Abstract
Limb girdle muscular dystrophy type 2A (LGMD2A) is caused by mutations in the calpain 3 gene. In a large family affected by LGMD2A with four severely affected members, three additional asymptomatic relatives had very high serum creatine kinase concentrations. All were homozygous for the R110X mutation and showed a total absence of calpain 3 in the muscle. Histological analysis of muscle in these three rare preclinical cases showed a consistent but unusual pattern, with isolated fascicles of degenerating fibres in an almost normal muscle. This pattern was also seen in one patient with early stage LGMD2A who had a P82L missense mutation and a partial deficiency of calpain 3 in the muscle, but was not seen in early stage patients affected by other forms of LGMD. These findings suggest that a peculiar pattern of focal degeneration occurs in calpainopathy, independently of the type of mutation or the amount of calpain 3 in the muscle.
Keywords: limb girdle muscular dystrophy, calpain 3, LGMD2A
The limb girdle muscular dystrophies (LGMDs) are a heterogeneous group of progressive disorders mainly affecting the pelvic and shoulder girdle musculature, ranging from a severe form with onset in the first decade and rapid progression, to a mild form with later onset and slower progression. The diagnostic criteria established by the ENMC-LGMD consortium include raised serum creatine kinase activity, myopathic electromyography, and muscle biopsy with features ranging from mildly myopathic to frankly dystrophic.1
The inheritance may be autosomal dominant (LGMD1) or recessive (LGMD2). During the past decade, at least 15 LGMD genes, five autosomal dominant (AD) and 10 autosomal recessive (AR), have been mapped. The AD forms are relatively rare and mutations in these genes are found in less than 10% of all patients with LGMD. The five forms of AD LGMD are: LGMD1A, at 5q22, which encodes the protein myotilin; LGMD1B at 1q11, which encodes lamin A/C; LGMD1C at 3p25, which encodes caveolin 3; LGMD1D at 6q23; and LGMD1E at 7q.
Among the AR forms, the four most severe forms have been mapped to 17q21, 4q12, 13q12, and 5q33, which encode glycoproteins of the sarcoglycan subcomplex of the dystrophin–glycoprotein complex, known as α sarcoglycan (α SG), β SG, γ SG, and δ SG, respectively. Mutations in these genes cause LGMD2C, LGMD2D, LGMD2E, and LGMD2F, respectively, and constitute a distinct subgroup of LGMDs, the sarcoglycanopathies. Among the clinically milder forms, the gene responsible for LGMD2A at 15q encodes calpain 3, LGMD2B at 2p encodes dysferlin, LGMD2G at 17q encodes telethonin, LGMD2H at 9q encodes TRIM 32, LGMD2I at 19q encodes the fukutin related protein, and LGMD2J at 2q encodes titin (LGMD gene/protein revisions in Zatz et al and Bushby and Beckmann1,2).
LGMD2A was first identified in a small community in the Reunion Island. Linkage to chromosome 15q was established in this community, and later confirmed in other populations.3,4 Subsequently, the mutated gene—the muscle specific neutral calcium dependent protease calpain 3 (CAPN3)—was identified.5 To date, more than 100 distinct pathogenic mutations have been identified in the CAPN3 gene, including nonsense, missense, deletions/insertions, and splice site mutations (Leiden database at http://www.dmd.nl/capn3_home.html). These mutations are spread diffusely over the entire length of the gene. Most of them represent private variations affecting single families, although certain mutations have been found in more than one family.6,7
CASE REPORT
Among more than 250 patients with LGMD submitted to muscle histological and protein analysis we identified one highly inbred family affected by LGMD2A in which there were four severely affected adult members and three still asymptomatic young individuals with high serum creatinine kinase concentrations (increased 10–12-fold; family 22 in Passos-Bueno and colleagues8). All affected individuals and all those with very high CK concentrations had a homozygous R110X mutation in the calpain 3 gene. Muscle protein analysis showed a total absence of calpain 3 in muscle (fig 1D).
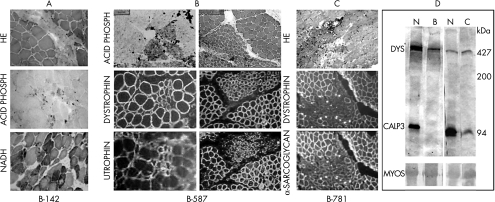
Figure 1.
