Abstract
Background/Aims: p33ING1b is a tumour suppressor protein involved in growth control and apoptosis. Suppression of p33ING1b expression is associated with the loss of cellular growth control and immortalisation, whereas its overexpression causes cell cycle arrest. Moreover, normal p33ING1b expression is essential for optimal function of p53. Acute lymphoblastic leukaemia (ALL) is the most common malignancy of childhood, accounting for one third of all childhood malignancies. A variety of cytogenetic abnormalities have been described but there is no single abnormality common to all cases. Deregulation of the TP53 pathway is a common genetic abnormality in human malignancies. However, TP53 mutations are uncommon in ALL. It is possible that alternative mechanisms of regulation of the TP53 apoptosis pathway, such as modulation of p33ING1b expression, may be important in ALL. The aim of this study was to assess the expression of p33ING1b in childhood ALL.
Methods: One hundred and forty five patients with childhood ALL were investigated in this immunohistochemical study of the expression of p33ING1b.
Results: Loss of nuclear expression of p33ING1b was seen in 78% of cases. This was associated with increased cytoplasmic expression of the protein. Kaplan Meier survival analysis demonstrated a trend towards a better prognosis for patients with tumours that had lost nuclear p33ING1b.
Conclusion: These results suggest that the loss of nuclear p33ING1b expression may be an important molecular event in the pathogenesis of childhood ALL.
Keywords: ING1, p33ING1b, p53, leukaemia
Garkavtsev et al were the first to describe the inhibitor of growth 1 (ING1) gene in 1996.1 Since then, it has been mapped to a locus on the long arm of chromosome 13 (13q33–34).2,3 The ING1 gene consists of three exons that encode a series of spliced mRNAs,4,5 which may be translated to produce three proteins: p47ING1a, p33ING1b, and p24ING1c. 6–8 The most widely expressed of these proteins appears to be p33ING1b. 7–9
Functional studies have shown that pro ducts of the ING1 gene are involved in the restriction of cell growth and proliferation, apoptosis, anchorage dependence, cellular senescence, the maintenance of genomic stability, and the modulation of cell cycle checkpoints.10–13 Suppression of p33ING1b is associated with the loss of cellular growth control and immortalisation, whereas its overexpression arrests cells in the G0/G1 phase of the cell cycle.
Co-immunoprecipitation studies have indicated that ING1 gene protein products interact with the TP53 tumour suppressor gene protein product p53, whereas co-transfection studies demonstrate that ING1 has the ability to modulate TP53 transactivation of the cyclin dependent kinase inhibitor WAF1.14 Extension of these preliminary findings suggested that the association of competent protein forms of each member of the ING1–TP53 complex is essential for optimum inhibition of cell growth and transactivation by TP53.14
We have shown by means of immunohistochemistry that p33ING1b is expressed in the nuclei of cells in almost all human tissue types, with cytoplasmic expression in a more restricted range of tissues. Recent studies have reported reduced ING1 mRNA expression in lymphoid malignancies, gastrointestinal tumours, and breast carcinomas.15–17 In contrast, mutations in ING1 appear to be extremely rare in breast carcinoma, colorectal carcinoma, head and neck tumours, squamous carcinoma, and other tumours.18–22
“Loss of p21WAF1 might lead to failure of cellular growth suppression, possibly through deregulation of the retinoblastoma gene”
Acute lymphoblastic leukaemia (ALL) is the most common malignancy of childhood, accounting for one third of all childhood malignancies.23 Approximately 2500 new cases of ALL are diagnosed each year in the USA.24 ALL occurs more frequently in individuals less than 15 years of age, and it is more common in males and white individuals.25 ALL is composed of different neoplastic subtypes of immature precursor lymphoblasts. For what was once a fatal disease, remission is now induced in 95% of cases of ALL and the five year survival rate is 70%.26,27 This incredible improvement in response is mainly the result of advances in multi adjuvant chemotherapy and supportive care.
About 90% of patients have a chromosomal abnormality in the leukaemic cells, such as hyperdiploidy (trisomy 12), deletions (13q), gene rearrangements (TAL1), and translocations (t12;21, t9;22, t9;14, and t4;11). A range of cell cycle inhibitors and regulators have been studied for their potential role in the development and progression of ALL, such as members of the INK gene family (p16INK4a, p15INK4b), the CIP/KIP family member p21WAF1, the retinoblastoma gene (RB), and the tumour suppressor p53.28–30 Loss of p16INK4a and p15INK4b expression are frequent in childhood ALL and result in deactivation of the RB gene. Moreover, loss of p21WAF1 might lead to failure of cellular growth suppression, possibly through deregulation of the RB gene.
