicated in auxin-induced cell elongation (6), and their transcripts appear to be constitutively unstable so that their abundance can be rapidly altered in response to transcriptional control by the plant hormone auxin (7). All unstable SAUR transcripts contain conserved DST sequences, so-called because of their location downstream of the coding region (5). A dimer of the DST element (8) or the 3′ end of the Arabidopsis thaliana SAUR-AC1 gene, which contains a DST element (7), are sufficient to destabilize reporter transcripts in plant cells. The DST sequence is approximately 45 nt in length and is comprised of three highly conserved domains separated by two variable regions. Mutagenesis studies have demonstrated that residues within two of the conserved domains, the ATAGAT and the GTA regions, are necessary for instability function (9). However, nothing is known about the genes required for DST-mediated mRNA decay.
Work in yeast has focused mainly on elucidating general, rather than sequence-specific, mRNA decay mechanisms. Through a combination of elegant molecular genetic and biochemical approaches, it is now clear that the major pathway responsible for the turnover of both unstable and stable mRNAs is the deadenylation-dependent decapping pathway (10). The initial event is shortening of the poly(A) tail, which is followed by decapping and subsequent 5′ to 3′ degradation of the body of the transcript. Genes encoding the proteins that catalyze the decapping (DCP1 and cofactor DCP2) and 5′ to 3′ exoribonuclease (XRN1) steps have been cloned (11). Analysis of mutant yeast strains that lack these genes was critical in establishing the steps in this pathway and in verifying the functions of proposed components of the mRNA decay machinery in vivo (11). Molecular genetic tools also have been extremely effective for the study of the accelerated decay of transcripts containing premature nonsense codons, not only in yeast (12) but also in Caenorhabditis elegans (13). Nonsense-mediated decay also occurs in mammalian (14) and plant (15) systems although it is unclear how many of the genetic determinants or the mechanisms will be the same as in yeast.
Aside from nonsense-mediated decay in C. elegans, genetic approaches are rarely applied to the study of mRNA stability in multicellular organisms, in which pathways mediated by specific cis-acting instability sequences have been the focus. Most studies aimed at understanding the cellular factors involved in these pathways have involved characterization of proteins that bind the instability sequences in vitro. Over the years, this approach has resulted in the identification and subsequent cloning of several genes that encode ARE-binding proteins (2). Transient transfections of overexpression constructs have begun to provide evidence for an in vivo role for some of these (16–18) but mutations affecting the corresponding endogenous genes that could aid in the interpretation of the overexpression data are generally lacking. One exception resulted from the fortuitous finding that mice deficient for the Zfp-36 gene product, tristetraprolin, are defective in rapid degradation of the tumor necrosis factor-α mRNA, an ARE-containing mRNA (19). Tristetraprolin was later found to be an ARE-binding protein (20), and more recently it has been shown that tristetraprolin-deficient mice are also defective in the deadenylation of the ARE-containing granulocyte–macrophage colony-stimulating factor mRNA (21). Deadenylation is thought to be the first step in the ARE-mediated mRNA degradation pathway (22). These studies highlight how a single mutant can provide important insights into rapid mRNA degradation pathways mediated by specific cis-regulatory elements in eukaryotes. However, further analysis of sequence-specific mRNA decay pathways would be improved by forward genetic approaches that are aimed at isolating multiple mutants defective in a given pathway. The recent isolation of mutant mammalian cell lines that stabilize green fluorescent protein-IL-3 reporter transcripts (23) emphasizes the potential of this approach, but the design of high-throughput mutant selections in genetically tractable transgenic systems would be advantageous for future analysis. To this end, we devised a selection approach in transgenic plants that facilitated the isolation of mutants of Arabidopsis with defects in DST-mediated mRNA degradation. The properties of these mutants indicate that they will provide powerful tools for analysis of the mechanism of DST-mediated mRNA degradation and that similar transgene-based selections should be applicable to sequence-specific decay pathways in other eukaryotes.
Materials and Methods
Gene Constructions and Plant Transformation.
p1493 and p1519 are pMON505 (24) derivatives and were constructed as described (9), with the following exceptions. The β-globin coding region was replaced by the hygromycin phosphotransferase (HPH) coding region, obtained as a BamHI fragment of pLG90 (25). In p1519, four tandem copies of the soybean SAUR 15A DST element were inserted into the 3′ UTR of the HPH and β-glucuronidase (GUS) genes between the coding sequence and the poly(A) signal.
Agrobacterium tumefaciens-mediated Arabidopsis transformation has been described (26). The Columbia accession was transformed with p1493. To avoid any hygromycin-resistant contamination of the parental line during large-scale selection experiments, Arabidopsis Columbia that lacks trichomes [gl1 Col-PRL, referred to in the text as WT (wild type) with respect to mRNA stability], was transformed with p1519.
Mapping of the 1519–31 and dst1 Loci.