Histological, histochemical, and immunohistochemical analysis of muscle biopsies from: (A) one adult patient affected by LGMD2A who was from the family with the R110X mutation, and who showed a typical dystrophic pattern; (B) one of the asymptomatic members from this family, who showed focal degeneration in the haematoxylin and eosin (HE) and the acid phosphatase reactions. The same bundle of fibres shows a patchy pattern of labelling for dystrophin and sarcoglycans, and strong sarcolemmal labelling for utrophin; (C) the patient with early stage LGMD2A who had the P82L mutation, showing focal degeneration; (D) multiplex western blot analysis for dystrophin (DYS) and calpain 3 (CALP3; using the antibody used by Anderson and colleagues9), showing the presence of a 427 kDa dystrophin band and the 94 kDa band of calpain 3 in normal controls (N), the absence of the 94 kDa calpain 3 band in patient B (preclinical), and partial deficiency in patient C. MYOS, myosin content in the Ponceau prestained blot for the evaluation of the amount of loaded muscle proteins.
A muscle biopsy from one affected relative showed typical dystrophic features (fig 1A). In the three preclinical patients, muscle histology revealed an almost normal histological pattern. However, an isolated fascicle of degenerating fibres was seen. Acid phosphatase staining was positive and there was patchy labelling for dystrophin and the sarcoglycan proteins, and an increased staining for utrophin in the plasma membrane from these degenerated fibres (fig 1B).
This pattern of focal degeneration was subsequently seen in one additional young patient with sporadic LGMD2A in the initial stages of the disease (fig 1C). She was identified through the deficiency of calpain 3 in the muscle, and DNA analysis detected a homozygous missense P82L mutation in the calpain 3 gene.
This histological pattern was not seen in other patients affected by muscular dystrophy who were studied by our group, or in preclinical or early stage cases of Xp21 muscular dystrophy, dysferlinopathies, sarcoglycanopathies, or telethoninopathy, and suggests that a different mechanism for the onset of degeneration may occur in patients with primary calpain 3 deficiency.
DISCUSSION
Calpain 3 is a proteolytic enzyme of the calpain superfamily whose specific role is not known. The protein is composed of 821 amino acids, organised into four domains, which include a cysteine proteolytic domain (II) and a calcium binding domain (IV). There are also three short specific inserted sequences located at the N-terminus domain I (NS), in the protease domain II (IS1), and between domains III and IV (IS2). In the IS2 region there is a titin binding site and a nuclear localisation signal.10
“Our results showed that the pattern of focal degeneration was not related to the amount of calpain 3 in the muscle because in the first family the deficiency was total, whereas in the second one it was only partial”
Various intracellular enzymes and transcription factors, in addition to cytoskeletal proteins, are processed by calpain, resulting in modification of their structures and activities. This suggests that calpain may play an important role in intracellular signal transduction.10 Complex proteolysis, promotion, and suppression of the network involving calpain 3 protein has been suggested in the response to muscle wasting. Thus, substrate processing by calpain 3 is essential for the proper function or maintenance of skeletal muscle.10 In this regard, preservation of the proteolytic domain of calpain 3 would be of utmost importance.
Our results showed that the pattern of focal degeneration was not related to the amount of calpain 3 in the muscle because in the first family the deficiency was total, whereas in the second one it was only partial (fig 1D). In addition, it is not related to a specific mutation, because it was seen as the consequence of two different mutations. The R110X mutation is located in the IIa protease domain of calpain 3, whereas the P82L mutation is located between the N-terminal first NS specific insertion sequence and the IIa protease domain. It is important to note that both mutations are at the N-terminal region of the protein, and possibly affect its proteolytic domain function.
It has also been suggested that calpain 3 deficiency may cause myonuclear apoptosis and a profound perturbation of the IκBα–nuclear factor-κB pathway in the earliest stages of the disease. Apoptotic myonuclei would be distributed in clusters.11 This would be consistent with our finding of clustered degenerated fibres grouped in possible clones.
Take home messages.
