Abstract
CstF-64 (cleavage stimulation factor-64), a major regulatory protein of polyadenylation, is absent during male meiosis. Therefore a paralogous variant, τCstF-64 is expressed in male germ cells to maintain normal spermatogenesis. Based on sequence differences between τCstF-64 and CstF-64, and on the high incidence of alternative polyadenylation in testes, we hypothesized that the RBDs (RNA-binding domains) of τCstF-64 and CstF-64 have different affinities for RNA elements. We quantified Kd values of CstF-64 and τCstF-64 RBDs for various ribopolymers using an RNA cross-linking assay. The two RBDs had similar affinities for poly(G)18, poly(A)18 or poly(C)18, with affinity for poly(C)18 being the lowest. However, CstF-64 had a higher affinity for poly(U)18 than τCstF-64, whereas it had a lower affinity for poly(GU)9. Changing Pro-41 to a serine residue in the CstF-64 RBD did not affect its affinity for poly(U)18, but changes in amino acids downstream of the C-terminal α-helical region decreased affinity towards poly(U)18. Thus we show that the two CstF-64 paralogues differ in their affinities for specific RNA sequences, and that the region C-terminal to the RBD is important in RNA sequence recognition. This supports the hypothesis that τCstF-64 promotes germ-cell-specific patterns of polyadenylation by binding to different downstream sequence elements.
Keywords: cleavage stimulation factor (CstF), male germ cell, mRNA processing, polyadenylation, RNA-binding domain (RBD), spermatogenesis
Abbreviations: CF Im, mammalian cleavage factor I; CPSF, cleavage and polyadenylation specificity factor; CstF, cleavage stimulation factor; DSE, downstream sequence element; GST, glutathione S-transferase; RBD, RNA-binding domain; RRM, RNA recognition motif; SELEX, systematic evolution of ligands by exponential enrichment; SV40, simian virus 40
INTRODUCTION
Polyadenylation is the process by which approx. 200 adenylate residues are added to 3′-ends of most eukaryotic pre-mRNAs [1,2]. This process is crucial for proper transcription, splicing, transport, translation and stability of mRNAs [3–12]. In mammals, polyadenylation requires at least five distinct components: the CPSF (cleavage and polyadenylation specificity factor), the CstF (cleavage stimulation factor), CF Im and CF IIm (mammalian cleavage factors I and II) and the PAP [poly(A) polymerase] [1,2]. Other proteins may also contribute to the efficiency or specificity of polyadenylation [13–19].
Two proteins of the polyadenylation machinery are known to bind to important RNA sequence elements near polyadenylation sites. The 160 kDa subunit of CPSF binds to the canonical AAUAAA element found usually 12–30 nt upstream of the cleavage site of a pre-mRNA [20,21], while CstF binds to a U- or G/U-rich DSE (downstream sequence element) through its 64 kDa subunit, CstF-64 [22–24]. CstF-64 contains an N-terminal RBD (RNA-binding domain) consisting of an RRM (RNA recognition motif) of the type found in the U1A small ribonucleoprotein [23,25–28], a CstF-77-binding domain [29], a repeated MEARA helical region [23,30] and a conserved C-terminal domain reported to bind the transcription factor PC-4 [31].
Meiotic and post-meiotic male germ cell pre-mRNAs have a higher incidence of non-AAUAAA polyadenylation signals than somatic cells [32,33]. There is also a much higher incidence of alternative polyadenylation in male germ cells [34,35]. We described earlier a germ-cell-expressed paralogue of CstF-64 that we designated τCstF-64 [36,37], which might contribute to polyadenylation of non-AAUAAA pre-mRNAs [32]. X-linked genes including CstF-64 are not expressed in meiotically dividing male germ cells due to the unique process of male X-inactivation [38–40]. To compensate for X chromosomal inactivation, many X-linked genes have autosomal paralogues that are expressed in male germ cells [41]. Similarly, τCstF-64 is on an autosome (Cstf2t in mice, CSTF2T in humans) and has diverged from the somatic CstF-64 [37,42]. Hence, we have hypothesized that τCstF-64 might be at least partially responsible for alternative polyadenylation and polyadenylation of non-AAUAAA pre-mRNAs in male germ cells [32].
The cloning and sequencing of the mouse and human τCstF-64 cDNAs showed that the translated sequence was similar to other known isoforms of CstF-64 (72.9 and 74% identical with mouse and human respectively, [37,42]). Alignment of CstF-64 and τCstF-64 showed that they contained common features such as similar RBDs, CstF-77 interaction domains, MEARA repeat regions, and C-terminal domains. The RRM-type RBDs of CstF-64 and τCstF-64 consist of two α-helices (α1 and α2; Figure 1) and four β-sheets (β1–β4; Figure 1). α-Helices α1 and α2 stabilize the RNA-binding β-sheet surface from behind (Figure 1B, [28]). Recently, Pérez Cañadillas and Varani [27] determined the solution structure of the CstF-64 RBD and found that it contained a third α-helix (α3) that is C-terminal to the core RBD. Those authors proposed a model in which α3 lay across the RNA-contact β-sheet surface of the CstF-64 RBD and precluded binding of RNA until moved by some signal from elsewhere in the molecule.
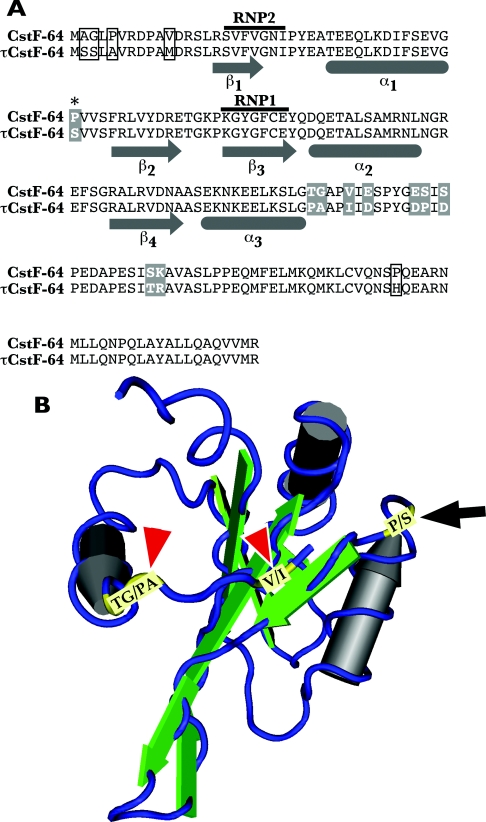
Figure 1. Amino acid differences of the RBD between the CstF-64 and τCstF-64 isoforms.