Mapping of the 1519–31 transgene locus and the dst1 (mutant defective in DST-mediated mRNA degradation) locus was carried out by using simple sequence length polymorphism (SSLP) markers (27). To map the 1519–31 locus, F3 families derived from a cross between 1519–31 and Landsberg erecta (gl1–1) (28) were scored for kanamycin resistance. Fifteen families that showed 100% kanamycin resistance were identified. Linkage was detected only to markers on chromosome II, the closest marker being nga1126 (position 51, 0 recombinants of 30 chromosomes analyzed).
To map the dst1 locus, leaves of F2 plants from a cross of dst1 (from the second backcross to p1519–31) with Landsberg erecta (gl1–1) were scored by Northern blot analysis for 3-fold or higher HPH-DST mRNA abundance compared with WT(1519–31). Thirty dst1/dst1 F2 plants were identified. Linkage was again detected with nga1126 (0 recombinants of 16 chromosomes analyzed) because of the necessity of the p1519 transgene for scoring the mutant phenotype. In addition, linkage was detected to markers on chromosome I as follows: nga59 (map position 1.6, 5 recombinants of 60 chromosomes analyzed), nga63 (map position 9.4, 4/60), nga248 (map position 40.1, 12/60), and nga128 (map position 81.4, 22/60).
Mutagenesis and Selection of Hygromycin-Resistant Plants.
Seeds homozygous at the 1519–31 locus (fourth generation of self-fertilized progeny after transformation) were treated with ethyl methanesulfonate (EMS) as described (29). The effectiveness of EMS mutagenesis was evaluated by scoring the occurrence of albinism and embryo lethality in M1 plants as described (29). M1 refers to the first mutagenized generation, and M2 refers to the progeny of self-fertilized M1 plants. M1 seeds were planted in groups of approximately 5,000, and M2 seed were collected from each group of M1 plants. A total of 52 groups of M2 seed from approximately 260,000 M1 plants were collected.
Putative mutants were selected on seed selection medium (30) supplemented with 1,000 μg/ml hygromycin and 50 μg/ml kanamycin and were defined as seedlings that had developed at least two true leaves after 3–4 weeks of selection.
Sequencing of DST Elements in dst1 and dst2.
Oligonucleotides complementary to the 3′ end of the HPH coding sequence (GATGCAGCTTGGGCGCAGGG) and the 5′ end of the E9 3′ UTR (CCCAATGCCATAAATACTCG) were used to amplify DST elements from genomic DNA extracted from dst1 and dst2 [F2 plants from the first backcross to WT (1519–31)] by PCR (Pfu, Stratagene) under standard cycling conditions. Products were cloned and multiple isolates were sequenced for each mutant.
RNA Analysis.
RNA extraction, Northern blot procedures, and quantitation of mRNA abundance using a Molecular Dynamics PhosphorImager were essentially as reported (8, 9) with exceptions noted below. Hybridization probes corresponding to the eIF4A (30), SAUR-AC1 (31), GUS (8), and HPH [BamHI fragment of pLG90 (25)] coding regions were labeled by using 32P dCTP (32).
Genetic Analysis.
The dst mutant phenotype was scored by analyzing HPH-DST mRNA abundance. Total RNA was isolated from two rosette leaves from individual plants (33). Northern blots containing 10 μg of total RNA were hybridized sequentially with radiolabeled HPH and eIF4A probes. The ratio of HPH/eIF4A signal was calculated for each plant in a given population and compared with that of WT(1519–31) plants analyzed in parallel.
Half-Life Analysis.
Approximately 500 1493–2 and 1519–31 seedlings were grown in 250 ml of liquid medium (23.2 g Gamborg's B-5 Medium/liter; pH = 5.0) for 12 days with gentle shaking in a 500-ml flask. Actinomycin D (75 μg/ml) was added, and samples were removed from the cultures and frozen in liquid nitrogen at 30-min intervals.
The half-life of the HPH mRNA in mutant and WT(1519–31) plants was measured in rosette leaves of mature plants (≈40 days old). This protocol was adapted from previous reports (34, 35). Rosette leaves from 12 F3 plants derived from the second backcross of dst1 and dst2 to WT(1519–31) were harvested in parallel with 12 WT(1519–31) rosette leaves. Leaves were incubated in buffer (15 mM sucrose/1 mM KCl/1 mM Pipes/1 mM sodium citrate, pH 6.5) for 30 min before addition of cordycepin (150 μg/ml). A brief (30 sec) vacuum was applied to each sample. Four leaves were harvested every 15 min for 60 min and frozen in liquid nitrogen. Northern blot analysis was performed as above. Standard methods were used to calculate mRNA half-life (1).
Results
Mutant Selection.
To identify mutations in the DST-mediated mRNA decay pathway, a mRNA abundance phenotype was engineered by using a transgenic approach. A binary vector was constructed (p1519) that contained a tetramer of the DST element inserted into the 3′ UTR of the HPH and GUS genes (Fig. 1A). HPH confers resistance to hygromycin and GUS expression is readily detected by incubation of plant tissue with substrates that are cleaved to colorimetric or fluorescent products (25, 36). WT (i.e., Columbia gl-1 plants that were WT with respect to mRNA degradation) plants harboring this construct were anticipated to have low HPH and GUS expression because of the impact of the DST element on mRNA turnover rates and therefore would have decreased hygromycin resistance and GUS activity. Conversely, a mutant with a defective trans-acting factor in the DST-mediated mRNA decay pathway would show increased hygromycin resistance and GUS activity. Plants transformed with a second binary vector (p1493, Fig. 1A) that was identical to p1519, except DST sequences were not included, served as a nondestabilized control throughout our experiments. Both constructs contained the nptII gene, to enable selection of transgenic plants, and were introduced via Agrabacterium tumefaciens-mediated transformation (26).
