Abstract
Background: Primary adenocarcinomas of the small intestine are rare, and the genetic mechanisms involved in their carcinogenesis remain unclear.
Aim: To examine the expression of candidate proteins in small intestinal adenocarcinomas by immunohistochemistry performed on tissue microarrays (TMAs).
Methods: Twenty seven primary sporadic small intestinal adenocarcinomas were analysed. The TMA technique was validated by comparing immunohistochemical labelling of hMLH1 and hMSH2 on TMAs and the tissue sections they derived from. The expression of Smad4, hMSH6, β catenin, and p53 was investigated and results compared with those obtained in 14 malignant ampullary tumours.
Results: TMA technology with threefold redundancy adequately represented the immunohistochemical pattern of small intestinal adenocarcinomas. Loss of hMLH1 expression, but not hMSH2 or hMSH6, was seen in two of 27 small intestinal adenocarcinomas. All ampullary tumours showed nuclear staining for hMSH2 and hMSH6. One case showed lack of immunostaining for hMLH1. Smad4 expression was absent in five small intestinal adenocarcinomas and two ampullary tumours. Overexpression of p53 was detected in the nuclei of 14 of the 27 small intestinal adenocarcinomas, and five of the 14 ampullary tumours. Nuclear or cytoplasmic expression of β catenin was present in all specimens.
Conclusion: Inactivation of the SMAD4/DPC4 gene seems to be involved in small intestinal adenocarcinoma tumorigenesis. Overexpression of p53 and abnormal expression of β catenin are two common events, unlike the loss of expression of the DNA mismatch repair proteins (hMLH1, hMSH2, and hMSH6). The carcinogenetic process appears to be similar in small intestinal adenocarcinomas and malignant ampullary tumours.
Keywords: SMAD4/DPC4, DNA mismatch repair proteins, β catenin, p53, tissue microarray
Primary adenocarcinomas of the small intestine are rare, although the small intestine constitutes almost 90% of the mucosal surface of the alimentary tract. The most frequent site for cancer is the duodenum, followed by the jejunum and the ileum.1–3 An increased risk for the development of small intestinal cancer is recognised in patients with Crohn’s disease,4–6 familial adenomatous polyposis (FAP), Peutz-Jeghers syndrome, coeliac disease,7 and cystic fibrosis.8,9 The genetic mechanisms involved in carcinogenesis of the small intestine remain unclear, mostly because of the small number of available cases.
Small intestinal adenocarcinoma occurs more frequently in hereditary non-polyposis colorectal cancer (HNPCC) syndrome,10,11 which is characterised by microsatellite instability (MSI). This involves mutations in the DNA mismatch repair (MMR) system, which recognises and repairs the DNA mismatches that occur during replication. This system comprises six genes, namely: hMLH1, hMSH2, hMSH3, hMSH6, hPMS1, and hPMS2, human homologues of the Escherichia coli MutS and MutL genes. The MSI phenotype can be demonstrated both by the polymerase chain reaction and by immunohistochemical study of hMLH1 and hMSH2 protein expression.12 Some studies have suggested a role for MSI in the pathogenesis of sporadic small intestinal carcinomas.13–16
“The genetic mechanisms involved in carcinogenesis of the small intestine remain unclear, mostly because of the small number of available cases”
Alterations in the p53 gene is the most common genetic event in sporadic small intestinal adenocarcinoma.17 In contrast, allelic losses of the APC (adenomatous polyposis coli) and DCC (deleted in colorectal cancer) genes are uncommon events.13,14,18 Recently, the SMAD4/DPC4 (deleted in pancreatic cancer 4) gene, which is a mediator of growth suppression through transforming growth factor β (TGFβ) signalling, has been shown to play a crucial role in small intestinal tumorigenesis.19 This prompted us to determine the immunohistochemical labelling of the SMAD4/DPC4 gene product in a series of sporadic small intestinal adenocarcinomas. To this end, we made use of tissue microarray (TMA) technology. We also studied the expression of hMLH1, hMSH2, hMSH6, β catenin, and p53 and compared the results with those obtained in malignant ampullary tumours. These data were correlated with standard clinicopathological features. Because no studies are available in the literature using TMAs in small intestinal adenocarcinomas, we first validated the technique by comparing the immunohistochemical labelling of hMLH1 and hMSH2 on TMA and the whole tissue sections from which they were derived.
MATERIALS AND METHODS
Patients and tumour samples
Formalin fixed, paraffin wax embedded tissues from 27 primary sporadic small intestinal adenocarcinomas were used for our present study. All specimens were obtained from surgical resection of the tumours. These samples included four adenocarcinomas complicating Crohn’s disease. Cases were selected from the files of the pathology departments of Saint-Antoine, Rothschild, and Beaujon hospitals, AP-HP, Paris, France, from 1988 to 2001. To ensure that the tumours were primary small intestinal adenocarcinomas, we excluded patients with pre-existing or coexisting extraintestinal malignancy. We also excluded cancers developing in patients with FAP and cancers occurring in patients fulfilling the Amsterdam criteria for a diagnosis of HNPCC syndrome. The two cases that occurred in this setting during the same period were used as controls for immunohistochemistry. Fourteen cases of malignant ampullary tumours were obtained from the pathology files of Saint-Antoine Hospital from 1994 to 2000. Patients with either FAP or HNPCC syndrome were again excluded in this last group.
