Abstract
The inhibitor of growth (ING) genes (ING1–4) probably descend from tumour suppressor genes. ING1 was the first to be identified and later isolated using an approach to detect genes whose expression is suppressed in cancer. The others were isolated through homology and similarity searches in human and mouse databases. All members contain a plant homeodomain involved in macromolecule recognition. Apart from the extensively studied ING1, little is known about the number of transcripts encoded by the other members or their gene structure. ING1 encodes several differentially spliced mRNAs, which may produce a family of proteins. The most widely expressed protein isoform is p33INGb1, which is involved in restriction of cell growth and proliferation, apoptosis, tumour anchorage independent growth, cellular senescence, maintenance of genomic stability, and modulation of cell cycle checkpoints. ING1 gene mutation is uncommon in cancer, although the subcellular localisation of p33INGb1 may have an effect on its function. The p33INGb1 cellular compartmental shift from the nucleus to the cytoplasm may cause loss of normal cellular function, and may play a central role in the pathogenesis of several cancers.
Keywords: ING1, ING2, p33ING1b, p53, tumour suppressors
Two major classes of tumour associated genes have been implicated in tumorigenesis: oncogenes and tumour suppressor genes. Inactivation, by loss or mutation, of tumour suppressor genes plays an essential role in the genesis of many tumours.1 Tumour suppressor proteins negatively regulate cell growth through a variety of mechanisms controlling the cell cycle.2 As long as these genetic elements are fully active, these genes serve as potent buffers against tumour progression, preventing the deregulation of normal growth control.
INHIBITOR OF GROWTH GENE FAMILY
The inhibitor of growth (ING) gene family, although newly recognised, is thought to be a part of this evolutionarily old family of putative tumour suppressor genes. The ING1 gene was the first member of this family to be identified and was later described by Garkavtsev and colleagues early in 1996.3 The family presently comprises the ING1, ING2 (ING1-L), ING3, and ING4 (ING2) genes.3–7 These genes are conserved between species including humans, mouse, yeast, and frog.7–12 In addition, these genes seem to maintain a high degree of homology with each other, which ranges from 32% to 76%.
CLONING AND ISOLATION STRATEGY
Limited numbers of tumour suppressors have been identified,1 mainly because of the recessive nature of tumour suppressors and the labourious identification methods involved. The ING1 gene was isolated using a new approach designed particularly for the detection of genes whose expression is suppressed in cancer cells; that is, tumour suppressor genes.3 The technique is based on subtractive hybridisation and the subsequent selection and isolation of transforming genetic suppressor element (GSE) fragments. These fragments are short cDNA sequences that are capable of promoting neoplastic transformation. GSE fragments are isolated from random cDNA expression libraries. A normal mammary epithelial cell line (184A1) and seven breast cancer cell lines (MCF-7, BT-474, Hs-578T, ZR-75, MD-MB-468, MD-MB-435, and BT-20) were used to prepare these cDNA libraries. These GSE fragments act effectively as oncogenes in gene transfection techniques through blocking the activation of tumour suppressors. Therefore, transfection and expression of the antisense sequences of the tumour suppressor gene would block protein production of this gene, hence, promoting cellular growth. In contrast, transfection and expression of the sense sequence of the tumour suppressor gene would block cellular growth.
The other members of the ING gene family were isolated through homology and similarity searches in cDNA library databases of human and mouse origin.
LOCATION, STRUCTURE, AND TRANSCRIPTS ENCODED BY MEMBERS OF THE ING GENE FAMILY
ING1 has been mapped to chromosome 13 and locus 13q33–34.13,14 ING2 (ING1L) has been mapped to chromosome 4 at position 4q35.1.4 ING3 has been mapped to chromosome 7 and locus 7q31.15 Finally, ING4 (ING2) has been mapped to chromosome X at position Xq13.1.16
The ING gene family belongs to a larger family of genes encoding proteins that contain structural motifs that are involved in the regulation of transcription. All members of this family contain a plant homeodomain (PHD) finger made of metal chelating residues arranged in the following order: four cysteine, one histidine, and three cysteine residues (C4HC3).17 This PHD finger domain spans 50–80 amino acids (aa).
“Although the ING1 gene has been extensively studied, little is known about the number of transcripts encoded by the other ING gene family members or their gene structure”
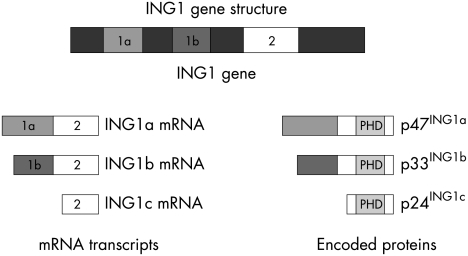
ING1 has been identified in humans and has been found to encode a series of differentially spliced mRNAs, with variable initiation sites and usage of three distinct exons (1a, 1b, and 2), as shown in fig 1.17–19 These mRNAs comprise ING1a, ING1b, and ING1c,5,20,21 which if translated would produce a family of proteins p47ING1a (422 aa and 46 451 Da), p33ING1b (279 aa and 31 843 Da), p27ING1b (235 aa and 27 000 Da), and p24ING1c (210 aa and 23 656 Da).5,18–21 However, the most widely expressed of these protein isoforms appears to be p33ING1b.5,6,22 All these proteins share a common exon (exon 2), which contains the PHD finger motif. The ING1 gene has been also identified in the mouse, yeast, and frog. Because of a cloning error, early investigations into the expression and the function of the ING1 gene products mistakenly used p24ING1c instead of p33ING1b.17,20,23
Figure 1.
The structure of the ING1 gene.