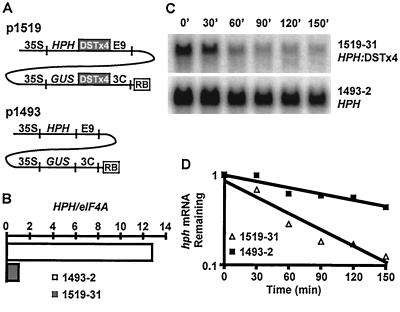
Figure 1.
The basis for the dst mutant selection. (A) Diagrams representing two T-DNAs, p1519 and p1493, are shown. 35S, cauliflower mosaic virus 35S promoter; E9, pea ribulose 1,5-bisphosphate carboxylase small subunit E9 polyadenylation signal; 3C pea ribulose 1,5-bisphosphate carboxylase small subunit 3C polyadenylation signal; RB, right border sequence of T-DNA (this T-DNA functions without a left border). The nptII gene is carried on both T-DNAs, but is not depicted. (B) Comparison of the steady-state abundance of the HPH transcripts in 1519–31 and 1493–2 seedlings. HPH mRNA abundance at time 0 (first lane) of the half-life determination shown in C was quantitated and normalized by using the abundance of the eIF4A message. Normalized HPH abundance is shown relative to 1519–31 seedlings. (C) Comparison of the stability of HPH transcripts in 1519–31 and 1493–2 seedlings after addition of actinomycin D. (D) Signals obtained from Northern blots were quantitated and plotted on a semilog scale. Linear regression analysis is shown for each set of points. Results shown in C and D are from one experiment that is representative of several.
Line 1519–31 was chosen for mutagenesis because segregation of kanamycin resistance (conferred by the nptII gene) and Southern blot analysis indicated that it harbored a single insert of the transfer DNA (T-DNA; data not shown). In addition, the abundance of the HPH-DST and GUS-DST messages in this line reflected the average abundance of these transcripts in a population of 1519 transformants. The T-DNA insertion in 1519–31 was mapped to chromosome II near the molecular marker nga1126 (map position 51). The line chosen to serve as the nondestabilized control in this study (p1493–2) had HPH and GUS expression levels that were representative of a pool of several p1493 transgenic lines and appeared to be a single locus insertion based on segregation of kanamycin resistance. Expression levels in p1493–2 and p1519–31 were stable and showed no evidence of gene silencing over multiple generations.
The DST tetramer resulted in an approximately 12.5-fold decrease in steady-state HPH mRNA abundance in 1519–31 seedlings relative to 1493–2 (Fig. 1B). Based on the known mRNA instability function of the DST sequence, this difference was presumed to be caused by decreased mRNA stability of the DST-containing transgene mRNA. To examine this, the decay of the HPH transcript encoded by 1519–31 and 1493–2 was compared in seedlings grown in liquid medium after the addition of the transcriptional inhibitor, actinomycin D (Fig. 1B). The 1519–31 HPH transcript, containing the DST tetramer, was degraded approximately three times faster than the 1493–2 HPH transcript that did not contain DST sequences (Fig. 1 C and D). The disparity between the effect of the DST sequence on the steady-state abundance and the half-life of the HPH transcript (12.5-fold vs. 3-fold) is most likely caused by dampening that is commonly observed when general inhibitors of transcription are used to measure mRNA decay rates (10, 35, 37–39). Nonetheless, it is clear that the DST element has reduced the half-life of the HPH mRNA in 1519–31.
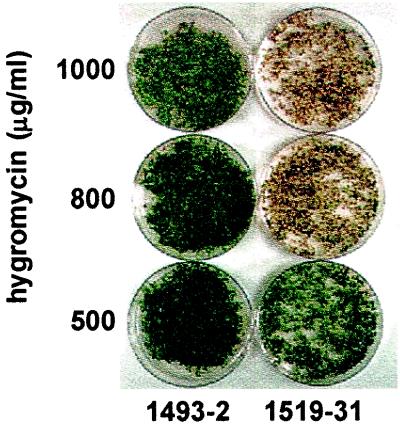
Destabilization of the HPH transcript by the DST element resulted in reduced resistance to hygromycin in 1519–31 seedlings relative to 1493–2 seedlings, particularly at higher concentrations of the antibiotic (Fig. 2). We hypothesized that mutants defective in DST-mediated mRNA degradation would have increased abundance of the HPH-DST transcript and therefore would have increased resistance to hygromycin relative to the parental line. Because we were interested in mutations affecting the DST pathway in trans, our goal was to identify mutants with increases in both HPH-DST and GUS-DST mRNA abundance.