Single representative blocks of tumour and adjacent non-neoplastic tissue were selected from each case for hMLH1 and hMSH2 immunohistochemical evaluation.
Construction and processing of small intestinal and ampullary cancers TMAs
Areas of invasive adenocarcinoma were identified on corresponding haematoxylin and eosin stained slides. Core biopsies of 0.6 mm in diameter were taken from each donor block and arrayed into a recipient paraffin block (35 mm × 22 mm × 5 mm) using a tissue microarrayer (Beecher Instruments, Sun Prairie, Wisconsin, USA), as described by Kononen et al.20 There were three cores for each tumour in the array, resulting in three histological spots on the corresponding slides. The positive control was normal small intestinal mucosa, which was represented in the array by five cores.
Immunohistochemistry
Both microarray and whole section slides were stained using the same technique. In brief, slides were dewaxed in xylene and rehydrated in graded alcohol dilutions. Endogenous peroxidase activity was blocked by incubation in H2O2 for 2 × 15 minutes. Antigen retrieval, consisting of microwave processing at 750 W and 150 W for 15 minutes each and pressure cooking in 0.01 M citrate buffer (pH 6.0), was applied. The slides were incubated with monoclonal antibodies against hMLH1 (clone G168–728; 1/70 dilution; PharMingen, San Diego, California, USA), hMSH2 (clone FE11; 1/100 dilution; Calbiochem, Cambridge, Massachusetts, USA), hMSH6 (clone 44; 1/100 dilution; Becton Dickinson, Lexington, North Carolina, USA), Smad4 (B-8; 1/300 dilution; Santa Cruz Biotechnology, Santa Cruz, California, USA), β catenin (E-5; 1/250 dilution; Santa Cruz Biotechnology), and p53 (clone BP53-12-1; 1/800 dilution; Biogenex, San Ramon, California, USA). Antibody reactions were detected with a biotinylated antimouse secondary antibody (30 minutes), followed by incubation with a final layer of peroxidase labelled streptavidin for 20 minutes. The bound antibody was detected with a Super Sensitive Detection kit (Biogenex). The slides were counterstained with haematoxylin.
Immunostaining of hMLH1, hMSH2, and hMSH6 in tumorous cells was evaluated as positive or negative. For these antibodies, a case was considered as negative on TMA if all three spots were negative. Smad4 labelling was scored as “diffusely positive”, “focally positive”, or “negative”. Positive labelling was defined as strong and uniform expression of Smad4 in the cytoplasm of cells, with at least focal expression of Smad4 in the nuclei. Negative cases contained cells with no cytoplasmic or nuclear expression of Smad4. Cases were regarded as focally positive when a few cells were labelled with the antibody to Smad4. For β catenin, membranous, cytoplasmic, or nuclear staining was evaluated as present or absent, separately for each specimen. Positivity for p53 was scored semiquantitatively in accordance with the positivity of tumorous cells: less than 10%, 10–50%, and more than 50%. Tumour cells with focal or diffuse positive staining (10–50% and more than 50%) were classified as overexpressing p53 protein. Two authors (MS and JFF) reviewed the slides without knowledge of each others results. Any disagreements were reviewed jointly to arrive at a consensus score.
Histopathology
Pathological variables were reviewed, including site (duodenum, jejunum, or ileum), differentiation (well, moderate, or poor), and T and N stage, according to American Joint Committee on Cancer criteria. Some of these parameters (differentiation and T and N stage) were also examined in malignant ampullary tumours.
Statistical analysis
Qualitative data were compared by means of the χ2 test using Yate’s correction when appropriate. A p value of ⩽ 0.05 was considered significant.
RESULTS
Clinicopathological features
The age of the patients ranged from 32 to 79 years (mean, 55.1). There were 15 men and 12 women. Ten tumours occurred in the duodenum, eight in the jejunum, and six in the ileum. The precise tumour site was unknown in three cases. Ten cancers were stage T4, 15 stage T3, one stage T2, and one stage T1. Eight tumours had no regional lymph node metastasis and 19 had metastasis. Fifteen adenocarcinomas were well differentiated, six were moderately differentiated, and six were poorly differentiated.
Validation of TMA technology and expression of MMR proteins
On whole tissue sections, a loss of expression of hMLH1 was seen in two of 27 small intestinal adenocarcinomas. There was no loss of hMSH2 expression.