(A) CstF-64 and τCstF-64 amino acid sequences. Constructs used in the present study end at amino acid 180 as shown. α-Helical and β-sheet regions are indicated; RNP1 and RNP2 indicate conserved sequence motifs in RRM-type RNA-binding proteins. Amino acids that differ between CstF-64 and τCstF-64 are boxed or boxed in grey; amino acids that were altered in the present study are boxed in grey, and amino acid 41 is indicated by an asterisk. (B) Three-dimensional representation of the CstF-64 RBD [27] highlighting the differences between CstF-64 and τCstF-64. α-Helices are grey cylinders, β-sheets are green ribbons, and the peptide backbone is a blue thread. Amino acids that differ are yellow. The black arrow indicates amino acid 41 (P/S), and amino acids 106, 107 and 110 (TG/PA and V/I) are indicated by red arrowheads.
Alignment of CstF-64 and τCstF-64 RBD sequences suggests that they have nearly identical three-dimensional structures. However, the core RBD of τCstF-64 differs from that of CstF-64 in that it has a serine residue at amino acid 41 rather than a proline (Figure 1A, [37,42]). The third α-helices (α3) in τCstF-64 and CstF-64 are the same, but there are a number of amino acid differences between CstF-64 and τCstF-64 immediately downstream of α3 (Figure 1A). These variations between τCstF-64 and CstF-64 suggest a potential for the two proteins to have differing affinities for specific RNA sequences.
Currently, no consensus ‘testis-specific polyadenylation signal’ has been determined [32,33]. This led us to consider an alternative approach for determining potential sequences based on the binding preferences of τCstF-64. In the present study, we examined the binding affinities of several ribopolymers to CstF-64 and τCstF-64. A UV cross-linking competition assay was used to determine binding of the CstF-64 and τCstF-64 RBDs to various RNA substrates. We found that CstF-64 and τCstF-64 had nearly identical affinities for each of the following substrates, poly(A), poly(G) and poly(C), with the affinity for poly(C) being significantly lower than for any of the other elements. However, we found that CstF-64 bound poly(U) more efficiently than poly(GU). On the other hand, it was the reverse for τCstF-64: poly(GU) was bound more readily than poly(U). When using mutants to examine specific amino acids that might contribute to such specificities, we saw no change in binding of CstF-64 to poly(U) by changing the Pro-41 of CstF-64 to a serine residue. However, changing nine amino acids in the region adjacent to α3 decreased slightly the affinity of CstF-64 for poly(U), suggesting that the region in or around α3 contributed to the different RNA-binding specificities of CstF-64 and τCstF-64. The different binding affinities of CstF-64 and τCstF-64 for different RNA elements are consistent with the hypothesis that τCstF-64 promotes germ cell-specific patterns of polyadenylation by binding to different sequence elements downstream of polyadenylation sites than does CstF-64.
EXPERIMENTAL
Recombinant protein purification
RBDs of mouse CstF-64 and mouse τCstF-64 were prepared as GST (glutathione S-transferase) fusion proteins. PCR was performed on previously described clones of mouse CstF-64 and mouse τCstF-64 cDNAs [37,42] with primers that inserted a BamHI site upstream of the ATG and an EcoRI site 550 bp downstream of the region that contained the RBDs (CstF-64: Primer 1, 5′-CCTCGGGGGGGATCCATGGCGGGTTTG-3′ and Primer 2, 5′-CAGCGCAATCTCAGAATTCACAATTCTC-3′; τCstF-64: Primer 3, 5′-GTCCTCTCGGGATCCATCATGTCGAGTTTG-3′ and Primer 4, 5′-GCAATCTCTGAATTCATGATTCTCATC-3′). These PCRs generated fragments of 579 bp and 580 bp for mouse CstF-64 RBD and τCstF-64 RBD respectively, which were cloned into pCR-TOPO II vector using the TOPO-TA cloning system (Invitrogen, Carlsbad, CA, U.S.A.). These constructs were used as the source to clone CstF-64 and τCstF-64 RBDs into the BamHI and EcoRI sites of pGEX-2TK expression vector (Amersham Biosciences, Piscataway, NJ, U.S.A.) ensuring they were in-frame with the GST tag. Mouse CstF-64 RBD and τCstF-64 RBD in pGEX-2TK plasmid DNAs were electroporated using a Cell-Porator® Escherichia coli pulser (Life Technologies, Gaithersburg, MD, U.S.A.) into BL21 (DE3) Star One Shot cells (Invitrogen). These cells were induced at mid-exponential phase by the addition of 0.25 mM (final concentration) IPTG (isopropyl β-D-thiogalactoside) at 37 °C for 5 h. Bacterial pellets were resuspended in lysis buffer (50 mM Tris/HCl, pH 7.5, 0.1 M NaCl, 1.0 mM EDTA, 0.5% Nonidet P40, 1 mM o-phenanthroline, 50 μM aprotinin and 1 mM EGTA), sonicated on ice, and cleared by centrifugation (158000 gav, 45 min and 4 °C; Beckman Ti45 rotor; 45000 rev./min). GST-tagged recombinant proteins were isolated in batch using the glutathione–Sepharose 4B system (Amersham Biosciences) according to the manufacturer's instructions. Protein concentration was estimated using the Bio-Rad Protein Assay reagent (Bio-Rad Laboratories, Hercules, CA, U.S.A.) and by comparison with known amounts of BSA isolated by SDS/12.5% PAGE and visualized using Coomassie Brilliant Blue R-250 (Fisher Scientific, Hampton, NH, U.S.A.).