Figure 2.
A titration of hygromycin resistance was performed on 1493–2 and 1519–31 seedlings. Approximately 500 seeds were surface-sterilized and plated on primary seed selection media containing 500, 800, or 1,000 μg/ml hygromycin.
To induce mutations in line 1519–31, seeds were subjected to EMS mutagenesis, and M2 seeds were collected from groups of M1 plants and plated on seed selection media containing 1,000 μg/ml hygromycin. Of ≈730,000 M2 seeds that were plated, 323 seedlings that appeared to exhibit increased resistance to hygromycin were transferred to soil for propagation. M3 seeds from these lines were plated on selective media, and approximately 20% of the putative mutants displayed heritable hygromycin resistance. In most of these lines, hygromycin resistance was not attributed to an increase in HPH-DST mRNA abundance and therefore may have been caused by a defect in hygromycin uptake. However, two mutants, derived from independent M1 groups, showed a heritable increase in HPH-DST mRNA abundance. These two mutants, referred to as dst because they are defective in DST-mediated mRNA degradation, also exhibited an increase in GUS-DST mRNA abundance.
Genetic Analysis of dst1 and dst2.
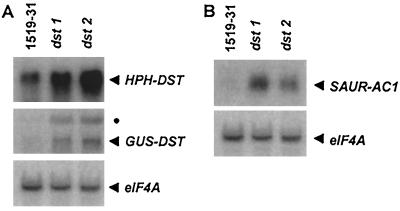
The abundance of the HPH-DST and GUS-DST transcripts was 3- to 4-fold higher in dst1 and dst2 compared with WT(1519–31) plants (Fig. 3A). In contrast, the abundance of the eIF4A transcript, used as a non-DST-containing control, was equivalent in mutants and WT(1519–31) (Fig. 3A). Other transcripts, encoding a subunit of RNA polymerase II (AtRPB15.9) and aconitase, that lack DST elements were also equally abundant in the mutants and WT(1519–31) (data not shown). Increased HPH-DST and GUS-DST mRNA abundance is a consistent phenotype that has been observed in the progeny of four backcrosses of dst1 and dst2 to WT(1519–31). Because both the HPH-DST and GUS-DST transcripts were elevated in these mutants, it is highly unlikely that the phenotype is caused by a mutation in one or more of the DST elements present in the 3′UTR of the HPH-DST transcript. To examine this possibility directly, the DST tetramer was amplified by PCR from genomic DNA extracted from dst1 and dst2 and was sequenced. The DST tetramers in both mutants were found to be unaltered (data not shown).
Figure 3.
(A) The abundance of DST-containing reporter transcripts, HPH-DST and GUS-DST, are elevated in dst1 and dst2. The higher molecular weight GUS transcript (●), which is approximately twice the expected size, is a consistent characteristic of 1519–31. It is most likely the result of a partial recognition of a cryptic downstream polyadenylation signal that is activated by incomplete integration of the T-DNA. We have measured the abundance of the lower molecular weight band (the expected size) throughout these experiments, although the two bands are coordinately regulated in the mutants. (B) The abundance of an endogenous DST-containing message, SAUR-AC1, is elevated in dst1 and dst2. Samples were not treated with exogenous auxin. Total RNA was prepared from leaves of WT(1519–31), dst1, and dst2 (F2 plants from the second backcross to p1519–31) and analyzed by Northern blot hybridization. Radiolabeled probes were prepared against the indicated transcripts and were used in sequential hybridizations of one Northern blot.
Genetic analysis indicated that dst1 and dst2 are incompletely dominant mutations in independent single genes. To arrive at this conclusion, the inheritance of the dst mutations was examined by measuring HPH-DST mRNA abundance in the rosette leaves of mature plants derived from crosses of dst1 and dst2 to WT(1519–31). Northern blots were used for these studies rather than relying on reporter protein activity because direct measurements were more sensitive to the differences in mRNA abundance observed between the mutants and WT(1519–31). Progeny of the second backcross to WT(1519–31) (F1) and the progeny of self-fertilizations of these plants (F2) were used in these studies as presented in Table 1. Ten F1 plants were examined for each mutant. HPH-DST mRNA abundance was on average 2.1 ± 0.22-fold higher in dst1 F1 and 2.8 ± 0.23-fold higher in dst2 F1 plants compared with WT(1519–31). These levels of HPH-DST mRNA abundance are intermediate between WT(1519–31) and mutant levels as would be expected if heterozygotes showed a partial mutant phenotype. Segregation of increased HPH-DST mRNA abundance in the F2 populations was also consistent with incomplete dominance. Of 46 F2 plants segregating dst1, nine were observed with high HPH-DST mRNA abundance [3.1 ± 0.17-fold higher than WT (1519–31)], 23 had intermediate HPH-DST mRNA abundance (1.7 ± 0.07) and 14 showed HPH-DST mRNA abundance that was similar to WT(1519–31) (0.9 ± 0.04). Segregation of the dst2 mutant phenotype was similar to that of dst1. Of 53 F2 plants analyzed, 13 fell into the mutant class (4.0 ± 0.16), 28 fell into the intermediate class (2.2 ± 0.11), and 12 were similar to WT(1519–31) (1.0 ± 0.04). Segregation in dst1 and dst2 F2 populations was most easily explained by a ratio of 1:2:1 (WT/intermediate/mutant) as would be expected for inheritance of a incompletely dominant single gene (Table 1).