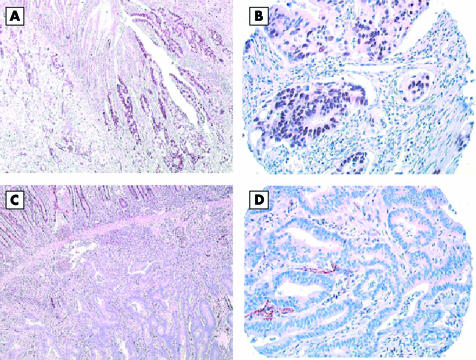
With the TMA technology, most tumours showed minimal intratumorous heterogeneity within the three spots of the array. With the hMLH1 and hMSH2 antibodies, 2.3% and 5.8%, respectively, of histospots were not interpretable. This was the result of loss of tissue on the TMA, no cancer in the spot, or no internal positive control. The result of hMLH1 and hMSH2 staining on microarrays was in agreement with the whole section analysis in all cases (fig 1A–D). There was no loss of hMSH6 expression.
Figure 1.
hMLH1 immunostaining of adenocarcinoma of the small intestine. One case positive on (A) large standard section and (B) tissue microarray section; one case negative on (C) large section and (D) tissue microarray.
All 14 malignant ampullary tumours showed nuclear staining for hMSH2 and hMSH6. One case showed a lack of immunostaining for hMLH1.
Expression of Smad4 protein
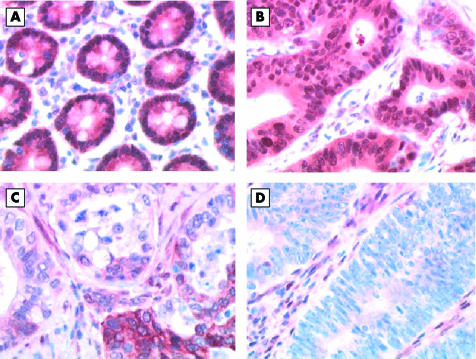
Both cytoplasmic and nuclear Smad4 labelling was seen in normal small intestinal epithelium (fig 2A). Of the 27 small intestinal adenocarcinomas, 21 were diffusely positive (fig 2B), one was focally positive (fig 2C), and five were negative (fig 2D). Of the four cases complicating Crohn’s disease, three were diffusely positive and one was negative. Of the 14 malignant ampullary tumours, 12 were diffusely positive and two were negative. Interestingly, the two small intestinal adenocarcinomas and the only malignant ampullary tumour that lost the expression of hMLH1 showed positive staining for Smad4.
Figure 2.
Smad4 immunostaining. (A) Normal small intestinal mucosa showing strong positivity. Adenocarcinoma of the small intestine: (B) strongly positive case, (C) case with mild focal positivity, and (D) negative case.
Expression of p53 protein
Overexpression of the p53 protein was detected in the nuclei of 14 of the 27 small intestinal adenocarcinomas, including three of the four cases complicating Crohn’s disease.
In malignant ampullary tumours, overexpression was seen in five of the 14 cases. There was no overexpression of p53 protein in tumours immunonegative for hMLH1.
On the contrary, overexpression of p53 protein was present in three of five small intestinal adenocarcinomas and in one of two malignant ampullary tumours with loss of staining for Smad4.
Expression of β catenin
In normal small intestinal epithelium, β catenin was expressed exclusively on the plasma membrane, with no nuclear positivity. Cytoplasmic positivity, without nuclear staining, was found in most of the small intestinal adenocarcinomas (25 of 27). Exclusive nuclear staining was seen in the two other tumours (7.4%). Cytoplasmic positivity with no nuclear staining was seen in 13 of the malignant ampullary tumours, and exclusive nuclear staining was seen in one case.
Comparison between small intestinal adenocarcinomas and malignant ampullary tumours
There was no difference between these two groups of tumours regarding the immunohistochemical results (hMLH1, hMSH2, hMSH6, Smad4, p53, or β catenin).
Relation between the immunohistochemical pattern and the clinicopathological features of the tumours
The results are presented in detail for Smad4 (table 1) and p53 (table 2). There was no significant relation between the expression of the proteins tested on immunohistochemistry and the clinicopathological features of the tumours (tumour site, T and N stage, and presence of Crohn’s disease).
Table 1.
Relation between Smad4 expression and clinicopathological features in a series of 27 small intestinal adenocarcinomas
| Smad4+ (n = 22)† | Smad4− (n = 5) | p Value | |
| Site* | NS | ||
| Duodenum | 9 (42.9%) | 1 (33.3%) | |
| Jejunum | 7 (33.3%) | 1 (33.3%) | |
| Ileum | 5 (23.8%) | 1 (33.3%) | |
| T stage | NS | ||
| T1 | 1 (4.5%) | 0 | |
| T2 | 1 (4.5%) | 0 | |
| T3 | 13 (59.1%) | 2 (40%) | |
| T4 | 7 (31.8%) | 3 (60%) | |
| N stage | NS | ||
| N0 | 7 (31.8%) | 1 (20%) | |
| N1 | 15 (68.2%) | 4 (80%) | |
| No Crohn’s disease | 19 (86.4%) | 4 (80%) | NS |
| Crohn’s disease | 3 (13.6%) | 1 (20%) |
*Only 24 tumours were considered for tumour site; †including cases that were diffusely and focally positive.