We have identified three potential nuclear localisation signals (NLS) in the common C-terminal region (exon 2) consisting of highly positively charged amino acids (such as lysine, proline, and arginine)—PNSKRS, PKEKK, and KKKKR—at amino acid positions 145–151, 176–181, and 185–190, respectively.22 Others have also found several NLS at amino acid positions 146–165 and 176–210.17,24 Furthermore, two nucleolar targeting sequences (NTS) were identified at amino acid positions 151–154 (RRQR) and 185–189 (KKKK).24
Although the ING1 gene has been extensively studied, little is known about the number of transcripts encoded by the other ING gene family members or their gene structure. ING2, which was previously called ING1L, was identified in humans by Shimada and colleagues and by Nagashima and colleagues.4,6 ING2 (ING1L) encodes a 280 aa protein of 32 800 Da (p33ING2), which shares 58.9% similarity with p33ING1b. ING3 has been identified in mice and was found to encode a putative 47 kDa protein.7 Finally, ING4, which was previously called ING2, was identified in mice by Jager and colleagues.5 ING4 (ING2) has been found to encode a 42 aa protein of 5 kDa, which shares 76% homology with all the ING1 gene protein members.5
TISSUE EXPRESSION AND SUBCELLULAR LOCALISATION
Northern blots have shown that ING1 mRNA is expressed as two bands of 2.2 and 2.5 kb in various human tissues.4 However, there is no information on which protein isoforms they represent. The first study to examine the mRNA expression of the different transcripts encoded by the ING1 gene was that of Jager and colleagues.5 They showed that various human tissues express ING1b, and ING1c, whereas ING1a expression was more restricted. We have shown that ING1b is abundantly expressed in all human normal and tumour cell lines studied.22 The first study to investigate the expression of the different protein isoforms was that of Boland and colleagues25; p33ING1b was broadly expressed and p24ING1c was more restricted. However, p47ING1a was only seen in brain cells that had been transfected with p47ING1a constructs and induced by isopropylthiogalactoside. Several studies have shown that protein products encoded by the ING1 gene are predominantly localised to the nucleus.13,26–28 Moreover, our study of a wide range of normal tissues has shown that the expression of nuclear p33ING1b is highly ubiquitous, with immunolabelling being seen in almost all normal cells and tissues, whereas the expression of cytoplasmic p33ING1b is more restricted.22
Northern blotting has also shown that ING2 (ING1L) is generally expressed in human tissues as two transcripts of 1.3 and 1.5 kDa.4 ING4 (ING2) mRNA is expressed in normal human tissues and in both breast cancer and melanoma cell lines.5
FUNCTIONAL STUDIES OF THE ING1 GENE
Functional studies have shown that both products of the ING1 gene, p33ING1b and p24ING1c, are involved in restriction of cell growth and proliferation, apoptosis, tumour anchorage independent growth, cellular senescence, maintenance of genomic stability, and modulation of cell cycle check points.13,29–32 Earlier studies mistakenly (as a result of a cloning error) used p24ING1c instead of p33ING1b for functional studies.17,20,23 Nevertheless, all previous functional studies were repeated and confirmed for p33ING1b.6,23,33
p33ING1b and the retinoblastoma protein 2 (pRb2) have limited homology, which raises the possibility that p33ING1b may interact with cell cycle regulators such as p53 and pRb.26 In human cells, coimmunoprecipitation studies have indicated that protein product(s) of the ING1 gene (principally p33ING1b) physically interact with the TP53 tumour suppressor gene protein product p53, whereas cotransfection studies confirmed the ability of ING1 to modulate p53 dependent transactivation of the kinase inhibitor p21WAF1.6,23,30 Extension of these preliminary findings suggested that the association of competent protein forms of each member of the ING1–TP53 complex is essential for optimum expression of the transactivational activity of TP53.30,34
Within the human body, at least some of the physiological tumour suppressor activities of p53 may occur independently of p33ING1b. Nevertheless, p33ING1b directly cooperates with p53 in growth regulation by modulating the ability of p53 to act as a transcriptional activator.30 Reduction of ING1 expression inhibits the growth suppressive activity of p53, suggesting that p33ING1b is essential for p53 function. The involvement of p33ING1b in the p53 signalling pathway indicates that ING1 is a potential tumour suppressor gene, the loss or inactivation of which may contribute to altered cell growth control, resistance to apoptosis, or establishment of an immortal tumour phenotype, even if wild-type p53 is retained. Therefore, if the loss of p33ING1b compromises the function of p53 only slightly, this may provide emerging cancer cells with a selective growth advantage, and even a small advantage might make cancer imminent.
“Cotransfection studies confirmed the ability of ING1 to modulate p53 dependent transactivation of the kinase inhibitor p21WAF1”
In mice, ING1 has been found to encode two protein products, which share 80–88% homology with human ING1 proteins.8 The bigger protein (p37) acts as a p53 cooperator and hence it behaves as a tumour suppressor. In contrast, the smaller protein (p31) acts as a p53 inhibitor, greatly lowering the cellular response to p53 apoptotic stimuli after DNA damage, therefore operating as an oncoprotein. Similarly, in the frog, ING variants had either an agonist or an antagonistic effect on apoptosis.7
The function of p33ING1b in providing checkpoint stability has been demonstrated by suppression of its normal function by antisense constructs.32 In these experiments, functional suppression was associated with the abolition of arrest at the G1–S, and S phase, in addition to the generation of a high number of chromosomal abnormalities and breaks in mitotic cells, resulting from improper DNA repair mechanisms.