Table 1.
Genetic analysis of mutants defective in DST-mediated mRNA degradation
| Cross | Class | HPH* | n | χ2; P† | |
|---|---|---|---|---|---|
| DST1/DST1 × dst1/dst1 | F1 | Int | 2.1 ± 0.22 | 10 | |
| DST2/DST2 × dst2/dst2 | F1 | Int | 2.8 ± 0.23 | 10 | |
| DST1/DST1 × dst1/dst1 | F2 | WT | 0.9 ± 0.04 | 14 | |
| Int | 1.7 ± 0.07 | 23 | 1.1; P > 0.1 | ||
| High | 3.1 ± 0.17 | 9 | |||
| DST2/DST2 × dst2/dst2 | F2 | WT | 1.0 ± 0.04 | 12 | |
| Int | 2.2 ± 0.11 | 28 | 0.2; P > 0.1 | ||
| High | 4.0 ± 0.16 | 13 |
Int, intermediate.
HPH mRNA abundance: (HPH/eIF4A) segregating class/(HPH/eIF4A)wt ± standard error.
χ2 calculated for 1∶2∶1 segregation of WT/Int/High HPH mRNA abundance DST/DST, WT(1519-31).
Crossing dst1 with dst2 yielded F1 plants with intermediate HPH-DST mRNA abundance, indicating that dst1 and dst2 are not allelic (data not shown). When the F2 plants from this cross were analyzed, 10/51 showed HPH-DST mRNA abundance similar to WT(1519–31). Had the dst mutations been in the same gene, or if they were tightly linked, no F2 plants with HPH-DST mRNA abundance similar to WT(1519–31) would have been expected. There was no class of F2 plants observed with an additive effect on HPH-DST mRNA abundance, as might be expected if dst1 and dst2 function independently to destabilize DST-containing messages. In addition, the finding that F1 plants from the dst1 × dst2 cross showed an intermediate mRNA abundance phenotype indicated a lack of additive interaction between these loci because the individual mutants crossed to WT(1519–31) showed the same phenotype in the F1.
To begin the characterization of the molecular basis of these mutant phenotypes, the dst1 locus was mapped by using molecular markers (27). This analysis showed that dst1 is located on chromosome I, near molecular markers nga59 and nga63.
Phenotypic Analysis of dst1 and dst.
If dst1 and dst2 are indeed mutations that disrupt the DST-mediated mRNA degradation pathway in trans, one would expect the abundance of endogenous DST-containing messages to be elevated in addition to the DST-containing transgene mRNAs. Therefore, the abundance of the SAUR-AC1 transcript, the only DST-containing message in Arabidopsis that has been examined at the level of mRNA stability (7), was analyzed in mutants and WT(1519–31) plants. As is clearly seen in Fig. 3B, the abundance of this message is elevated in dst1 and dst2, indicating that these mutations may elevate the abundance of any message destabilized by DST sequences.
Neither of the dst mutants had any obvious defects in development or morphology. Because expression of at least one auxin-responsive gene, SAUR-AC1, was altered in dst1 and dst2, mutant phenotypes associated with the production and/or perception of auxin were analyzed. dst1 and dst2 displayed normal root gravitropism as indicated by the wavy root assay (40) or by rotating vertical plates after germination of seedlings (41). Additionally, there was no loss or enhancement of apical dominance in these plants, nor were there alterations in root length or morphology, other phenotypes that have been associated with auxin response mutants (42).
Increased Stability of HPH-DST mRNA in dst1 and dst2 Compared with WT(1519–31).
The simplest explanation for the elevation of DST-containing transcripts in dst1 and dst2 is a defect that results in a decreased rate of DST-mediated mRNA degradation. This is likely because the DST sequence is the only regulatory element shared by the three transcripts that were elevated in the mutants. To test this hypothesis, we measured the kinetics of HPH-DST mRNA degradation in WT(1519–31), dst1, and dst2 plants.
The measurable differences in HPH-DST mRNA decay rates were anticipated to be minimal because the difference in HPH-DST abundance between dst1 or dst2 and WT(1519–31) was 3- or 4-fold, respectively (Fig. 3A, Table 1), and differences in mRNA abundance are often not fully reflected in differences in mRNA half-lives when measured by using general inhibitors of transcription (10, 35, 37–39), as mentioned earlier. For example, in the case of the HPH mRNA, we observed that the 12.5-fold increase in mRNA abundance of the HPH and HPH-DST mRNA was reflected as a 3-fold increase in measured mRNA stability (Fig. 1 B and C). These measurements indicate that differences in half-life measurements (3-fold − 1-fold = 2-fold difference) are dampened and only reflect about 17% of the expected difference based on steady-state mRNA abundance (12.5-fold − 1-fold = 11.5-fold difference; 2/11.5 = 0.17). Because the abundance of the HPH mRNA is about 3-fold higher in WT(1519–31) than mutants (3-fold − 1-fold = 2-fold difference) we would expect the measured mRNA half-life to be increased to 1.34 times that of WT(1519–31) [(2-fold difference × 0.17) +1].