Because TP53 mutations are uncommon in ALL at diagnosis,31 and deletion of chromosome 13q is common in ALL, it is possible that deregulation of the p53 apoptosis pathway by modulation of p33ING1b expression may be an important molecular mechanism in ALL. Therefore, we undertook our study to investigate the expression of p33ING1b in 145 patients with childhood ALL.
MATERIALS AND METHODS
Patients and tissues studied
The patients studied comprise a series of 145 cases of ALL diagnosed at the Royal Victoria Infirmary, Newcastle upon Tyne, UK, between 1985 and 1997. Bone marrow trephine biopsies were taken at diagnosis. Follow up data, including age, sex, presenting white blood cell (WBC) count and survival data were available for all patients. All patients were entered into clinical trials established by the Medical Research Council to assess the relative qualities of different treatment regimens for childhood ALL. These were UKALL X (n = 61), UKALL XI (n = 24), and UKALL XI’92 (n = 60). All patients received standard induction treatment containing daunorubicin and vincristine, with supplementary methotrexate in intensification blocks.32 Normal human bone marrow trephine specimens and normal human thymus were used as positive controls for immunohistochemistry.
Monoclonal antibodies used
Monoclonal antibodies used in our study were anti-p53 (NCL-DO7; Novocastra, Newcastle, UK) and anti-p33ING1b (GN1), which was generated in our laboratories.9 The GN1 monoclonal antibody was generated using a full length p33ING1b recombinant protein as an immunogen in the production and selection of specific monoclonal antibody secreting hybridoma cell lines.
Statistical analysis
To assess the relation between p33ING1b, p53, WBC count, sex, age, and CD10 status, several statistical tests were applied. Relations between the continuous variables were assessed by Spearman’s test for rank correlations. In addition, the continuous variables were categorised by splitting each into two groups (high and low) around the median, and then compared using Fisher’s exact test.
For survival analysis, Kaplan-Meier step graphs were assembled and survival differences compared using the log rank test. The data were analysed with respect to p33ING1b nuclear and cytoplasmic expression, sex, age, WBC count, ALL phenotype, and CD10 status. All analysis was carried out using SPSS statistical software.
Immunohistochemistry
Multiple 5 μm sections were cut from paraffin wax embedded, formalin fixed normal and tumour tissue blocks. All sections were mounted on slides pretreated with APES in the conventional manner. A standard streptavidin–biotin–peroxidase complex (Dako, Ely, Cambridgeshire, UK) method was used. First, sections were dewaxed in xylene and then rehydrated by immersion in 99%, 95%, and 70% ethanol. Endogenous peroxidases were blocked by pretreatment of sections with a 0.5% solution of hydrogen peroxide in methanol for 10 minutes; the sections were then washed in running tap water. Antigen retrieval was achieved by pressure cooking for one minute in citrate buffer (200mM citric acid, 500mM NaOH, pH 6.0). Sections were then transferred into Tris buffered saline (TBS; 140mM NaCl, 50mM Tris/HCl, pH 7.6) for five minutes. The sections were then blocked with 10% normal rabbit serum in TBS for 10 minutes. Excess serum was removed and sections were covered and incubated at room temperature with the appropriate primary monoclonal antibody at the appropriate dilution for 60 minutes (GN1 at a 1/200 dilution and NCL-DO7 at a 1/20 dilution). Sections were washed twice with TBS for five minutes each, then incubated with the secondary biotinylated rabbit antimouse antibody (Dako) at a 1/500 dilution for 45 minutes at room temperature. After this, the sections were washed twice for five minutes each with TBS, then covered with the tertiary streptavidin–biotin–peroxidase antibody (Dako) at a 1/100 dilution for 45 minutes at room temperature. After rinsing twice for five minutes each with TBS, the peroxidase activity was developed with 3,3‘-diaminobenzidine (Sigma, Poole, Dorset, UK) 1 mg/ml in sterile distilled water-H2O2 solution for five minutes. The sections were then washed in tap water, counterstained with haematoxylin, then dehydrated and mounted in DPX.