The growth inhibitory effect of ING1 can be suppressed by the SV40-Tag oncoprotein, a phenomenon that is also seen for pRb and p53.26 Repression of p33ING1 protein expression can extend the life span of normal fibroblasts in vitro, suggesting a relation between p33ING1 negative growth regulation and cellular senescence, in addition to raising the possibility of the usefulness of p33ING1b in gene therapy.26,34,35 In glioma cells, cotransfection of both p53 and p33ING1b considerably augmented apoptosis.36 This was glioma cell specific—no effect was seen in normal neurones—so that normal cells were not damaged.
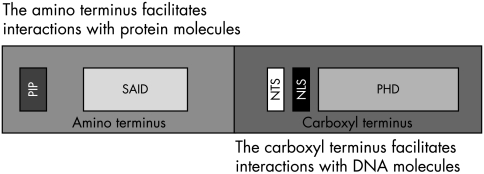
The C-terminal region of the p33ING1b molecule harbours the PHD motif, which is thought to act as a macromolecule recognition domain (fig 2).4,11 This facilitates the function of the PHD finger as both a regulator of transcription through interaction with RNA and DNA, and a regulator of chromatin remodelling through the targeting of nuclear recognition in chromatin structures. In contrast, the N-terminal region is more involved with protein interactions. This was first suggested because the N-terminus of p33ING1b shares homology with pRb2, and is responsible for the interaction with Sin3, a major component of the histone deacetylation complex 1 (HDAC1), through a 90 aa sequence located in the N-terminus of p33ING1b.11,12,23,37 Studies have shown that this interaction occurs through binding to SAP30 through a SAP30 interacting domain (SAID) in the N-terminus. Moreover, data from murine ING1 suggest that only p37, which is homologous to p33ING1b, and not p31, which is homologous to p24ING1c, coimmunoprecipitates with p53.8 Therefore, p33ING1b, through its interaction with Sin3–HDCA1, represses p53 control of the cell cycle.23 Moreover, p33ING1b binds the proliferating cell nuclear targeting antigen (PCNA) through a PCNA interacting protein domain (PIP) located in the N-terminus at amino acid positions 9–16.28
Figure 2.
The structure of the p33ING1b protein, showing the proliferating cell nuclear targeting antigen interacting protein domain (PIP), SAP30 interacting domain (SAID), nucleolar targeting signals (NTS), nuclear localisation signals (NLS), and plant homeodomain (PHD).
The notion that the ING1 gene encodes a chromatin remodelling protein is supported by the study of the Yng2 protein from Saccharomyces cervisiae.10 The Yng2 protein, which shares a PHD finger homology with human ING1, has been shown to interact with members of the histone acetyltransferase complex (Tra1).11 These interactions facilitate DNA transcription through the acetylation or the deacetylation of histones.12,37,38
In a mouse model, overexpression of ING1 mRNA and protein was seen in skin squamous cell carcinoma.39 Cheung and colleagues have reported high p33ING1 protein concentrations in squamous cell carcinoma of the skin after exposure to ultraviolet (UV) damage, which was independent of p53 status.40 In accordance with these results, we have shown that p33ING1b is retained in squamous cell carcinoma of the skin, cervix, oesophagus, and lung.22 Although the reason for this retention is not yet clear, p33ING1b may be playing a protective role by assisting in the induction of apoptosis; however, with high amounts of apoptotic stimuli only resistant cancer cells survive, which may lead to progression of the cancer.39 In contrast, in a study of 31 cases of squamous cell carcinoma, all cases showed reduced concentrations of ING1 protein products compared with normal tissue.41
“The notion that the ING1 gene encodes a chromatin remodelling protein is supported by the study of the Yng2 protein from Saccharomyces cervisiae”
Recent work provided evidence of the overexpression of p33ING1b after UV exposure in skin malignancies.40 This provides further support for its tumour suppressive function and also suggests a function in DNA repair after UV damage, or perhaps enhancement of apoptosis.24 It is well known that p53 protein production is enhanced by the same stimulant (UV), in accordance with the proposed cooperation of the molecule with p33ING1b.30 In addition, there was evidence of nuclear to nucleolar translocation of p33ING1b in normal skin fibroblast cell lines after exposure to UV rays.24 Others have shown a p33ING1b mediated role in the repair of UV damaged DNA.42
UPSTREAM REGULATORS AND DOWNSTREAM MODULATORS
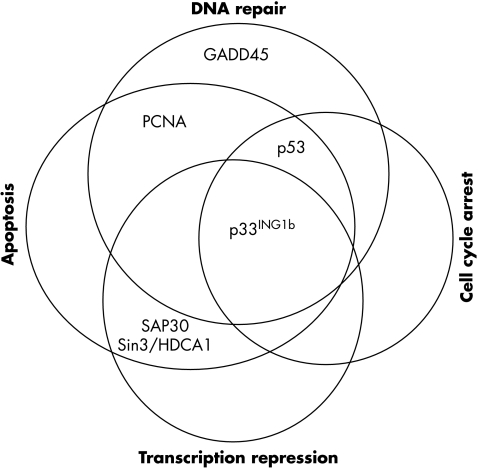
Originally, p53 was thought to be an upstream regulator of p33ING1b. However, it was found that tissue expression of p33ING1b is similar in both p53 preserved and p53 deficient mice.8 This proves that p33ING1b expression is independent of p53 status. To date, no upstream regulator of p33ING1b has been identified. However, several downstream modulators or effectors of function have been established (fig 3).
Figure 3.
Downstream modulators or effectors of p33ING1b function in vital cellular processes. GADD45, growth arrest and DNA damage; HDAC1, histone deacetylation complex 1; PCNA, proliferating cell nuclear targeting antigen.