RNA ligands
The four different RNA ribopolymers, poly(U)18, poly(A)18, poly(G)18 and poly(GU)9 (Table 1) were synthesized using the T7-MEGAshortscript High Yield Transcription kit (Ambion, Austin, TX, U.S.A.) as directed. Poly(C)18 was synthesized by Midland Chemicals (Midland, TX, U.S.A.). 32P-labelled poly(U)18 substrate was synthesized by including 2 μl of [32P]UTP (3000 Ci/mmol; PerkinElmer, Boston, MA, U.S.A.). RNAs were then phenol/chloroform ratio extracted followed by ethanol precipitation. Non-radiolabelled RNA ribopolymers were quantified spectrophotometrically by measuring absorbance at 260, 280 and 320 nm. Radiolabelled RNA substrate was washed in 10% (w/v) trichloroacetic acid after extraction and precipitation, then c.p.m. (counts per minute) were measured on a Wallac 1409 scintillation counter (PerkinElmer).
Table 1. Oligonucleotides used in RNA ligand preparation using the T7-MEGAshortscript High Yield Transcription kit.
| Strand | Sequence |
|---|---|
| Top | 5′-TAATACGACTCACTATAGGG-3′ |
| Bottom | |
| Poly(U)18 | 5′-AAAAAAAAAAAAAAAAAACCCTATAGTGAGTCGTATTA-3′ |
| Poly(G)18 | 5′-CCCCCCCCCCCCCCCTATAGTGAGTCGTATTA-3′ |
| Poly(A)18 | 5′-TTTTTTTTTTTTTTTTTTCCCTATAGTGAGTCGTATTA-3′ |
| Poly(GU)9 | 5′-AACAACAACAACAACAACCCCTATAGTGAGTCGTATTA-3′ |
Site-directed mutagenesis
Site-directed mutagenesis was used to create two mutant constructs of the GST–CstF-64 RBD fusion protein using the QuikChange II Site-Directed Mutagenesis kit (Stratagene, La Jolla, CA, U.S.A.). The first construct mutated amino acid 41 of CstF-64 RBD from a proline to a serine residue (‘CstF-64 P41S’; Table 2). A second construct included mutations for nine amino acids downstream of the RBD (T106P, G107A, V110I, E112D, E117D, S118D, S120D, S129T and K130R, ‘CstF-64 9AA mutant’; Table 2).
Table 2. Oligonucleotides used for site-directed mutagenesis.
Underlined nucleotide(s) indicate changes from the normal sequence.
| Mutation | Primers for mutation |
|---|---|
| P41S* | 5′-GAAACTAACAACTGACCCAACCTCAG-3′ |
| 5′-CTGAGGTTGGGTCAGTTGTTAGTTTC-3′ | |
| T106P† | 5′-GAAGAGCCTTGGCCCTGGTGCACCTG-3′ |
| (1) | 5′-CAGGTGCACCAGGGCCAAGGCTCTTC-3′ |
| G107A† | 5′-GGCCCTGCTGCACCTGTC-3′ |
| (4) | 5′-GACAGGTGCAGCAGGGCC-3′ |
| V110I, E112D† | 5′-CTGCACCTATCATAGATTCACCTTATG-3′ |
| (3) | 5′-CATAAGGTGAATCTATGATAGGTGCAG-3′ |
| E117D, S118D† | 5′-CCTTATGGAGATCCCATCAGCCCTG-3′ |
| (5) | 5′-CAGGGCTGATGGGATCTGGATAAGG-3′ |
| S120D† | 5′-GATCCCATCGACCCTGAGGATG-3′ |
| (6) | 5′-CATCCTCAGGGTCGATGGGATC-3′ |
| S129T, K130R† | 5′-GAATCCATTACCAGAGCAGTTGC-3′ |
| (2) | 5′-GCAACTGCTCTGGTAATGGATTC-3′ |
*In this pair of oligonucleotides GST–CstF-64 RBD at amino acid 41 are mutated from a proline residue to a serine to create CstF-64 P41S.
†This set of oligonucleotide pairs allow for the different mutations on GST–CstF-64 RBD to be introduced sequentially to obtain the CstF-64 9AA construct. The number beneath the mutation corresponds to the order in which the mutations were performed.
UV-cross-linking and substrate competition assay
The UV-cross-linking assay used 0.2 μg (4.2 pmol) of either GST–CstF-64 RBD or GST–τCstF-64 RBD with one of two amounts of 32P-labelled poly(U)18 substrate: 6.8 and 17 pmol. Simultaneously, competing amounts of one of five non-radioactive competitor RNAs [poly(U)18, poly(G)18, poly(A)18, poly(C)18 or poly(GU)9] were added in the range 0.84–168.2 pmol in a total reaction volume of 20 μl in Buffer D (20 mM Hepes, pH 7.9, 1.5 mM MgCl2, 10 mM KCl and 0.5 mM dithiothreitol) [43]. Binding of [32P]poly(U)18 was competed for by each non-radioactive ribopolymer; hence all affinities were measured relative to poly(U)18. Reactions were incubated at 30 °C for 30 min and then exposed to 1.0 J/cm2 of ultraviolet light in a CL-1000 UV cross-linker (UVP, Upland, CA, U.S.A.), followed by incubation with 10 units of RNase ONE™ (Promega, Madison, WI, U.S.A.) or RNase Cocktail™ (Ambion, Austin, TX, U.S.A.) at 37 °C for 15 min per reaction. Half of each reaction was boiled in SDS/PAGE loading buffer and separated by SDS/12.5%-PAGE. The gels were stained with Coomassie Brilliant Blue R-250, destained extensively in gel fix [20% (v/v) methanol and 10% (v/v) acetic acid], dried, and exposed to a storage phosphor screen (Molecular Dynamics, Sunnyvale, CA, U.S.A.) for band density quantification. Each condition was tested in triplicate and repeated at least three times.