NS, not significant.
Table 2.
Relation between p53 expression and clinicopathological features in a series of 27 small intestinal adenocarcinomas
| p53+ (n = 14) | p53− (n = 13) | p Value | |
| Site* | NS | ||
| Duodenum | 4 (33.3%) | 6 (50%) | |
| Jejunum | 5 (41.7%) | 3 (25%) | |
| Ileum | 3 (25%) | 3 (25%) | |
| T stage | NS | ||
| T1 | 0 | 1 (7.7%) | |
| T2 | 0 | 1 (7.7%) | |
| T3 | 10 (71.4%) | 5 (38.5%) | |
| T4 | 4 (28.6%) | 6 (46.1%) | |
| N stage | NS | ||
| N0 | 6 (42.9%) | 2 (15.4%) | |
| N1 | 8 (57.1%) | 11 (84.6%) | |
| No Crohn’s disease | 11 (78.6%) | 12 (92.3%) | NS |
| Crohn’s disease | 3 (21.4%) | 1 (7.7%) |
*Only 24 tumours were considered for tumour site.
DISCUSSION
Primary adenocarcinomas of the small intestine are rare compared with those occurring in the colon.1–3 The average annual incidence rate of this cancer using information collected in the USA by SEER (Surveillance, Epidemiology and End Results) between 1973 and 1990 was 3.7/million.2 Fifty percent of all small intestinal adenocarcinomas occur in the duodenum,1,2 especially in the second portion, in areas adjacent to the ampulla of Vater.21 This observation may implicate bile as a small bowel carcinogen at this site.21,22 Moreover, there is an association between cholecystectomy and small intestinal and proximal colon cancers, which decreases with increasing distance from the common bile duct.22 Other theories have been proposed to explain the relatively low incidence of this cancer, such as the rapid turnover of the epithelial cells, the rapid transit time reducing exposure to luminal carcinogens, an immunological protective local IgA mediated mechanism, and the relative sterility of enteric contents converting bile salts into carcinogens.23 Crohn’s disease is a well recognised risk factor for small intestinal carcinoma, but the anatomical distribution of small intestinal adenocarcinomas complicating Crohn’s disease is different from those appearing in the general population, with an ileal predominance.5,24
We used TMA technology to investigate the expression of proteins potentially involved in small intestinal carcinogenesis. This technology allowed us to process all our samples on a single slide, thus assuring identical experimental conditions. Nevertheless, the amount of tumour analysed was considerably reduced and might not be representative of the protein expression patterns of the entire tumour. Most validation studies published in the literature suggest that threefold redundancy is highly representative of colon cancer sections. Moreover, the validation study carried out in our laboratory on colorectal cancer showed that one core allowed appropriate classification of 96.7% to 97.1% of cases and three cores 98.8% to 99.6%.24a Therefore, we chose to analyse three spots for each tumour in our study. The results of the analysis of the three cores was similar to those from the whole tissue section in all cases for the expression of hMLH1 and hMSH2. We conclude that TMA technology with threefold redundancy adequately represents the immunohistochemical pattern in small intestinal adenocarcinoma.