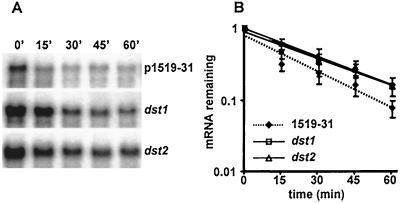
Three independent experiments were performed in which excised rosette leaves from WT(1519–31), dst1, and dst2 plants were incubated with a transcriptional inhibitor. Cordycepin was used to inhibit transcription in these experiments because a recent report showed that it was more effective than actinomycin D at blocking transcription in leaf tissue (35). Leaves were sampled every 15 min, and HPH mRNA abundance at each time point was measured by Northern blot analysis. HPH mRNA abundance was standardized relative to the abundance of the eIF4A transcript. Fig. 4A shows representative blots from one of these experiments. The amount of mRNA remaining at each time point was calculated, and the average of the three values for each time point is plotted in Fig. 4B.
Figure 4.
Analysis of HPH-DST mRNA stability in leaves from WT(1519–31), dst1, and dst2 plants after addition of cordycepin. (A) Representative Northern blots showing the HPH-DST signal over the 60-min time course in WT(1519–31), dst1, and dst2. (B) Average normalized HPH-DST signals (± SEM) from the three time courses are plotted on a semilog scale. Linear regression analysis is shown for each set of points.
The decay of the HPH-DST mRNA in both dst1 and dst2 (upper curves in Fig. 4B, lower two blots in Fig. 4A) was clearly slower than WT(1519–31). As expected, the differences in decay kinetics become particularly evident at the later time points (45 and 60 min). The average half-life of the HPH mRNA was calculated to be 1.33-fold greater in dst1 and 1.28-fold greater in dst2 compared with WT(1519–31). This difference between the half-life of the HPH mRNA in mutants and WT(1519–31) was almost identical to what was anticipated based on previous experiments that compared the stability of the HPH mRNA with the HPH-DST mRNA (Fig. 1 B–D). Based on these observations we conclude that the dst1 and dst2 mutations result in an increase in mRNA stability that results in increased abundance of DST-containing mRNAs.
Discussion
Using a transgene-based mutant selection strategy, we isolated mutants of Arabidopsis defective in the rapid mRNA degradation pathway mediated by the DST element. These mutants were selected by virtue of increased resistance to hygromycin that resulted from an increase in HPH-DST mRNA abundance and then were screened for increased GUS-DST mRNA to avoid cis mutations. The two dst mutants that exhibited these heritable phenotypes also elevated the abundance of an endogenous DST-containing transcript encoded by SAUR-AC1, further arguing that they each affected a trans-acting factor important for DST-mediated mRNA degradation in vivo. Analysis of HPH-DST mRNA decay kinetics in mutants versus WT(1519–31) indicated that increased mRNA abundance of DST-containing transcripts in dst1 and dst2 can be explained by an increase in message stability. Before this work, it was unclear whether a selection aimed at identifying the genetic determinants of sequence-specific mRNA decay in transgenic organisms would be successful. Therefore beyond identifying loci that provide experimental access to the DST-mediated mRNA decay machinery in plants, this work provides a valuable approach that may have similar impact in other eukaryotes.
The dst mutant selection had two interesting characteristics. First, the mutants were rare, having been isolated at a frequency of approximately 1/300,000. The stringency of the selection and secondary screens may have been partially responsible because they were designed to eliminate all mutants that did not elevate both HPH and GUS expression at the RNA level (e.g., hygromycin transport mutants). However, this cannot be the only explanation because in Arabidopsis, mutations in nonessential genes are typically expected at frequencies of ≈1/2,000 in similar EMS-mutagenized populations (43). Second, the dst mutations did not fully restore the abundance of the HPH-DST or GUS-DST transcripts to the levels found in plants with nondestabilized transcripts. The abundance of the HPH-DST mRNA was 3- to 4-fold higher in mutants compared with WT(1519–31) whereas the nondestabilized HPH message was greater than 10-fold higher in abundance than the HPH-DST mRNA. These observations indicate that dst1 and dst2 may be relatively weak alleles that allow partial function of the DST-mediated mRNA decay pathway. Functional redundancy among components that recognize the DST instability determinant may have contributed to the low frequency of sequence-specific decay mutants.