Data analysis
Signal density data from the UV-cross-linking assays were analysed using Microsoft Excel spreadsheet program (Microsoft, Redmond, WA, U.S.A.). Binding inhibition constants (Ki) were extrapolated from Dixon plots for competitive inhibition as follows:
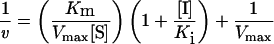 |
where v is the reaction velocity, S is the substrate concentration, I is the inhibitor concentration, Vmax is the rate of maximum turnover, Km is the substrate concentration at half-maximal velocity, and Ki is the competitive inhibition constant. This method is useful since it provides a graphical means of determining Ki for competitive inhibitors [44,45]. At the point of intersection, 1/v and [I] are the same for both lines (see Figure 3 and [44]). Since the inhibition is competitive, Vmax is also the same for both lines. Hence, the equation can be reduced to:
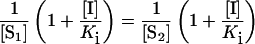 |
which is only true if either S1=S2 or [I]=−Ki. Thus [I] at the point of intersection equals −Ki. In this case, Ki is equivalent to Kd, and we will refer to these values henceforth as Kd [44]. ANOVA statistical analyses and post tests were conducted using GraphPad InStat version 3.0 (GraphPad Software, San Diego, CA, U.S.A.).
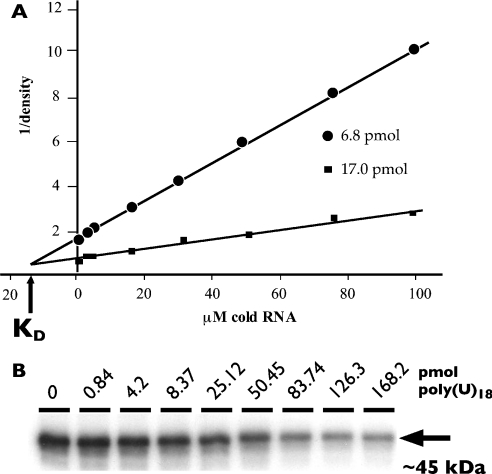
Figure 3. Competition/UV-cross-linking assay to determine relative affinities of RNA binding to the CstF-64 or τCstF-64 RBDs.
(A) Representative Dixon plot showing inhibition of UV cross-linking of the GST–CstF-64 RBD to 6.8 and 17 pmol of [32P]poly(U)18 by unlabelled poly(U)18. See the Experimental section for details. (B) Representative autoradiograph of GST–CstF-64 RBD binding to [32P]poly(U)18 (17 pmol) in the presence of increasing amounts (in pmol as indicated) of unlabelled poly(U)18. The arrow indicates the approximate mobility of the GST–CstF-64 RBD construct as determined by Coomassie Blue staining.
RESULTS
Because we were concerned that binding equilibria might change dramatically in a two-phase ‘partitioning’ assay such as electrophoretic mobility-shift assay [46], and that a filter binding assay might not have enough sensitivity to discriminate binding differences, we chose to establish a UV-cross-linking assay for the analysis of RNA binding to CstF-64 RBDs. The UV-cross-linking assay has the advantages of high sensitivity, the complexes could be ‘frozen’ by the UV-cross-linking step, thereby eliminating concerns over stability of the complex, and the technique has been well established in the analysis of CstF-64 (cf. [23,24,47,48]). We also chose to use the N-terminal RNA-binding portions of CstF-64 and τCstF-64 in the binding assays because the full-length protein inhibits binding of the RBD to RNA [23,24], the CstF-64 RBD region has been well characterized in other RNA-binding experiments [27,49,50], and the full-length proteins are not expressed well in bacteria ([24,37] and results not shown).
Conditions for the UV-cross-linking/binding assay
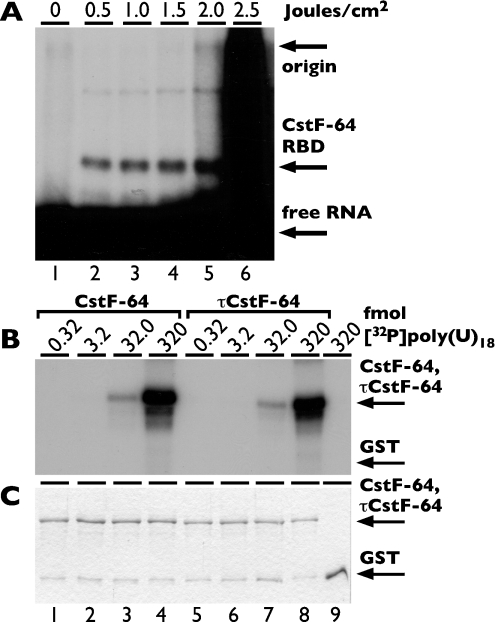
To determine optimal conditions of cross-linking, we incubated 0.2 μg of GST-fused CstF-64 RBD with 10000 c.p.m. of 32P-labelled SV40 (simian virus 40) late pre-mRNA substrate [24] at 30 °C for 30 min and then submitted the mixture to the UV fluxes indicated (Figure 2A). Mixtures were then separated by SDS/10%-PAGE and visualized by autoradiography. In the absence of UV light, no 32P-labelled GST–CstF-64 RBD was detected (Figure 2A, lane 1). Lanes 2–4 showed radioactive material at approx. 45 kDa, consistent with efficient cross-linking of 32P-labelled RNA to the GST–CstF-64 RBD (lanes 2–4). However, there was little evidence of slower migrating radioactive material in those lanes (lanes 2–4), indicating minimal higher order cross-linking. At higher amounts of UV flux (lanes 5 and 6), there was considerable slower migrating radioactivity, probably resulting from protein–protein cross-linking and other higher order events. Based on these results, we performed all subsequent experiments at 1.0 J/cm2 UV flux.
Figure 2. Conditions of UV cross-linking and RNA binding.
(A) GST–CstF-64 RBD was bound to 32P-labelled SV40 late pre-mRNA [24], exposed to increasing amounts of UV light from 0 J/cm2 (lane 1) to 2.5 J/cm2 (lanes 2–6), and separated by SDS/10% PAGE, dried, and analysed by autoradiography. Arrows indicate the mobility of GST–CstF-64 RBD (∼45 kDa) and free RNA. (B) Increasing amounts of [32P]poly(U)18 from 0.32 to 320 fmol was incubated with 0.2 μg of either GST–CstF-64 RBD (lanes 1–4), GST–τCstF-64 RBD (lanes 5–8), or GST (lane 9). After a 30 min incubation, the material was irradiated with 1.0 J/cm2 of UV light and processed for analysis as indicated in the Experimental section. Arrows indicate the mobilities of GST–CstF-64 RBD, GST–τCstF-64 RBD (∼45 kDa) and GST (∼26 kDa). (C) Coomassie Blue staining of SDS/PAGE from (B), indicating mobilities of GST–CstF-64 RBD, GST–τCstF-64 RBD and GST. Note that the Coomassie-stained product at approx. 26 kDa in lanes 1–8 is a breakdown product of the GST-fusion proteins.