Scoring
Tissue scoring was carried out by two observers, one an experienced histopathologist. Tissues were visualised by conference light microscopy, the cell type was identified, and the proportions and intensities of nuclear and cytoplasmic staining for p33ING1b were estimated. In the case of p53, only nuclear staining was assessed. Intensity was assessed on a 4 point scale, as follows: 0, negative; 1, weak; 2, intermediate; and 3, strong staining. The percentage of cells staining was assessed on a 6 point scale, as follows: 1, 0–4%; 2, 5–19%; 3, 20–39%; 4, 40–59%; 5, 60–79%; and 6, 80–100%. The scores for intensity and proportion were then multiplied to give a composite score from 0–18, according to the method of Detre et al.33
RESULTS
General
The series comprised 89 boys and 56 girls between the ages of 0 and 14 years. One hundred and thirty four patients were classed as CD10 positive at diagnosis, whereas 31 were negative. The presenting WBC count for the whole series ranged from 1.4 to 1000 × 109/litre, with a mean value of 48.94 × 109/litre. There were 115 patients with B cell ALL (B-ALL), 16 with T cell ALL (T-ALL), nine with pre-B-ALL, three with non-B non-T-ALL, and two patients were classed as biphenotypic.
p33ING1b expression
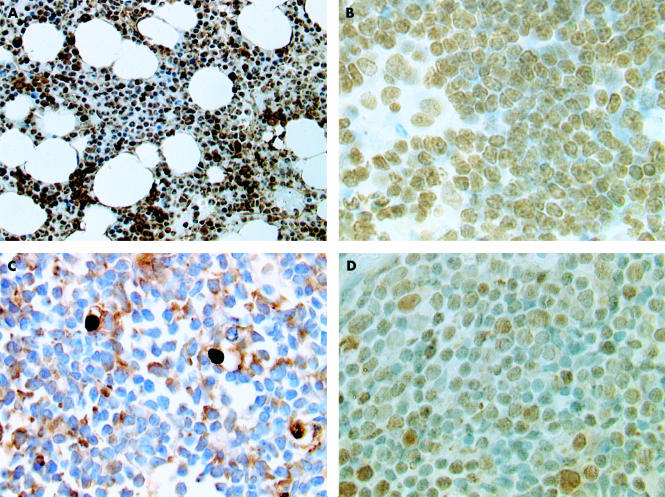
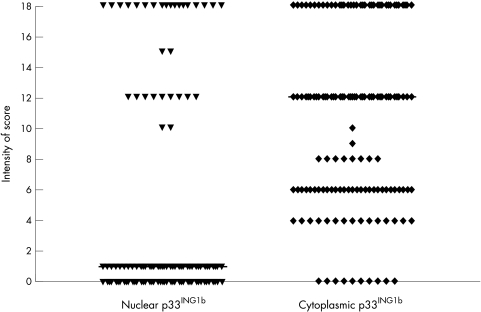
Analysis of normal tissue showed strong uniform expression of p33ING1b in the nuclei of normal cells in bone marrow trephines (fig 1A). Strong nuclear expression was also seen in terminal deoxytransferase positive precursor lymphoblastic cells in normal thymus and bone marrow (fig 1B). There was no cytoplasmic p33ING1b expression in the normal tissue. In contrast, p33ING1b was lost from the nuclei in 111 of the 145 patients with ALL (figs 1C,2), and replaced in all 111 cases by increased cytoplasmic expression of p33ING1b.
Figure 1.
The immunohistochemical patterns of expression of p33ING1b in normal tissue and acute lymphoblastic leukaemia (ALL). (A) Normal bone marrow showing nuclear expression of p33ING1b in almost all cells. (B) Normal thymus showing p33ING1b expression in precursor lymphoid cells. (C) ALL bone marrow trephine showing loss of p33ING1b from tumour nuclei with increase in cytoplasmic expression. (D) ALL bone marrow trephine showing p53 overexpression.
p53 expression
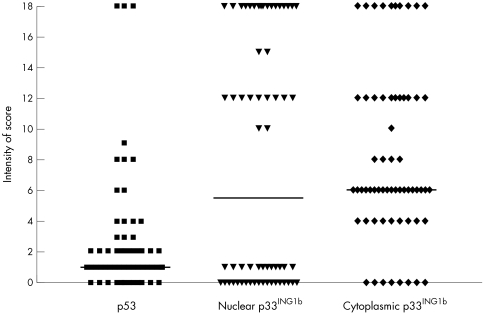
A sample of 68 cases was selected randomly, 34 of which were positive for p33ING1b, whereas the remaining 34 cases were negative. They were then assessed for the expression of p53 (fig 3). Most cases showed absent or weak labelling; only three cases in the series showed strong labelling (fig 1D).