Perhaps it is by necessity rather than coincidence that both dst mutations were found to be incompletely dominant (Table 1). The DST element is comprised of three conserved domains that are separated by two variable regions (8). Two of these conserved domains are essential for instability function (9), but it is not known whether they function as independent targets or as a single target for cellular factors that recognize the DST element. Incompletely dominant mutant phenotypes could arise if defective DST-recognition factors no longer recognize the DST sequence, but still bind interacting partner proteins required to mediate mRNA decay. Conversely, an incompletely dominant phenotype could result if a defective DST-binding protein retained the ability to bind DST but lacked the ability to interact with or recruit proteins required for mRNA degradation. Another alternative would be that a dst gene product encodes a DST-specific ribonuclease that binds the DST sequence but has decreased RNase activity, thereby providing partial protection to mRNAs bound by the mutant version.
dst1 and dst2 may be rare and weak alleles because regulation of the abundance of DST-containing mRNAs is an essential function in Arabidopsis and stronger alleles were lethal. This may also explain why neither of the dst mutants had an obvious developmental or morphological mutant phenotype. Thus far, the DST element has been implicated experimentally only in the regulation of the SAUR genes (7). Although the function of the SAUR gene products is unknown, the expression patterns of these genes suggest an early role in auxin-induced cell elongation (6). Because auxin has a central role in many aspects of plant physiology, there may be a narrow tolerance for misexpression of auxin response genes such as the SAURs. Alternatively, the lack of apparent auxin-related phenotypes in the mutants may be explained by redundancy among the approximately 60 members of the Arabidopsis SAUR gene family, some of which may not encode unstable mRNAs or contain DST elements (Thomas Guilfoyle, personal communication). It is also likely that the stability of other plant transcripts is regulated by the DST element as well. DNA microarray experiments that compare the abundance of thousands of mRNAs in dst mutants and WT should prove useful in identification of new transcripts that are regulated by the DST pathway.
Few approaches designed to isolate mutations in genes encoding trans-acting factors involved in sequence-specific mRNA decay pathways have been reported. Mutant screens in yeast and C. elegans, although quite successful, have focused on general (5′ to 3′, 3′ to 5′ or nonsense-mediated) decay pathways. Aside from the present study, the only approach that has led to the isolation of sequence-specific decay mutants was the screen conducted in human HT1080 fibrosarcoma cells for stabilized green fluorescent protein-IL-3 mRNA cited in the introduction (23). Because the latter system is not amenable to most genetic approaches, cloning the corresponding genes may be challenging. By using a selection rather than a screen and working with transgenic Arabidopsis, our approach represents an important advance that has distinct advantages for future analysis. Most notably, isolation of the dst mutants should facilitate the cloning of trans-acting factor genes involved in DST-mediated mRNA degradation through map-based procedures available in Arabidopsis. The ability to study the roles of the DST genes in intact plants via crosses to other mutants and introduction of specialized transgenes will offer additional potential for addressing mechanistic questions. Finally, based on the work presented here, similar selections for mutants defective in rapid mRNA degradation mediated by cis-regulatory elements should be feasible in other eukaryotic model organisms if selectable markers that facilitate detection of small expression differences can be engineered.
Acknowledgments
We thank Dr. Ambro van Hoof, Dr. Michael Thomashow, and James Kastenmayer for critical reading of the manuscript and Dr. Joanne Chory for providing pLG90. We also thank Dr. Michael Sullivan and Debrah Thompson for construction of p1519 and p1493 and Jonathan Vogel and Linda Danhof for technical assistance. This work was funded by grants from the United States Department of Energy (FG0291-ER20021), the United States Department of Agriculture (9801498 and 2000–01491), and the McKnight Foundation (to P.J.G.). M.A.P.-A. received postdoctoral fellowships from North Atlantic Treaty Organization-Spain and the Ministerio de Educación y Ciencia, Spain.
Abbreviations
- DST
downstream element
- dst
mutant defective in DST-mediated mRNA degradation
- ARE
AU-rich element
- UTR
untranslated region
- SAUR
small auxin up RNA
- EMS
ethyl methanesulfonate
- HPH
hygromycin phosphotransferase
- GUS
β-glucuronidase
- WT
wild type
- T-DNA
transfer DNA
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.240354097.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.240354097
References
- 1.Ross J. Microbiol Rev. 1995;59:423–450. doi: 10.1128/mr.59.3.423-450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen C Y A, Shyu A B. Trends Biochem Sci. 1995;20:465–470. doi: 10.1016/s0968-0004(00)89102-1. [DOI] [PubMed] [Google Scholar]