In order to establish the linear range of substrate concentrations, we tested binding of both GST–CstF-64 RBD and GST–τCstF-64 RBD to a range of 32P-labelled poly(U)18 substrates (Figure 2B). The amount of [32P]poly(U)18 used ranged from 0.32 to 320 fmol. Signal from UV-cross-linked RNA increased linearly for both GST–CstF-64 RBD (lanes 1–4) and GST–τCstF-64 RBD (lanes 5–8). GST alone showed no RNA cross-linking even at 320 fmol of [32P]poly(U)18 (lane 9). Figure 2(C) shows Coomassie Blue staining of the gel to indicate amounts of protein loaded.
We also tested binding of [32P]poly(U)18 to GST–CstF-64 RBD with a range of 2–320 pmol of unlabelled poly(U)18 in a competition assay [results not shown, but see Figure 3 and Supplementary Figure S.1 (http://www.BiochemJ.org/bj/401/bj4010651add.htm)]. Based on these results, we chose 6.8 and 17 pmol of [32P]poly(U)18 as the appropriate concentrations to use in competition assays for Dixon analysis of the Ki because they flanked the apparent Kd [44].
CstF-64 and τCstF-64 have different affinities for RNA elements
Although CstF-64 and τCstF-64 share a great deal of sequence identity, variations in and around the RBD may account for the differences in binding to RNA elements (Figure 1). To measure these differences, we designed a competition and UV-cross-linking assay to determine the relative affinities of GST-fusion proteins containing the CstF-64 and τCstF-64 RBDs for simple ribopolymers [poly(U), poly(A), poly(G), poly(C) and poly(GU)]. These ribopolymers were chosen as simple models of more complex sequences that are often found downstream of authentic polyadenylation sites [33,51]. Recombinant CstF-64 and τCstF-64 RBDs were incubated with either 6.8 or 17 pmol of 32P-labelled poly(U)18. To determine relative affinity for different ribopolymers, increasing amounts of non-radioactive poly(U)18, poly(A)18, poly(G)18, or poly(C)18 were included in the incubations as competitors. To separate bound from unbound RNA, the mixtures were allowed to equilibrate, and then cross-linked by exposure to UV light, treated with ribonuclease and separated by SDS/PAGE (Figure 3B). Radioactivity bound by either the CstF-64 or τCstF-64 RBD was quantified, and the binding inhibition constant (Ki) was determined by Dixon plot analysis [44]. Figure 3(A) shows an example plot comparing inhibition of [32P]poly(U)18 binding and cross-linking to the CstF-64 RBD by unlabelled poly(U)18. Using this assay, we were able to measure Ki, which is equivalent to the dissociation constant (Kd) relative to poly(U) under the conditions described here [52]. We will henceforth refer to the values obtained as Kd values.
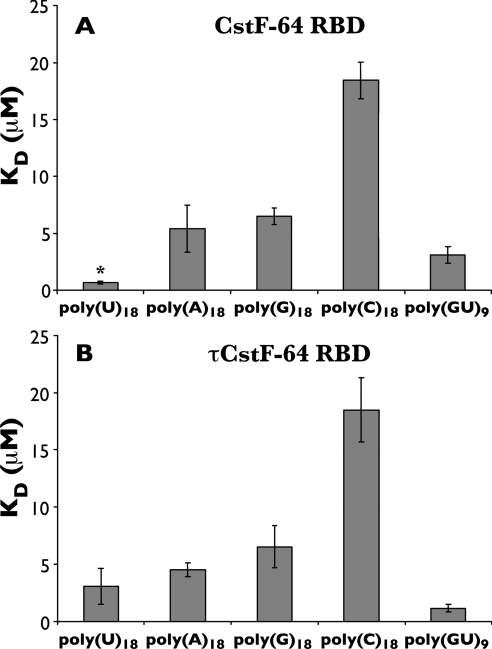
When CstF-64 was incubated with each of the different competitor ribopolymers ([poly(U)18, poly(A)18, poly(G)18 or poly(C)18] in the presence of substrate [32P]poly(U)18, CstF-64 had the highest affinity for poly(U)18 (Kd=0.66±0.13 μM) compared with the other ribopolymer competitors tested (Figure 4A). This is consistent with several previous studies that suggested that CstF-64 bound to U-rich elements [24,27,50,53,54]. When tested for the same ribopolymer competitors, τCstF-64 did not bind as strongly to poly(U) (Kd=3.04±1.56 μM) as did CstF-64 (Figure 4B). When poly(G)18 and poly(A)18 were tested for binding to τCstF-64, they showed similar binding affinities as CstF-64 (Kd≈4.5–6.5 μM) (Figure 4B). Finally, both CstF-64 and τCstF-64 bound poly(C)18 with the weakest affinity (Kd≈18.5 μM) (Figures 4A and 4B).
Figure 4. RNA-binding affinities of CstF-64 and τCstF-64 for ribopolymers [poly(U)18, poly(A)18, poly(G)18, poly(C)18 and poly(GU)9].
(A) Kd values (micromolar) relative to poly(U)18 of the GST–CstF-64 RBD for poly(U)18 (Kd=0.66±0.13 μM), poly(A)18 (Kd=5.4±2.05 μM), poly(G)18 (Kd=6.51±0.74 μM), poly(C)18 (Kd=18.44±1.62 μM) or poly(GU)9 (Kd=3.09±0.74 μM). Poly(U)18 (P<0.001) and poly(C)18 (P<0.001) bindings were statistically significant according to ANOVA analysis with Tukey–Kramer multiple comparisons post-test. (B) τCstF-64 has preferential binding to poly(GU)9 compared with the other polymers [poly(U)18, Kd=3.04±1.56 μM; poly(A)18, Kd=4.49±0.61 μM; poly(G)18, Kd=6.52±1.85 μM; poly(C)18, Kd=18.49±2.82 μM; poly(GU)9, Kd=1.14±0.34 μM]. Binding to poly(C)18 was statistically significantly different from all other values according to ANOVA analysis (P<0.001).