Few studies have investigated the mechanisms of epithelial carcinogenesis in the small bowel. A particular pathway involved in cancer development and progression is characterised by inactivation of the DNA mismatch repair system. This pattern, which leads to DNA replication errors, is characteristic of tumours arising in the setting of HNPCC syndrome, with an underlying germline mutations in the hMSH2 or hMLH1 genes.25 Patients with HNPCC syndrome are at an increased risk for extracolonic cancers, including small intestinal carcinomas,10,11 with a cumulative risk of this particular tumour type estimated at 1%.11 Cancers developing in HNPCC syndrome are characterised by MSI, which also occurs in 10–15% of sporadic colorectal tumours and in sporadic extracolonic cancers, especially in tumours within the HNPCC spectrum (stomach, endometrium, and urinary tract). Some series studied the frequency of MSI in sporadic small intestine adenocarcinomas.13–16,19 In these studies, MSI status had been determined by polymerase chain reaction amplification of microsatellite markers: Hibi et al found MSI in five of 11 cases,15 Keller et al in one of five cases,16 Wheeler et al in one of 21 cases,14 Rashid et al in two of 15 cases,13 and Bläker et al in two of 17 cases.19 Because immunohistochemistry for hMLH1 and hMSH2 proteins provides a sensitive and specific method for screening for MSI, we studied the expression of these two proteins. We found a loss of expression of hMLH1 protein in two of 27 sporadic small intestinal carcinomas, although there was no loss of hMSH2 or hMSH6. These results are consistent with the report of Wheeler et al,14 who also performed immunohistochemistry of MMR proteins in small intestinal adenocarcinomas: all tumours stained positive for hMLH1 and hMSH2. Therefore, it seems that MSI and loss of expression of MMR proteins is a relatively rare event in sporadic small intestinal adenocarcinomas, with a frequency equal to or even lower than that seen in sporadic colorectal cancers. Because we only saw a loss of hMLH1 expression in our cases, we could hypothesise that the inactivation of the hMLH1 gene was the result of hypermethylation of the promoter, as has been demonstrated in colorectal cancer.26
“Microsatellite instability and loss of expression of mismatch repair proteins is a relatively rare event in sporadic small intestinal adenocarcinomas, with a frequency equal to or even lower than that seen in sporadic colorectal cancers”
MSI has been described as a feature of some ulcerative colitis associated colorectal cancers.27 However, it is only rarely involved in this setting: Cawkwell et al found neither MSI by PCR nor loss of hMLH1 and hMSH2 expression among 33 colorectal carcinomas complicating ulcerative colitis.28 MSI also seems to be very unusual in cancers complicating Crohn’s disease, even rarer than in those occurring in ulcerative colitis.29 However, it should be noted that these studies only included colorectal carcinomas. In our limited study of four cases, there was no loss of hMLH1 and hMSH2 protein expression.
The molecular genetic alterations present in most sporadic colorectal adenocarcinomas are well known, and include activating mutations of the ras protooncogene,30 mutation and deletion of the p53 gene on chromosome 17p, deletion of the DCC gene on chromosome 18q, and mutation of the APC gene on chromosome 5q.31 These last events characterise the loss of heterozygosity pathway of colorectal carcinogenesis. Alterations in the p53 gene are the most common genetic event in sporadic small intestinal adenocarcinomas.17,18 Our results are in agreement with these observations; indeed, overexpression of the p53 gene product was present in 14 of the 27 tumours. These studies, together with our findings, support the hypothesis that p53 plays a major role in the progression of small intestinal adenocarcinomas. In contrast, allelic losses of the APC and DCC genes are uncommon events in small intestinal adenocarcinoma.13–15,17 Recently, a high frequency of loss at chromosome 18q21 has been shown in small intestinal adenocarcinomas.19 18q21 is the location of three putative tumour suppressor genes, DCC,32 SMAD4,33 and SMAD2.34 DNA sequencing for SMAD4/DPC4 revealed sequence alterations in four of 17 small intestinal carcinomas.19 The SMAD4 gene, which is often mutated in pancreatic35 and colon36 carcinomas, plays a key role in the TGFβ signal transduction pathway. SMAD4 can be inactivated by intragenic mutation in one allele combined with loss of the other allele (loss of heterozygosity) or by homozygous deletions.37 Both genetic alterations lead to a complete absence of Smad4 labelling in the nucleus and the cytoplasm. Because loss of Smad4 protein expression has been demonstrated as a sensitive and specific indicator for SMAD4 gene alterations,38,39 we used this technique to determine the status of SMAD4 in 27 sporadic small intestinal adenocarcinomas, including four cases complicating Crohn’s disease. We found a loss of immunostaining for the Smad4 protein in five of the 27 tumours. No case with loss of Smad4 expression also lost hMLH1, hMSH2, or hMSH6 expression. These data are similar to those obtained in colorectal cancer.40 No colorectal cancers with MSI showed loss of Smad4 protein or SMAD4 gene mutation, although many of the MSI positive cell lines did carry TGFβIIR changes, which is particularly vulnerable in the case of dysfunction of the MMR system.41 On the contrary, the loss of Smad4 protein expression is often associated with overexpression of the p53 protein. Our study using TMA technology suggests a role for the SMAD4 gene in small intestinal adenocarcinoma tumorigenesis, in accordance with the results obtained by Bläker et al.19
β Catenin, a protein involved in cell adhesion and in intracellular signalling, is regulated by the APC protein, which facilitates its degradation.42 When the APC gene is mutated, β catenin accumulates in the cytoplasm and in the nucleus. The translocation of β catenin from the cell membrane to the cytoplasm or nucleus is thought to play a role in the development of colorectal cancers.43 This translocation is mostly caused by mutations in the APC gene. However, it seems that a relatively large number of colon cancers have β catenin mutations, with a wild-type APC gene.44 In our study, nuclear or cytoplasmic expression of β catenin was present in all specimens, suggesting that it plays a role in small intestinal carcinogenesis. These data are in agreement with a previous report.19
Take home messages.