Cotransfection studies confirmed the ability of p33ING1b to modulate p53 dependent transactivation of the kinase inhibitor p21WAF1, which causes the cell cycle to arrest in the G1 phase.30 Moreover, optimal function of both members of the p33ING1b–p53 complex is necessary for several important cellular process, including restriction of cell growth, proliferation, apoptosis, cellular senescence, maintenance of genomic stability, and modulation of cell cycle checkpoints.13,29–32
Because p33ING1b was found to be a member of the Sin3–HDAC1 complex,23,43 it was thought that p33ING1b modulates transcription repression through members of the Sin–HDAC1 complex, which include SAP30 and pRb1. Studies have shown that this interaction occurs through binding to SAP30 via a SAID domain in the N-terminus. Transcription repression probably occurs through the repression of p53 responsive genes that halt cell cycle progression.
UV was found to induce the binding of p33ING1b to PCNA.28 This complex is responsible for enhancing DNA repair over DNA replication steps or, if the former fails, augmenting apoptosis to eliminate damaged cells. Moreover, a physical association was detected between p33ING1b and the GADD45 protein, which is associated with DNA repair mechanisms, after damage to melanoma cell lines induced by UV.42
Recently, it was found that p33ING1 regulates the expression of certain genes, including cyclin B1 and the protooncogene DEK.44 The importance of this regulation is not yet clear, although this process is dependent on the presence of wild-type p53.
PROPOSED MECHANISMS OF MALFUNCTION OF THE ING1 GENE
Several mechanisms of malfunction of the ING1 gene have been proposed. These include: gene malfunctions (mutations, rearrangements, loss of heterozygosity (LOH), homozygous loss, and DNA CpG island hypermethylation), reduced mRNA expression, reduced protein expression, and protein malformations.
The human ING1 tumour suppressor gene has been mapped to the subtelomeric region of the long arm of chromosome 13 (13q33–34).14 Both the RB (13q14) and the BRCA-2 (13q12) genes are located close to this locus.45,46 High rates of 13q LOH have been detected in a variety of tumours, including those of the oesophagus, colorectum, kidney, urinary bladder, breast, ovary, lung, lymphoid cells, and head and neck.47–55 However, recent studies have indicated that mutations in ING1 appear to be extremely rare in breast carcinomas, ovarian carcinomas, lymphoid malignancies, myeloid leukaemia, gastric carcinomas, colorectal carcinomas, squamous cell carcinoma of different origins, and melanoma (table 1).18,41,56–62 We have also shown that mutation of the ING1b gene is uncommon.22 Nevertheless, we have found six notable base pair differences between the cDNA corresponding to the ING1b mRNA that we produced and the previously published sequence. Others have since confirmed this.5,18–20 In contrast, reduced ING1 mRNA values have been seen in lymphoid malignancies, gastrointestinal tumours, and breast carcinomas.35,57,63
Table 1.
ING1 gene structural mutations in human cancers
| Codon | Position | Mutation | Change | Coding | Rate | Origin | Tissue type | First author (year) |
| 89 | N-terminus | Silent | G/A | Val | – | Skmel-110 | Melanoma | Campos (2002) |
| 95 | N-terminus | Missense | CCC/TCC | Pro/Ser | 1/422 | Patient | Breast and ovarian ca | Toyama (1999) |
| 101 | N-terminus | Silent | C/T | Asp | – | Skmel-110 | Melanoma | Campos (2002) |
| 166 | C-terminus | Silent | G/A | Arg | 1/442 | Patient | Breast and ovarian ca | Toyama (1999) |
| 172 | C-terminus | Missense | GAG/AAG | Glu/Lys | – | HCT116 | Colon carcinoma | Oki (1999), Ito (2002) |
| 173 | C-terminus | Silent | G/A | Ser | 3/23 | Patient | Head and neck SCC | Gunduz (2000) |
| 188 | NLS/NTS | Silent | G/A | Ser | 18/442 | Patient | Breast and ovarian ca | Toyama (1999) |
| 188 | NLS/NTS | Silent | G/A | Ser | – | MKN28 | Gastric carcinoma | Oki (1999) |
| 188 | NLS/NTS | Silent | G/A | Ser | – | MKN45 | Gastric carcinoma | Oki (1999) |
| 188 | NLS/NTS | Silent | G/A | Ser | 14/71 | Patient | Oral SCC | Krishnamurthy (2001) |
| 192 | C-terminus | Missense | GCC/GAC | Ala/Asp | 1/23 | Patient | Head and neck SCC | Gunduz (2000) |
| 214 | PHD | Missense | GCG/GAG | Ala/Glu | 1/31 | Patient | ESCC | Chen (2001) |
| 215 | PHD | Missense | TGC/TCC | Cys/Ser | 1/23 | Patient | Head and neck SCC | Gunduz (2000) |
| 216 | PHD | Missense | AAC/AGC | Asx/Ser | 1/23 | Patient | Head and neck SCC | Gunduz (2000) |
| 219 | PHD | Silent | C/T | Pro | 1/31 | Patient | Oesophageal SCC | Chen (2001) |
| 223 | PHD | Silent | C/T | Asn | 1/31 | Patient | Oesophageal SCC | Chen (2001) |
| 228 | PHD | Silent | T/C | Cys | 1/442 | Patient | Breast and ovarian ca | Toyama (1999) |
| 233 | PHD | Missense | GTC/ATC | Val/Leu | 1/31 | Patient | Oesophageal SCC | Chen (2001) |
| 236 | PHD | Missense | GGG/GTG | Gly/Val | 1/31 | Patient | Oesophageal SCC | Chen (2001) |
| 239 | PHD | Silent | G/C | Ser | – | Skmel-24 | Melanoma | Campos (2002) |
| 244 | PHD | Silent | T/C | Asn | – | Skmel-24 | Melanoma | Campos (2002) |
| 247 | PHD | Silent | C/A | Pro | – | Skmel-24 | Melanoma | Campos (2002) |
| 253 | PHD | Silent | T/C | Cys | – | Skmel-24 | Melanoma | Campos (2002) |
| 257 | PHD | Silent | G/T | Arg | – | Skmel-24 | Melanoma | Campos (2002) |
| 260 | PHD | Missense | AAC/AGC | Asn/Ser | – | Skmel-24 | Melanoma | Campos (2002) |
| 270 | PHD | Silent | A/G | Lys | – | Skmel-24 | Melanoma | Campos (2002) |
| 270 | PHD | Missense | AAG/AAT | Lys/Asn | 1/31 | Patient | Oesophageal SCC | Chen (2001) |
| 272 | PHD | Silent | A/G | Lys | – | Skmel-24 | Melanoma | Campos (2002) |
ca, carcinoma; NLS, nuclear localisation signal; NTS, nucleolar targeting sequence; PHD, plant homeodomain; SCC, squamous cell carcinoma.