Figure 3.
Scattered graph plot showing patterns of expression of p53 and p33ING1b in 68 patients with acute lymphoblastic leukaemia (ALL). Three cases showed strong staining, and most cases showed weak or intermediate staining for p53. The horizontal lines correspond to the median.
Relations between variables
The data were analysed both as continuous and categorical variables. A positive correlation was observed between p33ING1b nuclear expression and p53 expression (r = 0.349; p = 0.0035), and inverse correlations were found between p33ING1b nuclear and cytoplasmic labelling (r = −0.445; p < 0.0001), and between p33ING1b cytoplasmic labelling and p53 labelling (r = −0.313; p = 0.0092). No other associations were found. Moreover, there was no difference in the proportion of cases showing loss of p33ING1b in T-ALL compared with B-ALL, although it was found that all cases classed as pre-B-ALL or biphenotypic were p33ING1b negative (table 1).
Table 1.
The distribution of p33ING1b expression according to ALL subtype
| Type of ALL | Total cases | p33ING1b +ve | p33ING1b -ve |
| B-ALL | 115 | 28 | 87 |
| T-ALL | 16 | 3 | 13 |
| Pre-B-ALL | 9 | 0 | 9 |
| Non-B, non-T | 3 | 1 | 2 |
| Biphenotypic | 2 | 0 | 2 |
There was no difference in the proportion of cases showing loss of p33ING1b in T-ALL compared with B-ALL, although all cases classed as pre-B-ALL or biphenotypic were p33ING1b negative.
ALL, acute lymphoblastic leukaemia; B-ALL, B cell acute lymphoblastic leukaemia; T-ALL, T cell acute lymphoblastic leukaemia.
Survival analysis
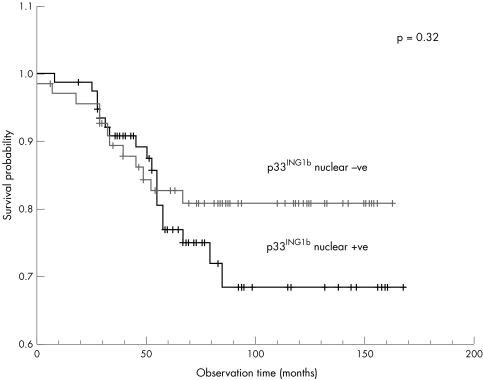
Considering the series as a whole, low WBC count (p = 0.007), CD10 positivity (p = 0.029), age 4–6 at presentation (p = 0.07), and female sex (p = 0.09) were associated with a better survival. There was a trend towards improved survival in patients in whom ALL was assessed as nuclear p33ING1b negative (p = 0.32; fig 4).
Figure 4.
Kaplan Meier survival analysis for patients with acute lymphoblastic leukaemia (ALL) (n = 145) according to p33ING1b nuclear expression. A trend towards better survival was seen in patients who had lost p33ING1b nuclear expression (p = 0.32).