It appears that inactivation of the SMAD4/DPC4 gene is involved in small intestinal adenocarcinoma tumorigenesis
Overexpression of p53 and abnormal expression of β catenin are two common events in small intestinal adenocarcinoma
The loss of expression of the DNA mismatch repair proteins (hMLH1, hMSH2, and hMSH6) is rare in small intestinal adenocarcinomas
It appears that the process of tumorigenesis is similar in small intestinal adenocarcinomas and malignant ampullary tumours
We found no significant differences in the expression of hMLH1, hMSH2, hMSH6, Smad4, p53, or β catenin proteins between small intestinal adenocarcinomas and malignant ampullary tumours. Similar to colorectal carcinoma, there is an adenoma–adenocarcinoma sequence in malignant ampullary tumours,45 and K-ras mutations seem to be involved in the progression of the disease.46 Molecular events in sporadic ampullary neoplasms also include p53 gene inactivation, alterations of the SMAD4 gene, and chromosome 5p21 allelic losses.47 Mutations of the APC gene are not frequent events.48,49 Some studies have reported a frequency of about 20% for MSI in this type of cancer.50–52
In conclusion, our immunohistochemical study, which was the first to make use of TMA for small intestinal adenocarcinomas and malignant ampullary tumours, shows that TMA technology with threefold redundancy adequately represents the immunohistochemical pattern of these two types of cancer. The inactivation of the SMAD4/DPC4 gene seems to be involved in small intestinal adenocarcinoma tumorigenesis. Overexpression of the p53 protein and translocation of β catenin to the nucleus/cytoplasm are two common events. In contrast, loss of expression of the proteins of the MMR system is less prominent in the pathogenesis of sporadic small intestinal adenocarcinomas. Finally, the carcinogenetic process appears to be similar in small intestinal adenocarcinomas and malignant ampullary tumours, as suggested by Scarpa and Zamboni.53
Acknowledgments
We acknowledge the financial help of the “Association Charles Debray”.
Abbreviations
APC, adenomatous polyposis coli
DCC, deleted in colorectal cancer
DPC4, deleted in pancreatic cancer 4
FAP, familial adenomatous polyposis
HNPCC, hereditary non-polyposis colorectal cancer
MMR, mismatch repair
MSI, microsatellite instability
TGFβ, transforming growth factor β
TMA, tissue microarray
REFERENCES
- 1.DiSario JA, Burt RW, Vargas H, et al. Small bowel cancer: epidemiological and clinical characteristics from a population-based registry. Am J Gastroenterol 1994;89:699–701. [PubMed] [Google Scholar]
- 2.Chow JS, Chen CC, Ahsan H, et al. A population-based study of the incidence of malignant small bowel tumours: SEER, 1973–1990. Int J Epidemiol 1996;25:722–8. [DOI] [PubMed] [Google Scholar]
- 3.Lien GS, Mori M, Enjoji M. Primary carcinoma of the small intestine. A clinicopathologic and immunohistochemical study. Cancer 1988;61:316–23. [DOI] [PubMed] [Google Scholar]
- 4.Petras RE, Mir-Madjlessi SH, Farmer RG. Crohns disease and intestinal carcinoma. A report of 11 cases with emphasis on associated epithelial dysplasia. Gastroenterology 1987;93:1307–14. [PubMed] [Google Scholar]
- 5.Michelassi F, Testa G, Pomidor WJ, et al. Adenocarcinoma complicating Crohn’s disease. Dis Colon Rectum 1993;36:654–61. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein D, Rogers A. Malignancy in Crohn’s disease. Am J Gastroenterol 1996;91:434–40. [PubMed] [Google Scholar]
- 7.Arber N, Hibshoosh H, Yasui W, et al. Abnormalities in the expression of cell cycle-related proteins in tumors of the small bowel. Cancer Epidemiol Biomarkers Prev 1999;8:1101–5. [PubMed] [Google Scholar]
- 8.Sheldon CD, Hodson ME, Carpenter LM, et al. A cohort study of cystic fibrosis and malignancy. Br J Cancer 1993;68:1025–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neglia JP, FitzSimmons SC, Maisonneuve P, et al. The risk of cancer among patients with cystic fibrosis. N Engl J Med 1995;332:494–9. [DOI] [PubMed] [Google Scholar]
- 10.Lynch HT, Smyrk TC, Watson P, et al. Genetics, natural history, tumor spectrum, and pathology of hereditary nonpolyposis colorectal cancer: an updated review. Gastroenterology 1993;104:1535–49. [DOI] [PubMed] [Google Scholar]