Chen and colleagues have shown reduced concentrations of p33ING1b in squamous carcinoma.41 We found reduced amounts or loss of nuclear expression of p33ING1b in some tumours, notably melanoma, seminoma, papillary thyroid carcinoma, invasive breast carcinoma, colorectal adenocarcinoma, and acute lymphoblastic leukaemia, which was associated with a concomitant upregulation in cytoplasmic p33ING1b expression.22,64–67
“The loss of nuclear p33ING1b or translocation from the nucleus to the cytoplasm would probably result in the loss of its tumour suppressor activity”
p33ING1b contains several important structural motifs that indicate that it functions in the nucleus (PHD finger, NLS, and NTS), in addition to its ability to bind and modulate the transcriptional activity of p53, which depends on nuclear localisation. Therefore, the loss of nuclear p33ING1b or translocation from the nucleus to the cytoplasm would probably result in the loss of its tumour suppressor activity.22,64,65 Optimal functioning of p53 is dependent on p33ING1b, and therefore loss of nuclear p33ING1b would be predicted to compromise p53 function.
Most of the previously presented studies suggest that gene mutation is not the mechanism whereby p33ING1b is involved in tumorigenesis. Rather, it appears that the modulation of p33ING1b mRNA levels or switching between transcribed protein products may be crucial p33ING1b alterations. Therefore, ING1 gene alterations are consistent with class II tumour suppressor genes.28,41,68
CLINICAL APPLICATIONS OF THE ING1 GENE AND ITS PROTEIN PRODUCTS
The two main applications of the ING1 gene would be its value in diagnosis and its therapeutic possibilities. We, and others, have found clear evidence that some tumours show reduced or even complete loss of nuclear expression of p33ING1b compared with their normal counterparts, which is associated in some instances with enhanced cytoplasmic expression of p33ING1b.22,41,64–67 Activated melanocytic lesions in particular showed this phenomenon. The loss of nuclear p33ING1b expression and enhanced cytoplasmic expression in melanomas suggests a potential diagnostic use of p33ING1b in the area of melanocytic neoplasia, although the finding of similar p33ING1b alterations in Spitz naevi would slightly limit its value.64 Furthermore, the almost universal complete loss of p33ING1b in acute lymphoblastic leukaemia suggests its possible use in the distinction between this and other lymphomas.65 ING1 gene immunogenecity in patients with breast cancer has raised the possibility of the potential value of ING1 in diagnosis and vaccine based treatment.5 The role of ING1 in augmenting apoptosis in glioma cells after cotransfection with p53, which spared damage to normal cells, provides evidence of its potential use in gene therapy targeting specific tumour cells.36,68 Several studies have indicated the potential use of p33ING1b in immunotherapy in conjunction with p53, and that p33ING1b status needs to be determined before the induction of trials.33,34 Class II tumour suppressor genes offer great therapeutic possibilities because they are present as non-mutated wild-type alleles in cancer cells.
FUTURE STUDIES
There are two main areas of interest. First, the functions of the ING1 gene and the other members of the ING gene family should be investigated further, in addition to the transcripts and proteins encoded by each. Second, studies should be undertaken to identify potential modulators and effectors of function of the ING gene family, which might be involved in tumorigenesis.
Certainly, mutations of the ING1 gene have been found to be a rare event. However, studies to determine the degree of alteration of the other members of the ING gene family are still pending. Transfection studies followed by in vitro translation of mRNAs of each member of the ING gene family would shed light on the biological behaviour of each transcript. The use of the yeast two hybrid system to investigate the exact role of the ING gene family protein members and their PHD finger domains through studying interactions with other protein macromolecules would be of great value. The development of knockout mice lacking the ING1 gene or any of the other ING gene family members would be of great advantage in determining which processes, if any, are affected by its absence. This would also help in the search for the upstream regulators. The role of the ING1 gene in gene therapy is still to be determined fully in several tumour types.
Take home messages.
The ING1 gene is a class II tumour suppressor gene that probably encodes a chromatin remodelling protein
ING1 gene mutation is rare in cancer
The transfer of p33ING1b from the nucleus to the cytoplasm may cause loss of normal cellular function of the protein
This event may play a central role in the development, progression, and the pathogenesis of several cancers
The ING1 gene may be of value in diagnosis and for gene therapy
CONCLUSION
We conclude that ING1 gene mutation is an uncommon event in cancer. Furthermore, we propose that the cellular compartment shift of p33ING1b from the nucleus to the cytoplasm may cause loss of normal cellular function of the protein. This event may play a central role in the development, progression, and pathogenesis of several cancers.