Previously, the DSE has been described as either U-rich or GU-rich [24,27,50,54–57]. Our results in Figure 4(B) showed that CstF-64 RBD bound with highest affinity to poly(U)18, as did τCstF-64 RBD, although its binding was weaker than that of CstF-64. To test the affinity with which elements that contain guanosine might bind to either CstF-64 or τCstF-64, we tested a polymer of (GU)9. We determined a binding affinity of CstF-64 for poly(GU)9 (Kd=3.09 μM±0.74; Figure 4A), slightly lower than previously reported by Deka et al. [49]. More interestingly, we found that τCstF-64 bound poly(GU)9 even more efficiently (Kd=1.14 μM ±0.34) than did CstF-64 (Figure 4B), suggesting differences between the paralogues with regard to their affinity for both U-rich and GU-rich sequences.
Changes in amino acid 41 do not affect RNA binding
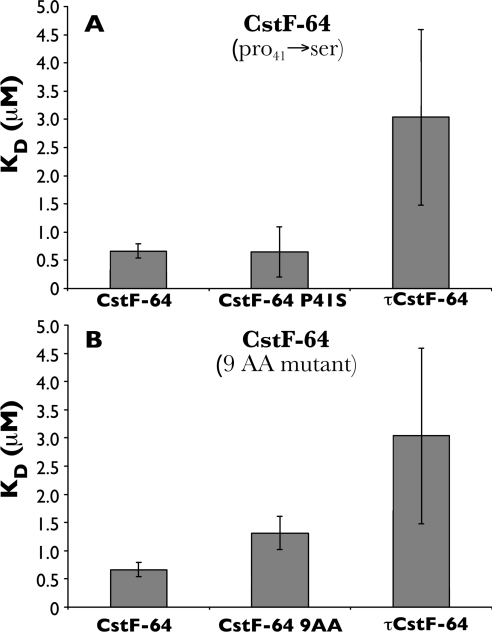
The CstF-64 and τCstF-64 RBDs differ at amino acid 41 where CstF-64 has a proline residue and τCstF-64 has a serine residue. To test whether this particular amino acid difference is an important determinant of RNA binding, site-directed mutagenesis was conducted on the CstF-64 construct to mutate the proline residue at position 41 to a serine residue. We saw no significant change in binding to poly(U)18 in the presence of substrate [32P]poly(U)18 upon changing Pro-41 (Figure 5A; CstF-64, Kd=0.66±0.13 μM) to a serine residue (Figure 5A; CstF-64 P41S, Kd=0.64±0.45 μM). This indicates that the amino acid at position 41 does not contribute significantly to the differences in RNA binding seen between CstF-64 and τCstF-64.
Figure 5. Contributions of different amino acids to CstF-64 and τCstF-64 binding preferences to poly(U).
(A, B) Kd values of GST–CstF-64 RBD and mutants for poly(U)18. (A) Amino acid 41 does not contribute to poly(U)18 RNA-binding specificity. When the GST–CstF-64 RBD construct is mutated at amino acid 41 from a proline residue to a serine residue (CstF-64 P41S), there is no change in binding towards poly(U)18 (Kd=0.64±0.45 μM; no statistical difference is noted according to ANOVA analysis). (B) Amino acids downstream of the C-terminal α-helix contribute to RNA-binding specificity. When the GST–CstF-64 RBD construct was mutated at nine amino acids C-terminal to α3-helix (CstF-64 9AA), there was a decrease in binding to poly(U)18 (Kd=1.31±0.29 μM). This changes the binding of CstF-64 RBD to resemble that of τCstF-64 for poly(U)18.
Elements downstream of the RBD may affect RNA-binding specificity
In addition to the proline→serine mutation at position 41, CstF-64 and τCstF-64 also differ at nine amino acids downstream of α3 of the RBD (Figure 1). This newly described helix is thought to occlude the RBD, but may not participate directly in RNA selection [49]. To determine whether these amino acids affect affinity for poly(U), site-directed mutagenesis was used to create a GST–CstF-64 RBD construct with mutations at these nine amino acids [CstF-64(9AA); Table 2B] so that it resembled τCstF-64 at these positions. The binding affinity of the CstF-64(9AA) mutant was tested as previously described (see Figures 3A and 3B) for poly(U)18, as this substrate, showed the highest affinity and most significant difference in binding to normal CstF-64 and τCstF-64. We found that the binding affinity of CstF-64(9AA) for poly(U)18 was lower (Kd=1.31±0.29 μM) than that of the wild-type CstF-64 (CstF-64, Kd=0.66±0.13 μM), and began to approach that of τCstF-64 (Figure 5B, Kd=3.04±1.56 μM). This suggested that at least part of the discrimination of CstF-64 and τCstF-64 for poly(U) is determined by the region downstream of the third α-helix (α3).
DISCUSSION
We are interested in understanding the role of τCstF-64 in polyadenylation during spermatogenesis. More specifically, we are studying this process in male germ cells because they offer a unique system wherein the mitotic cells express CstF-64, while the meiotic and post-meiotic cells express the paralogous τCstF-64. Both paralogues are essential for spermatogenesis to proceed in an uninterrupted manner (B. Dass, R. A. Hess, K. Carnes, S. Tardif, H. M. Weitlauf and C. C. MacDonald, unpublished work). Recent reports indicate that the testis has both a higher incidence of alternative polyadenylation, suggesting differences in polyadenylation site choice, and a greater use of non-AAUAAA polyadenylation signals, suggesting differences in the polyadenylation machinery [33]. This led us to study whether τCstF-64 contributes to either of these differences in polyadenylation. Therefore, as a first step, we proposed to examine the RNA-binding capabilities of τCstF-64 to simple RNA substrates and compare them with the binding capabilities of CstF-64. To test the hypothesis that CstF-64 and τCstF-64 bound to different RNA elements, we used a UV-cross-linking/competition assay for five ribopolymers and then used a Dixon plot to analyse the necessary parameters to determine Kd in this system [44,52]. Thus we obtained the Kd values of GST–CstF-64 RBD and GST–τCstF-64 RBD for poly(U) and the Kd relative to poly(U)18 for poly(A)18, poly(G)18, poly(C)18 and poly(GU)9.