- 3.Ohme-Takagi M, Taylor C B, Newman T C, Green P J. Proc Natl Acad Sci USA. 1993;90:11811–11815. doi: 10.1073/pnas.90.24.11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McClure B A, Guilfoyle T J. Plant Mol Biol. 1989;12:517–524. doi: 10.1007/BF00036966. [DOI] [PubMed] [Google Scholar]
- 5.McClure B A, Hagen G, Brown C S, Gee M A, Guilfoyle T J. Plant Cell. 1989;1:229–239. doi: 10.1105/tpc.1.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McClure B A, Guilfoyle T J. Science. 1989;243:91–93. doi: 10.1126/science.11540631. [DOI] [PubMed] [Google Scholar]
- 7.Gil P, Green P J. EMBO J. 1996;15:1678–1686. [PMC free article] [PubMed] [Google Scholar]
- 8.Newman T C, Ohme-Takagi M, Taylor C B, Green P J. Plant Cell. 1993;5:701–714. doi: 10.1105/tpc.5.6.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan M L, Green P J. RNA. 1996;2:308–315. [PMC free article] [PubMed] [Google Scholar]
- 10.Caponigro G, Parker R. Microbiol Rev. 1996;60:233–249. doi: 10.1128/mr.60.1.233-249.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He W, Parker R. Methods. 1999;17:3–10. doi: 10.1006/meth.1998.0701. [DOI] [PubMed] [Google Scholar]
- 12.Hilleren P, Parker R. RNA. 1999;5:711–719. doi: 10.1017/s1355838299990519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Culbertson M R. Trends Genet. 1999;15:74–80. doi: 10.1016/s0168-9525(98)01658-8. [DOI] [PubMed] [Google Scholar]
- 14.Sun X, Maquat L E. RNA. 2000;6:1–8. doi: 10.1017/s1355838200991660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Hoof A, Green P J. Plant J. 1996;10:415–424. doi: 10.1046/j.1365-313x.1996.10030415.x. [DOI] [PubMed] [Google Scholar]
- 16.Fan X H C, Steitz J A. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peng S S Y, Chen C Y A, Xu N H, Shyu A B. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loflin P, Chen C Y A, Shyu A B. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carballo E, Lai W S, Blackshear P J. Science. 1998;281:1001–1005. doi: 10.1126/science.281.5379.1001. [DOI] [PubMed] [Google Scholar]
- 20.Lai W S, Carballo E, Strum J R, Kennington E A, Phillips R S, Blackshear P J. Mol Cell Biol. 1999;19:4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carballo E, Lai W S, Blackshear P J. Blood. 2000;95:1891–1899. [PubMed] [Google Scholar]
- 22.Shyu A-B, Belasco J G, Greenberg M E. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 23.Stoecklin G, Ming X F, Looser R, Moroni C. Mol Cell Biol. 2000;20:3753–3763. doi: 10.1128/mcb.20.11.3753-3763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers S G, Klee H J, Horsch R B, Fraley R T. Methods Enzymol. 1987;153:253–277. [Google Scholar]
- 25.Gritz L, Davies J. Gene. 1983;25:179–188. doi: 10.1016/0378-1119(83)90223-8. [DOI] [PubMed] [Google Scholar]
- 26.Bariola P A, MacIntosh G C, Green P J. Plant Physiol. 1999;119:331–342. doi: 10.1104/pp.119.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bell C J, Ecker J R. Genomics. 1994;19:137–144. doi: 10.1006/geno.1994.1023. [DOI] [PubMed] [Google Scholar]
- 28.Koornneef M, Dellaert L W, van der Veen J H. Mutat Res. 1982;93:109–123. doi: 10.1016/0027-5107(82)90129-4. [DOI] [PubMed] [Google Scholar]
- 29.Lightner J, Caspar T. Methods Mol Biol. 1998;82:91–103. [PubMed] [Google Scholar]
- 30.Taylor C B, Bariola P A, DelCardayré S B, Raines R T, Green P J. Proc Natl Acad Sci USA. 1993;90:5118–5122. doi: 10.1073/pnas.90.11.5118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gil P, Liu Y, Orbovic V, Verkamp E, Poff K L, Green P J. Plant Physiol. 1994;104:777–784. doi: 10.1104/pp.104.2.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Feinberg A P, Vogelstein B. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 33.Verwoerd T C, Dekker B M, Hoekema A. Nucleic Acids Res. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seeley K A, Byrne D H, Colbert J T. Plant Cell. 1992;4:29–38. doi: 10.1105/tpc.4.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holtorf H, Schöb H, Kunz C, Waldvogel R, Meins F., Jr Plant Cell. 1999;11:471–483. doi: 10.1105/tpc.11.3.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jefferson R A, Kavanagh T A, Bevan M A. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shyu A-B, Greenberg M E, Belasco J G. Genes Dev. 1989;3:60–72. doi: 10.1101/gad.3.1.60. [DOI] [PubMed] [Google Scholar]
- 38.Belasco J G, Brawerman G. In: Control of Messenger RNA Stability. Belasco J, Brawerman G, editors. San Diego: Academic; 1993. pp. 475–493. [Google Scholar]
- 39.Zhang S, Sheng J, Liu Y, Mehdy M C. Plant Cell. 1993;5:1089–1099. doi: 10.1105/tpc.5.9.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okada K, Shimura Y. Aust J Plant Physiol. 1992;19:439–448. [Google Scholar]
- 41.Bell C J, Maher E P. Mol Gen Genet. 1990;220:289–293. [Google Scholar]
- 42.Boerjan W, Cervera M T, Delarue M, Beeckman T, Dewitte W, Bellini C, Caboche M, Van Onckelen H, Van Montagu M, Inze D. Plant Cell. 1995;7:1405–1419. doi: 10.1105/tpc.7.9.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Haughn G, Somerville C R. In: Biotechnology in Agricultural Chemistry. LeBaron H M, Mumma R O, Honeycutt R C, Duesing J H, editors. Washington, DC: Am. Chem. Soc.; 1987. pp. 98–107. [Google Scholar]