Abbreviations
aa, amino acids
GSE, genetic suppressor element
HDAC1, histone deacetylation complex 1
ING, inhibitor of growth
LOH, loss of heterozygosity
NLS, nuclear localisation signal
NTS, nucleolar targeting sequence
PCNA, proliferating cell nuclear targeting antigen
PHD, plant homeodomain
PIP, proliferating cell nuclear targeting antigen interacting protein domain
pRb, retinoblastoma protein
SAID, SAP30 interacting domain
UV, ultraviolet
REFERENCES
- 1.Hussain SP, Hofseth LJ, Harris CC. Tumor suppressor genes: at the crossroads of molecular carcinogenesis, molecular epidemiology and human risk assessment. Lung Cancer 2001;34(suppl 2):S7–15. [DOI] [PubMed] [Google Scholar]
- 2.Uhlmann EJ, Gutmann DH. Tumor suppressor gene regulation of cell growth: recent insights into neurofibromatosis 1 and 2 gene function. Cell Biochem Biophys 2001;34:61–78. [DOI] [PubMed] [Google Scholar]
- 3.Garkavtsev I, Kazarov A, Gudkov A, et al. Suppression of the novel growth inhibitor p33(ING1) promotes neoplastic transformation. Nat Genet 1996;14:415–20. [DOI] [PubMed] [Google Scholar]
- 4.Shimada Y, Saito A, Suzuki M, et al. Cloning of a novel gene (ING1L) homologous to ING1, a candidate tumor suppressor. Cytogenet Cell Genet 1998;83:232–5. [DOI] [PubMed] [Google Scholar]
- 5.Jager D, Stockert E, Scanlan MJ, et al. Cancer-testis antigens and ING1 tumor suppressor gene product are breast cancer antigens: characterization of tissue-specific ING1 transcripts and a homologue gene. Cancer Res 1999;59:6197–204. [PubMed] [Google Scholar]
- 6.Nagashima M, Shiseki M, Miura K, et al. DNA damage-inducible gene p331NG2 negatively regulates cell proliferation through acetylation of p53. Proc Natl Acad Sci U S A 2001;98:9671–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner MJ, Gogela-Spehar M, Skirrow RC, et al. Expression of novel ING variants is regulated by thyroid hormone in the Xenopus laevis tadpole. J Biol Chem 2001;276:47013–20. [DOI] [PubMed] [Google Scholar]
- 8.Zeremski M, Hill JE, Kwek SSS, et al. Structure and regulation of the mouse ing1 gene—three alternative transcripts encode two PHD finger proteins that have opposite effects on p53 function. J Biol Chem 1999;274:32172–81. [DOI] [PubMed] [Google Scholar]
- 9.Ha S, Lee S, Chung M, et al. Mouse ING1 homologue, a protein interacting with A1, enhances cell death and is inhibited by A1 in mammary epithelial cells. Cancer Res 2002;62:1275–8. [PubMed] [Google Scholar]
- 10.Nagy PL, Griesenbeck J, Kornberg RD, et al. A trithorax-group complex purified from Saccharomyces cerevisiae is required for methylation of histone H3. Proc Natl Acad Sci U S A 2002;99:90–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Loewith R, Meijer M, Lees-Miller SP, et al. Three yeast proteins related to the human candidate tumor suppressor p33(ING1) are associated with histone acetyltransferase activities. Mol Cell Biol 2000;20:3807–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choy JS, Tobe BT, Huh JH, et al. Yng2p-dependent NuA4 histone H4 acetylation activity is required for mitotic and meiotic progression. J Biol Chem 2001;276:43653–62. [DOI] [PubMed] [Google Scholar]
- 13.Garkavtsev I, Demetrick D, Riabowol K. Cellular localization and chromosome mapping of a novel candidate tumor suppressor gene (ING1). Cytogenet Cell Genet 1997;76:176–8. [DOI] [PubMed] [Google Scholar]
- 14.Zeremski M, Horrigan SK, Grigorian IA, et al. Localization of the candidate tumor suppressor gene ING1 to human chromosome 13q34. Somat Cell Mol Genet 1997;23:233–6. [DOI] [PubMed] [Google Scholar]
- 15.Kawaji H, Schonbach C, Matsuo Y, et al. Exploration of novel motifs derived from mouse cDNA sequences. Genome Res 2002;12:367–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nolte D, Muller U. Human O-G1cNAc transferase (OGT): genomic structure, analysis of splice variants, fine mapping in Xq13.1. Mamm Genome 2002;13:62–4. [DOI] [PubMed] [Google Scholar]
- 17.Cheung KJ, Li G. The tumor suppressor ing1: structure and function. Exp Cell Res 2001;268:1–6. [DOI] [PubMed] [Google Scholar]
- 18.Gunduz M, Ouchida M, Fukushima K, et al. Genomic structure of the human ING1 gene and tumor-specific mutations detected in head and neck squamous cell carcinomas. Cancer Res 2000;60:3143–6. [PubMed] [Google Scholar]
- 19.Baranova AV, Ivanov DV, Makeeva NV, et al. Genomic organization of a tumor growth inhibitor gene ING1. Mol Biol 2000;34:232–6. [PubMed] [Google Scholar]
- 20.Garkavtsev I. Suppression of the novel growth inhibitor p33(ING1) promotes neoplastic transformation. Nat Genet 1999;23:373. [DOI] [PubMed] [Google Scholar]