DISCUSSION
Suppression of p33ING1b has been associated with the loss of cellular growth suppression control and resistance to apoptosis, whereas its overexpression has been linked with the inhibition of cell proliferation and enhanced apoptosis.10 The involvement of p33ING1b in the p53 signalling pathway provides a potential mechanism by which it may further function as a tumour suppressor. Loss of p33ING1b has been shown to contribute to altered cell growth control and resistance to apoptosis.12 Nuclear localisation of p33ING1b was first shown by Garkavtsev et al,2 and in our laboratory we have subsequently shown that p33ING1b is expressed in the nuclei of most normal human cells and tissues.9
In our study, p33ING1b was lost from tumour cell nuclei in 78% of bone marrow trephines taken from patients with ALL at presentation. There was no significant difference in the proportion of cases of different phenotypes that showed loss of p33ING1b. Loss of nuclear p33ING1b expression was associated with a higher cytoplasmic expression. Our results suggest a cellular compartment shift from the nucleus to the cytoplasm in ALL. p33ING1b contains several important structural motifs, such as nuclear and nucleolar localisation signals and a PHD domain, which indicate that it functions in the nucleus.14,22,34 In addition, its ability to bind and modulate the transcriptional activity of p53 depends on nuclear localisation. Therefore, it seems likely that translocation of p33ING1b from the nucleus to the cytoplasm would result in the loss of its tumour suppressor activity. The optimal function of p53 is dependent upon p33ING1b and therefore loss of nuclear p33ING1b would be predicted to compromise p53 function. This may be a factor in the deregulation of growth control that occurs in acute lymphoblastic leukaemia cells. It is possible that impaired function of p33ING1b represents an alternative mechanism for abrogation of functional p53 in ALL. This hypothesis is supported by our finding of a weak but significant correlation between p33ING1b and p53 expression, because p53 over expression is associated with TP53 gene mutations. In our study, p53 over expression was demonstrated in three of 68 cases. This is in keeping with the incidence of TP53 gene mutations in ALL at diagnosis.31 However, this would clearly require further analysis including TP53 sequencing because the relation between p53 immunostaining and mutations is not absolute.
“Our results suggest a cellular compartment shift from the nucleus to the cytoplasm in acute lymphoblastic leukaemia”
We are presently investigating a range of other types of tumours and an equivalent pattern of p33ING1b cellular compartment shift is evident in melanoma, thyroid, colon, and breast carcinomas.35 Others have reported changes in the subcellular localisation of p33ING1b, with a shift from the nucleus to nucleolus seen in HS68 fibroblast cell lines after exposure to UV light.34 Thus, it appears that reduced nuclear expression of p33ING1b may be important in several other human neoplasms.36 We have also cloned cDNAs corresponding to each of the reported ING1 mRNA isoforms into eukaryotic expression vectors and are currently investigating their independent functions.
The analysis of a variety of known prognostic factors in this series indicated that age 4–6 years, female sex, a low WBC count at presentation, and CD10 positivity were associated with a good outcome. Survival curves illustrating these data have been published previously.37 These results are consistent with other published studies.25 We found that 79.64% of patients with tumours that had lost nuclear p33ING1b survived compared with 78% of patients with tumours that had not lost nuclear p33ING1b. However, this was not significantly different in a log rank test (p = 0.32).
Take home messages.
Nuclear expression of p33ING1b was lost in 78% of cases of childhood acute lymphoblastic leukaemia (ALL) and was associated with increased cytoplasmic expression of the protein
Patients with tumours that had lost nuclear p33ING1b demonstrated a trend towards a better prognosis on Kaplan Meier survival analysis
Thus, the loss of nuclear p33ING1b expression may be an important molecular event in the pathogenesis of childhood ALL
Our study has shown that 78% of ALL cases studied showed loss of nuclear expression of p33ING1b, which was associated with cytoplasmic expression. The shift in the subcellular localisation (nuclear to cytoplasmic) may cause the loss of normal cellular function of the protein. These results support the view that p33ING1b may function as a tumour suppressor, and suggest that the loss of nuclear p33ING1b may be an important molecular event in the pathogenesis of childhood acute lymphoblastic leukaemia.
Figure 2.
Scattered graph plot showing the patterns of p33ING1b nuclear and cytoplasmic expression in 145 cases of childhood acute lymphoblastic leukaemia (ALL). Seventy eight per cent of patients with ALL had lost p33ING1b expression in the nuclei, and this was associated with an increased cytoplasmic p33ING1b expression. The horizontal lines correspond to the median.