Using this assay, we found that the CstF-64 RBD bound to synthetic poly(U)18 RNA with much greater efficiency than to other synthetic ribohomopolymers (Figure 4A). The CstF-64 RBD bound with similar efficiency to poly(G) and poly(A), and bound quite weakly to poly(C). Binding of the CstF-64 RBD to a poly(GU) ribopolymer was intermediate between those of poly(U) and poly(G) [Figure 4A, poly(GU)9]. From these binding affinities, we might predict that DSEs in natural polyadenylation sites would be U-rich. Use of U-rich DSEs has been supported in some studies [24,53,54], while in other studies GU-rich elements were implicated [56,58,59]. When in vitro RNA SELEX (systematic evolution of ligands by exponential enrichment) selection–amplification experiments were performed by two different groups, both found evidence supporting binding to GU-rich elements, although with a notable U bias [50,57]. More recently, Varani and co-workers [27,49] noted that binding of the CstF-64 RBD to a GU-rich synthetic RNA was enhanced by inclusion of a UU dinucleotide. Similarly, Chou et al. [53] suggested that a “four out of five base uridylate tract” was sufficient to restore efficient polyadenylation to an SV40 late mRNA polyadenylation signal. Therefore a UU dinucleotide is most likely an important part, although not necessarily the only part, of the DSE.
Because of the very weak binding to poly(C), another prediction might be that both the CstF-64- and τCstF-64-binding sites would have a paucity of C residues, and that C residues could delimit binding sequences. These results are supported by solution data from Pérez Cañadillas and Varani [27] using the isolated CstF-64 RBD. In further support, SELEX experiments using GST–CstF-64 RBDs to select RNA aptamers resulted in RNAs that were U-rich and GU-rich, flanked by C residues [50]. In contrast, in similar SELEX experiments using purified CstF complex (containing all three subunits) from calf thymus or HeLa cells RNA aptamers rich in G, U and C were selected [57]. This suggests that the binding specificity of the intact CstF complex depends on more than the simple binding affinity of the CstF-64 RBD for RNA, and could be different when the intact CstF-64 is in complex with the other polyadenylation factors.
It seemed that the binding affinity of CstF-64 for any of the ribopolymers was relatively low (micromolar range). Nevertheless, results of Deka et al. [49] support this range (Kd≈7–14 μM). This suggests that the affinity of CstF-64 for RNA probably is enhanced during the substrate recognition step of polyadenylation by other interacting factors. This notion is supported by older observations that the interaction of CstF and CPSF was co-operative, and that CstF-64 binding required the presence of CPSF ([22] and C. C. MacDonald, unpublished work). It is further likely that the proximity of polyadenylation factors to the nascent pre-mRNA during transcription [60,61] contributes greatly to the specificity of substrate recognition by increasing the local presentation of RNA elements to the CstF and CPSF. We therefore have great enthusiasm for the model suggested by Pérez Cañadillas and Varani [27] that a feature of CstF-64, possibly the third α-helix (α3), blocks casual RNA binding, but that the block is relieved when both CstF and CPSF interact with each other and with an authentic pre-mRNA substrate.
Early anecdotal references pointed out that the canonical AAUAAA polyadenylation signal was often not present in male germ cell mRNAs [62–67]. More than likely, this is evidence of a larger incidence of alternative polyadenylation during spermatogenesis [32,34–36]. A possible mechanism for this was revealed in a recent study of RNA sequence elements near sites of polyadenylation of mRNAs that were expressed during different stages of male germ cell development [33]. This study revealed significant differences in polyadenylation control elements between premeiotic and post-meiotic male germ cells, further supporting the view that some of the mechanisms of polyadenylation might be altered in those cells. We believe that differences in the RBDs of CstF-64 and τCstF-64 might account for some of these differences [32,37]. As detailed, when compared with each other, the CstF-64 and τCstF-64 RBDs bound with equal affinity to poly(G), poly(A) and poly(C) (Figure 4). There were differences, however, in affinities for poly(U): CstF-64 bound four to five times more avidly to poly(U) than did τCstF-64 (Figure 4). In fact, the Kd of τCstF-64 for poly(U) was not significantly different than its Kd for poly(G) or poly(A), suggesting that it had little selectivity for poly(U) over these two other ribohomopolymers. In contrast, the binding affinities of CstF-64 and τCstF-64 to poly(GU) were reversed: τCstF-64 bound poly(GU) nearly three times as well as did CstF-64, suggesting that poly(GU) might be a candidate for τCstF-64-specific DSEs in germ cell mRNAs.
According to Allain and colleagues [28,68], even though they are distant from the RNA-binding surface, loops of RRM-type RBDs may contribute to additional RNA contact for extra specificity. Therefore we looked at amino acid sequences in or around the RBDs that might contribute to the differences in binding to poly(U). When we compared amino acid sequence for CstF-64 and τCstF-64 there was a single difference (proline→serine) at amino acid 41 in the region between α1-helix and β2-sheet (Figure 1). This difference is conserved in orthologues of τCstF-64 cloned from 15 mammalian species ([42,69] and B. Dass, unpublished work). However, site-directed mutagenesis on the CstF-64 RBD at this location (CstF-64 RBD-P41S, Figure 1) did not display any change in binding to poly(U)18 (Figure 5A). This suggests that position 41 in the RBD does not contribute greatly to the specificity of binding of either CstF-64 or τCstF-64 to poly(U).