- 21.Saito A, Furukawa T, Fukushige S, et al. p24/ING1-ALT1 and p47/ING1-ALT2, distinct alternative transcripts of p33/ING1. J Hum Genet 2000;45:177–81. [DOI] [PubMed] [Google Scholar]
- 22.Nouman GS, Angus B, Lunec J, et al. Comparative assessment of expression of the inhibitor of growth 1 gene (ING1) in normal and neoplastic tissues. Hybridoma 2002;21:1–10. [DOI] [PubMed] [Google Scholar]
- 23.Skowyra D, Zeremski M, Neznanov N, et al. Differential association of products of alternative transcripts of the candidate tumor suppressor ING1 with the mSin3/HDAC1 transcriptional corepressor complex. J Biol Chem 2001;276:8734–9. [DOI] [PubMed] [Google Scholar]
- 24.Scott M, Boisvert FM, Vieyra D, et al. UV induces nucleolar translocation of ING1 through two distinct nucleolar targeting sequences. Nucl Acids Res 2001;29:2052–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boland D, Olineck V, Bonnefin P, et al. A panel of CAb antibodies recognize endogenous and ectopically expressed ING1 protein. Hybridoma 2000;19:161–5. [DOI] [PubMed] [Google Scholar]
- 26.Garkavtsev I, Riabowol K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33(ING1) candidate tumor suppressor. Mol Cell Biol 1997;17:2014–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garkavtsev I, Boland D, Mai J, et al. Specific monoclonal antibody raised against the p33(ING1) tumor suppressor. Hybridoma 1997;16:537–40. [DOI] [PubMed] [Google Scholar]
- 28.Scott M, Bonnefin P, Vieyra D, et al. UV-induced binding of ING1 to PCNA regulates the induction of apoptosis. J Cell Sci 2001;114:3455–62. [DOI] [PubMed] [Google Scholar]
- 29.Helbing CC, Veillette C, Riabowol K, et al. A novel candidate tumor suppressor, ING1, is involved in the regulation of apoptosis. Cancer Res 1997;57:1255–8. [PubMed] [Google Scholar]
- 30.Garkavtsev I, Grigorian IA, Ossovskaya VS, et al. The candidate tumour suppressor p33(ING1) cooperates with p53 in cell growth control. Nature 1998;391:295–8. [DOI] [PubMed] [Google Scholar]
- 31.Garkavtsev I, Hull C, Riabowol K. Molecular aspects of the relationship between cancer and aging: tumor suppressor activity during cellular senescence. Exp Gerontol 1998;33:81–94. [DOI] [PubMed] [Google Scholar]
- 32.Turovets NA, Agapova LS, Kopnin PB, et al. Inactivation of p33(ING1) tumor suppressor affects the function of the cell cycle “checkpoints” and stability of the genome. Russian J of Genet 2000;36:305–12. [PubMed] [Google Scholar]
- 33.Shimada H, Liu TL, Ochiai T, et al. Facilitation of adenoviral wild-type p53-induced apoptotic cell death by overexpression of p33(ING1) in T. Tn human esophageal carcinoma cells. Oncogene 2002;21:1208–16. [DOI] [PubMed] [Google Scholar]
- 34.Oren M. Tumor suppressors. Teaming up to restrain cancer. Nature 1998;391:233–4. [DOI] [PubMed] [Google Scholar]
- 35.Tokunaga E, Maehara Y, Oki E, et al. Diminished expression of ING1 mRNA and the correlation with p53 expression in breast cancers. Cancer Lett 2000;152:15–22. [DOI] [PubMed] [Google Scholar]
- 36.Shinoura N, Muramatsu Y, Nishimura M, et al. Adenovirus-mediated transfer of p33(ING1) with p53 drastically augments apoptosis in gliomas. Cancer Res 1999;59:5521–8. [PubMed] [Google Scholar]
- 37.Nourani A, Doyon Y, Utley RT, et al. Role of an ING1 growth regulator in transcriptional activation and targeted histone acetylation by the NuA4 complex. Mol Cell Biol 2001;21:7629–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nourani A, Doyon Y, Utley R, et al. The yeast nua4 acetyltransferase complex contains an ing1 family growth regulator and is important for pho5 expression. Yeast 2001;18:S63–3. [Google Scholar]
- 39.Vladimir S. Spiegelman TJS. The role of novel tumor suppressor ING1 in mouse carcinogenesis. Toxicol Pathol 1999;27:729–30. [Google Scholar]
- 40.Cheung KJJ, Bush JA, Jia W, et al. Expression of the novel tumour suppressor p33(ING1) is independent of p53. Br J Cancer 2000;83:1468–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen L, Matsubara N, Yoshino T, et al. Genetic alterations of candidate tumor suppressor ING1 in human esophageal squamous cell cancer. Cancer Res 2001;61:4345–9. [PubMed] [Google Scholar]
- 42.Cheung KJ, Mitchell D, Lin P, et al. The tumor suppressor candidate p33(ING1) mediates repair of UV-damaged DNA. Cancer Res 2001;61:4974–7. [PubMed] [Google Scholar]
- 43.Kuzmichev A, Zhang Y, Erdjument-Bromage H, et al. Role of the Sin3–histone deacetylase complex in growth regulation by the candidate tumor suppressor p33(ING1). Mol Cell Biol 2002;22:835–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi M, Seki N, Ozaki T, et al. Identification of the p33(ING1)-regulated genes that include cyclin B1 and proto-oncogene DEK by using cDNA microarray in a mouse mammary epithelial cell line NMuMG. Cancer Res 2002;62:2203–9. [PubMed] [Google Scholar]