- 11.Aarnio M, Mecklin JP, Aaltonen LA, et al. Life-time risk of different cancers in hereditary non-polyposis colorectal cancer (HNPCC) syndrome. Int J Cancer 1995;64:430–3. [DOI] [PubMed] [Google Scholar]
- 12.Lindor N, Burgart LJ, Leontovich O, et al. Immunohistochemistry versus microsatellite instability testing in phenotyping colorectal tumors. J Clin Oncol 2002;20:1043–8. [DOI] [PubMed] [Google Scholar]
- 13.Rashid A, Hamilton SR. Genetic alterations in sporadic and Crohn’s-associated adenocarcinomas of the small intestine. Gastroenterology 1997;113:127–35. [DOI] [PubMed] [Google Scholar]
- 14.Wheeler J, Warren BF, McC Mortensen NJ, et al. An insight into the genetic pathway of adenocarcinoma of the small intestine. Gut 2002;50:218–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hibi K, Kondo K, Akiyama S, et al. Frequent genetic instability in small intestinal carcinomas. Jpn J Cancer Res 1995;86:357–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller G, Rotter M, Vogelsang H, et al. Microsatellite instability in adenocarcinomas of the upper gastrointestinal tract. Relation to clinicopathological data and family history. Am J Pathol 1995;147:593–600. [PMC free article] [PubMed] [Google Scholar]
- 17.Nishiyama K-I, Yao T, Yonemasu H, et al. Overexpression of p53 protein and point mutation of K-ras genes in primary carcinoma of the small intestine. Oncol Rep 2002;9:293–300. [PubMed] [Google Scholar]
- 18.Arai M, Shimizu S, Imai Y, et al. Mutations of the Ki-ras, p53 and APC genes in adenocarcinomas of the human small intestine. Int J Cancer 1997;70:390–5. [DOI] [PubMed] [Google Scholar]
- 19.Bläker H, von Herbay A, Penzel R, et al. Genetics of adenocarcinomas of the small intestine: frequent deletions at chromosome 18q and mutations of the SMAD4 gene. Oncogene 2002;21:158–64. [DOI] [PubMed] [Google Scholar]
- 20.Kononen J, Bubendorf L, Kallioniemi A, et al. Tissue microarrays for high-throughput molecular profiling of tumor specimens. Nat Med 1998;4:844–7. [DOI] [PubMed] [Google Scholar]
- 21.Ross RK, Hartnett NM, Bernstein L, et al. Epidemiology of adenocarcinomas of the small intestine: is bile a small bowel carcinogen? Br J Cancer 1991;63:143–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lagergren J, Ye W, Ekbom A. Intestinal cancer after cholecystectomy: is bile involved in carcinogenesis? Gastroenterology 2001;121:542–7. [DOI] [PubMed] [Google Scholar]
- 23.Arber N, Neugut AI, Weinstein IB, et al. Molecular genetics of small bowel cancer. Cancer Epidemiol Biomarkers Prev 1997;6:745–8. [PubMed] [Google Scholar]
- 24.Lashner BA. Risk factors for small bowel cancer in Crohn’s disease. Dig Dis Sci 1992;37:1179–84. [DOI] [PubMed] [Google Scholar]
- 24a.Jourdan F, Sebbagh N, Comperat E, et al. Tissue microarray technology: validation in colorectal carcinoma and analysis of p53, hMLH1, and hMLH2 immunohistochemical expression. Virchows Arch 2003;443:115–21. [DOI] [PubMed] [Google Scholar]
- 25.Liu B, Parsons R, Papadopoulos N. Analysis of mismatch repair genes in hereditary non-polyposis colorectal cancer patients. Nat Med 1996;2:169–74. [DOI] [PubMed] [Google Scholar]
- 26.Kang GH, Shim YH, Ro JY. Correlation of methylation of the hMLH1 promoter with lack of expression of hMLH1 in sporadic gastric carcinomas with replication error. Lab Invest 1999;79:903–9. [PubMed] [Google Scholar]
- 27.Suzuki H, Harpaz N, Tarmin L. Microsatellite instability in ulcerative colitis-associated colorectal dysplasias and cancers. Cancer Res 1994;54:4841–4. [PubMed] [Google Scholar]
- 28.Cawkwell L, Sutherland F, Murgatroyd H, et al. Defective hMSH2/hMLH1 protein expression is seen infrequently in ulcerative colitis associated colorectal cancers. Gut 2000;46:367–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noffsinger A, Kretschmer S, Belli J, et al. Microsatellite instability is uncommon in intestinal mucosa of patients with Crohn’s disease. Dig Dis Sci 2000;45:378–84. [DOI] [PubMed] [Google Scholar]
- 30.Bos JL, Fearon ER, Hamilton SR, et al. Presence of ras gene mutations in human colorectal cancers. Nature 1987;327:293–7. [DOI] [PubMed] [Google Scholar]
- 31.Kim H, Jen J, Vogelstein B, et al. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am J Pathol 1994;145:148–56. [PMC free article] [PubMed] [Google Scholar]
- 32.Cho KR, Oliner JD, Simons JW, et al. The DCC gene: structural analysis and mutations in colorectal carcinomas. Genomics 1994;19:525–31. [DOI] [PubMed] [Google Scholar]