Abbreviations
ALL, acute lymphoblastic leukaemia
B-ALL, B cell acute lymphoblastic leukaemia
ING1, inhibitor of growth 1
RB, retinoblastoma gene
T-ALL, T cell acute lymphoblastic leukaemia
TBS, Tris buffered saline
WBC, white blood cell
REFERENCES
- 1.Garkavtsev I, Kazarov A, Gudkov A, et al. Suppression of the novel growth inhibitor p33 (ING1) promotes neoplastic transformation. Nat Genet 1996;14:415–20. [DOI] [PubMed] [Google Scholar]
- 2.Garkavtsev I, Demetrick D, Riabowol K. Cellular localization and chromosome mapping of a novel candidate tumor suppressor gene (ING1). Cytogenet Cell Genet 1997;76:176–8. [DOI] [PubMed] [Google Scholar]
- 3.Zeremski M, Horrigan SK, Grigorian IA, et al. Localization of the candidate tumor suppressor gene ING1 to human chromosome 13q34. Somat Cell Mol Genet 1997;23:233–6. [DOI] [PubMed] [Google Scholar]
- 4.Baranova AV, Ivanov DV, Makeeva NV, et al. Genomic organization of a tumor growth inhibitor gene ING1. Mol Biol 2000;34:232–6. [PubMed] [Google Scholar]
- 5.Gunduz M, Ouchida M, Fukushima K, et al. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res 2000;60:3143–6. [PubMed] [Google Scholar]
- 6.Garkavtsev I. Suppression of the novel growth inhibitor p33 (ING1) promotes neoplastic transformation. Nat Genet 1999;23:373–73. [DOI] [PubMed] [Google Scholar]
- 7.Jager D, Stockert E, Scanlan MJ, et al. Cancer-testis antigens and ING1 tumor suppressor gene product are breast cancer antigens: characterization of tissue-specific ING1 transcripts and a homologue gene. Cancer Res 1999;59:6197–204. [PubMed] [Google Scholar]
- 8.Saito A, Furukawa T, Fukushige S, et al. p24/ING1-ALT1 and p47/ING1-ALT2, distinct alternative transcripts of p33/ING1. J Hum Genet 2000;45:177–81. [DOI] [PubMed] [Google Scholar]
- 9.Nouman GS, Angus B, Lunec J, et al. Monoclonal antibodies raised to products of the ING1 tumour suppressor gene locus. J Pathol 2000;192:26A.10951396 [Google Scholar]
- 10.Garkavtsev I, Riabowol K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33(ING1) candidate tumor suppressor. Mol Cell Biol 1997;17:2014–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helbing CC, Veillette C, Riabowol K, et al. A novel candidate tumor suppressor, ING1, is involved in the regulation of apoptosis. Cancer Res 1997;57:1255–8. [PubMed] [Google Scholar]
- 12.Garkavtsev I, Hull C, Riabowol K. Molecular aspects of the relationship between cancer and aging: tumor suppressor activity during cellular senescence. Exp Gerontol 1998;33:81–94. [DOI] [PubMed] [Google Scholar]
- 13.Turovets NA, Agapova LS, Kopnin PB, et al. Inactivation of p33(ING1) tumor suppressor affects the function of the cell cycle “checkpoints” and stability of the genome. Genetika 2000;36:305–12. [PubMed] [Google Scholar]
- 14.Garkavtsev I, Grigorian IA, Ossovskaya VS, et al. The candidate tumour suppressor p33(ING1) cooperates with p53 in cell growth control. Nature 1998;391:295–8. [DOI] [PubMed] [Google Scholar]
- 15.Ohmori M, Nagai M, Tasaka T, et al. Decreased expression of p33ING1 mRNA in lymphoid malignancies. Am J Hematol 1999;62:118–19. [DOI] [PubMed] [Google Scholar]
- 16.Oki E, Maehara Y, Tokunaga E, et al. Reduced expression of p33 (ING1) and the relationship with p53 expression in human gastric cancer. Cancer Lett 1999;147:157–62. [DOI] [PubMed] [Google Scholar]
- 17.Tokunaga E, Maehara Y, Oki E, et al. Diminished expression of ING1 mRNA and the correlation with p53 expression in breast cancers. Cancer Lett 2000;152:15–22. [DOI] [PubMed] [Google Scholar]
- 18.Toyama T, Iwase H, Watson P, et al. Suppression of ING1 expression in sporadic breast cancer. Oncogene 1999;18:5187–93. [DOI] [PubMed] [Google Scholar]
- 19.Sarela AI, Farmery SM, Markham AF, et al. The candidate tumour suppressor gene, ING1, is retained in colorectal carcinomas. Eur J Cancer 1999;35:1264–7. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez-Cespedes M, Okami K, Cairns P, et al. Molecular analysis of the candidate tumor suppressor gene ING1 in human head and neck tumors with 13q deletions. Genes Chromosomes Cancer 2000;27:319–22. [DOI] [PubMed] [Google Scholar]