- 45.Takahashi R. Role of the RB gene in carcinogenesis. Hum Cell 1993;6:260–5. [PubMed] [Google Scholar]
- 46.Wooster R, Neuhausen SL, Mangion J, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12–13. Science 1994;265:2088–90. [DOI] [PubMed] [Google Scholar]
- 47.Motomura K, Nishisho I, Takai S, et al. Loss of alleles at loci on chromosome 13 in human primary gastric cancers. Genomics 1988;2:180–4. [DOI] [PubMed] [Google Scholar]
- 48.Huang TH, Quesenberry JT, Martin MB, et al. Loss of heterozygosity detected in formalin-fixed, paraffin-embedded tissue of colorectal carcinoma using a microsatellite located within the deleted in colorectal carcinoma gene. Diagn Mol Pathol 1993;2:90–3. [PubMed] [Google Scholar]
- 49.Anglard P, Tory K, Brauch H, et al. Molecular analysis of genetic changes in the origin and development of renal cell carcinoma. Cancer Res 1991;51:1071–7. [PubMed] [Google Scholar]
- 50.Cairns P, Proctor AJ, Knowles MA. Loss of heterozygosity at the RB locus is frequent and correlates with muscle invasion in bladder carcinoma. Oncogene 1991;6:2305–9. [PubMed] [Google Scholar]
- 51.Borg A, Zhang QX, Olsson H, et al. Chromosome 1 alterations in breast cancer: allelic loss on 1p and 1q is related to lymphogenic metastases and poor prognosis. Genes Chromosomes Cancer 1992;5:311–20. [DOI] [PubMed] [Google Scholar]
- 52.Shenson DL, Gallion HH, Powell DE, et al. Loss of heterozygosity and genomic instability in synchronous endometrioid tumors of the ovary and endometrium. Cancer 1995;76:650–7. [DOI] [PubMed] [Google Scholar]
- 53.Reissmann PT, Koga H, Takahashi R, et al. Inactivation of the retinoblastoma susceptibility gene in non-small-cell lung cancer. The lung cancer study group. Oncogene 1993;8:1913–19. [PubMed] [Google Scholar]
- 54.Mitelman F, Heim S. Chromosome abnormalities in cancer. Cancer Detect Prev 1990;14:527–37. [PubMed] [Google Scholar]
- 55.Maestro R, Dolcetti R. Oncogenes and tumor suppressor genes in head and neck squamous cell carcinomas. Acta Otorhinolaryngol Ital 1996;16:21–6. [PubMed] [Google Scholar]
- 56.Ito K, Kinjo K, Nakazato T, et al. Expression and sequence analyses of p33(ING1) gene in myeloid leukemia. Am J Hematol 2002;69:141–3. [DOI] [PubMed] [Google Scholar]
- 57.Oki E, Maehara Y, Tokunaga E, et al. Reduced expression of p33(ING1) and the relationship with p53 expression in human gastric cancer. Cancer Lett 1999;147:157–62. [DOI] [PubMed] [Google Scholar]
- 58.Sanchez-Cespedes M, Okami K, Cairns P, et al. Molecular analysis of the candidate tumor suppressor gene ING1 in human head and neck tumors with 13q deletions. Genes Chromosomes Cancer 2000;27:319–22. [DOI] [PubMed] [Google Scholar]
- 59.Campos EI, Cheung KJ, Murray A, et al. The novel tumour suppressor gene ING1 is overexpressed in human melanoma cell lines. Br J Dermatol 2002;146:574–80. [DOI] [PubMed] [Google Scholar]
- 60.Toyama T, Iwase H, Watson P, et al. Suppression of ING1 expression in sporadic breast cancer. Oncogene 1999;18:5187–93. [DOI] [PubMed] [Google Scholar]
- 61.Sarela AI, Farmery SM, Markham AF, et al. The candidate tumour suppressor gene, ING1, is retained in colorectal carcinomas. Eur J Cancer 1999;35:1264–7. [DOI] [PubMed] [Google Scholar]
- 62.Krishnamurthy J, Kannan K, Feng J, et al. Mutational analysis of the candidate tumor suppressor gene ING1 in Indian oral squamous cell carcinoma. Oral Oncol 2001;37:222–4. [DOI] [PubMed] [Google Scholar]
- 63.Ohmori M, Nagai M, Tasaka T, et al. Decreased expression of p33ING1 mRNA in lymphoid malignancies. Am J Hematol 1999;62:118–19. [DOI] [PubMed] [Google Scholar]
- 64.Nouman GS, Anderson JJ, Mathers ME, et al. Nuclear to cytoplasmic compartment shift of the p33ING1b tumour suppressor protein is associated with malignancy in melanocytic lesions. Histopathology 2002;40:360–6. [DOI] [PubMed] [Google Scholar]
- 65.Nouman GS, Anderson JJ, Wood KW, et al. Loss of nuclear expression of the inhibitor of growth p33ING1b in childhood acute lymphoblastic leukaemia. J Clin Pathol 2002;55:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ahmed IAM, Nouman GS, Lunec J, et al. Expression of p33/ING1 gene products in colorectal cancer and its correlation with survival. Gastroenterology 2001;120:1548. [Google Scholar]
- 67.Nouman GS, Anderson JJ, Crosier S, et al. p33(ING1b) tumour suppresser protein expression in thyroid lesions determined by immunohistochemistry. J Pathol 2001;195:37A. [Google Scholar]
- 68.Sager R. Expression genetics in cancer: shifting the focus from DNA to RNA. Proc Natl Acad Sci U S A 1997;94:952–5. [DOI] [PMC free article] [PubMed] [Google Scholar]