Histological analysis of muscle in preclinical and early stage cases of limb girdle muscular dystrophy (LGMD) 2A showed a consistent but unusual pattern, with isolated fascicles of degenerating fibres in an almost normal muscle
This pattern was not seen in early stage patients affected by other forms of LGMD
A peculiar pattern of focal degeneration appears to occur in calpainopathy, independently of the type of mutation or the amount of calpain 3 in the muscle
In conclusion, histological alterations in rare preclinical and early stage LGMD2A muscle biopsies suggest that in calpainopathy a peculiar pattern of focal degeneration occurs in the earliest stages of the disease.
Acknowledgments
Antibodies were kindly given by: Dr LVB Anderson, Dr J Chamberlain (antidystrophin, 303–8); Dr G Morris, and Dr Nt Man (anti-utrophin); Dr C Bonnemann and Dr LM Kunkel (anti-β-SG); Dr E McNally (anti-γ-SG); and Dr V Nigro (anti-δ-SG), to whom we are very grateful. We also wish to thank Dr S Iannaconne, for important suggestions and critical reading of the manuscript; M Canovas, for the excellent technical assistance; and Dr RCM Parvanello, Dr I Pavanello Filho, C da Silva Costa, V Palhares, and all the physicians who referred affected patients. This work was supported by grants from Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP-CEPID), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), PRONEX, IAEA, and Associação Brasileira de Distrofia Muscular (ABDIM).
Abbreviations
AD, autosomal dominant
AR, autosomal recessive
LGMD, limb girdle muscular dystrophy
NS, N-terminus domain I
SG, sarcoglycan
REFERENCES
- 1.Zatz M, Vainzof M, Passos-Bueno MR. Limb-girdle muscular dystrophy: one gene with different phenotypes, one phenotype with different genes. Curr Opin Neurol 2000;13:511–17. [DOI] [PubMed] [Google Scholar]
- 2.Bushby K, Beckmann J. the 105th ENMC workshop—pathogenesis in the non-sarcoglycan limb-girdle muscular dystrophies, Naarden, 12–14 April 2002. Neuromuscul Disord 2003;13:80–90. [DOI] [PubMed] [Google Scholar]
- 3.Beckmann JS, Richard I, Hillaire D, et al. A gene for limb-girdle muscular dystrophy maps to chromosome 15 by linkage. C R Acad Sci III 1991;312:141–8. [PubMed] [Google Scholar]
- 4.Passos-Bueno MR, Richard I, Vainzof M, et al. Evidence of genetic heterogeneity in the autosomal recessive adult forms of limb-girdle muscular dystrophy following linkage analysis with 15q probes in Brazilian families. J Med Genet 1993;30:385–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Richard I, Broux O, Allamand V, et al. Mutations in the proteolytic enzyme calpain 3 cause limb-girdle muscular dystrophy type 2A. Cell 1995;81:27–40. [DOI] [PubMed] [Google Scholar]
- 6.Richards I, Roudaut C, Saenz A, et al. Calpainopathy—a survey of mutations and polymorphisms. Am J Hum Genet 1999;64:1524–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Paula f, Vainzof M, Passos-Bueno MR, et al. Clinical variability in calpainopathy: what makes the difference? Eur J Hum Genet 2002;10:825–32. [DOI] [PubMed] [Google Scholar]
- 8.Passos-Bueno MR, Moreira ES, Marie S, et al. Main clinical features for the three mapped autosomal recessive limb-girdle muscular dystrophies and estimated proportion of each form in 13 Brazilian families. J Med Genet 1996;33:97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson LVB, Davison K, Moss JÁ, et al. Characterization of monoclonal antibodies to calpain 3 and protein expression in muscle from patients with limb-girdle muscular dystrophy type 2A. Am J Pathol 1998;153:1169–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sorimachi H, Beckmann JS. Defects of non-lysosomal proteolysis: calpain 3 deficiency. In: Karpati G, ed. Structural and molecular basis of skeletal muscle disease. Basel: ISN Neuropathology Press, 2002:148–53.
- 11.Baghdiguian S, Martin M, Richard I, et al. Calpain 3 deficiency is associated with myonuclear apoptosis and profound perturbation of the IκBa;NF-κB pathway in limb-girdle muscular dystrophy type 2. Nat Med 1999;5:503–11. [DOI] [PubMed] [Google Scholar]