- 21.Krishnamurthy J, Kannan K, Feng J, et al. Mutational analysis of the candidate tumor suppressor gene ING1 in Indian oral squamous cell carcinoma. Oral Oncol 2001;37:222–4. [DOI] [PubMed] [Google Scholar]
- 22.Nouman GS, Anderson JJ, Angus B, et al. Sequencing of ING1 tumour suppressor gene cDNAs generated from mRNA recovered from normal and neoplastic cell lines. J Pathol 2000;192:26A.10951396 [Google Scholar]
- 23.Morgan RJ, Jr, Doroshow JH, Venkataraman K, et al. High-dose infusional doxorubicin and cyclophosphamide: a feasibility study of tandem high-dose chemotherapy cycles without stem cell support. Clin Cancer Res 1997;3:2337–45. [PubMed] [Google Scholar]
- 24.Sandler DP, Ross JA. Epidemiology of acute leukemia in children and adults. Semin Oncol 1997;24:3–16. [PubMed] [Google Scholar]
- 25.Donadieu J, Auclerc MF, Baruchel A, et al. Critical study of prognostic factors in childhood acute lymphoblastic leukaemia: differences in outcome are poorly explained by the most significant prognostic variables. Fralle group. French acute lymphoblastic leukaemia study group. Br J Haematol 1998;102:729–39. [DOI] [PubMed] [Google Scholar]
- 26.Kersey JH. Fifty years of studies of the biology and therapy of childhood leukemia. Blood 1997;90:4243–51. [PubMed] [Google Scholar]
- 27.Coustan-Smith E, Behm FG, Sanchez J, et al. Immunological detection of minimal residual disease in children with acute lymphoblastic leukaemia. Lancet 1998;351:550–4. [DOI] [PubMed] [Google Scholar]
- 28.Deng C, Zhang P, Harper JW, et al. Mice lacking p21cip1/waf1 undergo normal development, but are defective in G1 checkpoint control. Cell 1995;82:675–84. [DOI] [PubMed] [Google Scholar]
- 29.Steele RJ, Thompson AM, Hall PA, et al. The p53 tumour suppressor gene. Br J Surg 1998;85:1460–7. [DOI] [PubMed] [Google Scholar]
- 30.Matsuo Y, Sugimoto A, Harashima A, et al. Myeloperoxidase positive acute lymphoblastic leukemia cell lines, NALM-30, NALM-31 and NALM-32, carrying Philadelphia chromosome with biphenotypic characteristics. Hum Cell 1998;11:221–30. [PubMed] [Google Scholar]
- 31.Wada M, Bartram CR, Nakamura H, et al. Analysis of p53 mutations in a large series of lymphoid hematologic malignancies of childhood. Blood 1993;82:3163–9. [PubMed] [Google Scholar]
- 32.Richards S, Burrett J, Hann I, et al. Improved survival with early intensification: combined results from the Medical Research Council childhood ALL randomised trials, UKALL X and UKALL XI. Medical Research Council working party on childhood leukaemia. Leukemia 1998;12:1031–6. [DOI] [PubMed] [Google Scholar]
- 33.Detre S, Saclani Jotti G, Dowsett M. A “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol 1995;48:876–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott M, Boisvert FM, Vieyra D, et al. UV induces nucleolar translocation of ING1 through two distinct nucleolar targeting sequences. Nucleic Acids Res 2001;29:2052–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nouman GS, Angus B, Lunec J, et al. Comparative assessment of expression of the inhibitor of growth 1 (ING1) gene in normal amd neoplastic tissues. Hybridoma and Hybridomics 2002;21:1–10. [DOI] [PubMed] [Google Scholar]
- 36.Seiter K, Feldman EJ, Sreekantaiah C, et al. Secondary acute myelogenous leukemia and myelodysplasia without abnormalities of chromosome 11q23 following treatment of acute leukemia with topoisomerase II-based chemotherapy. Leukemia 2001;15:963–70. [DOI] [PubMed] [Google Scholar]
- 37.Lodge AJ, Hall AG, Reid MM, et al. Topoisomerase IIα and IIβ expression in childhood acute lymphoblastic leukaemia: relation to prognostic factors and clinical outcome. J Clin Pathol 2001;54:31–6. [DOI] [PMC free article] [PubMed] [Google Scholar]