- 33.Hahn SA, Schutte M, Hoque AT, et al. DPC4, a candidate tumor suppressor gene at human chromosome 18q21.1. Science 1996;271:350–3. [DOI] [PubMed] [Google Scholar]
- 34.Riggins GJ, Thiagalingam S, Rozenblum E, et al. Mad-related genes in the human. Nat Genet 1996;13:347–9. [DOI] [PubMed] [Google Scholar]
- 35.Hahn SA, Hoque AT, Moskaluk CA, et al. Homozygous deletion map at 18q21.1 in pancreatic cancer. Cancer Res 1996;56:490–4. [PubMed] [Google Scholar]
- 36.Thiagalingam S, Lengauer C, Leach FS, et al. Evaluation of candidate tumour suppressor genes on chromosome 18 in colorectal cancers. Nat Genet 1996;13:343–6. [DOI] [PubMed] [Google Scholar]
- 37.Hruban RH, Offerhaus GJA, Kern SE, et al. Tumor-suppressor genes in pancreatic cancer. J Hepatobiliary Pancreat Surg 1998;5:383–91. [DOI] [PubMed] [Google Scholar]
- 38.Wilentz RE, Iacobuzio-Donahue CA, Argani P, et al. Loss of expression of Dpc4 in pancreatic intraepithelial neoplasia: evidence that DPC4 inactivation occurs late in neoplastic progression. Cancer Res 2000;60:2002–6. [PubMed] [Google Scholar]
- 39.Wilentz RE, Su GH, Dai JL, et al. Immunohistochemical labelling for dpc4 mirrors genetic status in pancreatic adenocarcinomas: a new marker of DPC4 inactivation. Am J Pathol 2000;156:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salovaara R, Roth S, Loukola A, et al. Frequent loss of SMAD4/DPC4 protein in colorectal cancers. Gut 2002;51:56–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woodford-Richens KL, Rowan AJ, Gorman P, et al. SMAD4 mutations in colorectal cancer probably occur before chromosomal instability, but after divergence of the microsatellite instability pathway. Proc Natl Acad Sci U S A 2001;98:9719–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rubinfield B, Albert I, Porfiri E, et al. Loss of β-catenin regulation by the APC tumour supressor protein correlates with loss of structure due to common mutations of the gene. Cancer Res 1997;57:4624–30. [PubMed] [Google Scholar]
- 43.Hao X, Tomlinson I, Ilyas M, et al. Reciprocity between membranous and nuclear expression of β-catenin in colorectal tumors. Virchows Arch 1997;431:167–72. [DOI] [PubMed] [Google Scholar]
- 44.Sparks AB, Morin PJ, Vogelstein B, et al. Mutational analysis of the APC/β-catenin/Tcf pathway in colorectal cancer. Cancer Res 1998;58:1130–4. [PubMed] [Google Scholar]
- 45.Seifert E, Schulte F, Stolte M. Adenoma and carcinoma of the duodenum and papilla of Vater: a clinicopathologic study. Am J Gastroenterol 1992;87:37–42. [PubMed] [Google Scholar]
- 46.Zhao B, Kimura W, Futakawa N, et al. p53 and p21/Waf1 protein expression and K-ras codon 12 mutation in carcinoma of the papilla of Vater. Am J Gastroenterol 1999;94:2128–34. [DOI] [PubMed] [Google Scholar]
- 47.Moore PS, Orlandini S, Zamboni G, et al. Pancreatic tumours: molecular pathways implicated in ductal cancer are involved in ampullary but not in exocrine nonductal or endocrine tumorigenesis. Br J Cancer 2001;84:253–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Achille A, Scupoli MT, Magalini AR, et al. APC gene mutations and allelic losses in sporadic ampullary tumours: evidence of genetic difference from tumours associated with familial adenomatous polyposis. Int J Cancer 1996;68:305–12. [DOI] [PubMed] [Google Scholar]
- 49.Achille A, Baron A, Zamboni G, et al. Chromosome 5 allelic losses are early events in tumours of the papilla of Vater and occur at sites similar to those of gastric cancer. Br J Cancer 1998;78:1653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Achille A, Biasi MO, Zamboni G, et al. Cancers of the papilla of Vater: mutator phenotype is associated with good prognosis. Clin Cancer Res 1997;3:1841–7. [PubMed] [Google Scholar]
- 51.Imai Y, Tsurutani N, Oda H, et al. Genetic instability and mutation of the TGF-beta-receptor-II gene in ampullary carcinomas. Int J Cancer 1998;76:407–11. [DOI] [PubMed] [Google Scholar]
- 52.Imai Y, Inoue T, Ishikawa T. Mutations of the human Mut S homologue 6 gene in ampullary carcinoma and gastric cancer. Int J Cancer 1998;78:576–80. [DOI] [PubMed] [Google Scholar]
- 53.Scarpa A, Zamboni G. Genomic anomalies in pancreatic tumors other than common adenocarcinoma. Ann N Y Acad Sci 1999;880:179–90. [DOI] [PubMed] [Google Scholar]