Another series of differences between CstF-64 and τCstF-64 lay C-terminal to α3 described by Pérez Cañadillas and Varani [27]; these nine amino acids are similarly conserved in τCstF-64 orthologues from 15 species (B. Dass, unpublished work). Upon testing the affinity of the CstF-64 RBD with the nine amino acid mutations for poly(U) (CstF-64 RBD-9AA; Figure 5B), we found that these changes lowered the affinity of the CstF-64 RBD for poly(U)18, causing it to more closely resemble the τCstF-64 RBD in affinity. This suggests to us that the region C-terminal to α3 has a direct effect on the specificity of RNA binding. Pérez Cañadillas and Varani [27] proposed that α3 was not directly involved in RNA binding, but instead served as a ‘gate’ to occlude RNA binding unless caused to move. Our own data support at least part of this model: while it likely serves as a gate, amino acids C-terminal to α3 also affect RNA discrimination. We do not yet have a model for which protein–RNA contacts are necessary for this discrimination.
These data begin to address one of the mechanisms that might contribute to the high incidence of alternative polyadenylation described in male germ cell mRNAs. Since CstF-64 and τCstF-64 are expressed in different temporal patterns during spermatogenesis [40] and have different RNA-binding specificities (the present study), they may recognize different DSEs of pre-mRNA polyadenylation signals of male germ cell pre-mRNAs. Our recent work suggests that there are a complex series of changes to 3′-end elements used as spermatogenesis proceeds [33]. Some of these changes include a reduction in the specificity of DSEs. Furthermore, 3′-untranslated regions in germ cell mRNAs were found to be notably shorter than in somatic cells. Since we saw a reduction in the discrimination of τCstF-64 for certain sequences such as poly(U), this suggests a model in which τCstF-64 might be more promiscuous in its choice of binding sites, thus binding to sites more 5′ in the nascent transcript.
Online data
Acknowledgments
We acknowledge the NIH (National Institutes of Health; 2 R01 HD37109-04) and a minority student supplement to R.R.M. We also acknowledge Daniel Hardy, James Hutson and the members of the MacDonald laboratory for their discussions and contributions, S. Sridhara, Robert Shaw and J. Andrew Hockert for comments on this paper, Joel Graber for sharing results before publication, and an anonymous reviewer for suggesting a more consistent method of data analysis.
References
- 1.Zhao J., Hyman L., Moore C. Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edmonds M. A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid Res. Mol. Biol. 2002;71:285–389. doi: 10.1016/s0079-6603(02)71046-5. [DOI] [PubMed] [Google Scholar]
- 3.Logan J., Falck-Pedersen E., Darnell J. E., Jr, Shenk T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse β maj-globin gene. Proc. Natl. Acad. Sci. U.S.A. 1987;84:8306–8310. doi: 10.1073/pnas.84.23.8306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Connelly S., Manley J. L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988;2:440–452. doi: 10.1101/gad.2.4.440. [DOI] [PubMed] [Google Scholar]
- 5.Edwalds-Gilbert G., Prescott J., Falck-Pedersen E. 3′ RNA processing efficiency plays a primary role in generating termination-competent RNA polymerase II elongation complexes. Mol. Cell. Biol. 1993;13:3472–3480. doi: 10.1128/mcb.13.6.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vagner S., Vagner C., Mattaj I. W. The carboxyl terminus of vertebrate poly(A) polymerase interacts with U2AF 65 to couple 3′-end processing and splicing. Genes Dev. 2000;14:403–413. [PMC free article] [PubMed] [Google Scholar]
- 7.Huang Y., Carmichael G. C. Role of polyadenylation in nucleocytoplasmic transport of mRNA. Mol. Cell. Biol. 1996;16:1534–1542. doi: 10.1128/mcb.16.4.1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson R. J., Standart N. Do the poly(A) tail and 3′ untranslated region control mRNA translation? Cell. 1990;62:15–24. doi: 10.1016/0092-8674(90)90235-7. [DOI] [PubMed] [Google Scholar]
- 9.Sachs A. B., Sarnow P., Hentze M. W. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 10.Wickens M., Anderson P., Jackson R. J. Life and death in the cytoplasm: messages from the 3′ end. Curr. Opin. Genet. Dev. 1997;7:220–232. doi: 10.1016/s0959-437x(97)80132-3. [DOI] [PubMed] [Google Scholar]
- 11.Decker C. J., Parker R. Mechanisms of mRNA degradation in eukaryotes. Trends Biochem. Sci. 1994;19:336–340. doi: 10.1016/0968-0004(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 12.Decker C. J., Parker R. Diversity of cytoplasmic functions for the 3′ untranslated region of eukaryotic transcripts. Curr. Opin. Cell Biol. 1995;7:386–392. doi: 10.1016/0955-0674(95)80094-8. [DOI] [PubMed] [Google Scholar]
- 13.Gunderson S. I., Beyer K., Martin G., Keller W., Boelens W. C., Mattaj L. W. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994;76:531–541. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 14.Lutz C. S., Alwine J. C. Direct interaction of the U1 snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 15.Lutz C. S., Murthy K. G., Schek N., O'Connor J. P., Manley J. L., Alwine J. C. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 16.Bagga P. S., Arhin G. K., Wilusz J. DSEF-1 is a member of the hnRNP H family of RNA-binding proteins and stimulates pre-mRNA cleavage and polyadenylation in vitro. Nucleic Acids Res. 1998;26:5343–5350. doi: 10.1093/nar/26.23.5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunderson S. I., Polycarpou-Schwarz M., Mattaj I. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol. Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 18.Veraldi K. L., Arhin G. K., Martincic K., Chung-Ganster L. H., Wilusz J., Milcarek C. hnRNP F influences binding of a 64-kilodalton subunit of cleavage stimulation factor to mRNA precursors in mouse B cells. Mol. Cell. Biol. 2001;21:1228–1238. doi: 10.1128/MCB.21.4.1228-1238.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Venkataraman K., Brown K. M., Gilmartin G. M. Analysis of a noncanonical poly(A) site reveals a tripartite mechanism for vertebrate poly(A) site recognition. Genes Dev. 2005;19:1315–1327. doi: 10.1101/gad.1298605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jenny A., Keller W. Cloning of cDNAs encoding the 160 kDa subunit of the bovine cleavage and polyadenylation specificity factor. Nucleic Acids Res. 1995;23:2629–2635. doi: 10.1093/nar/23.14.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murthy K. G. K., Manley J. L. Characterization of the multisubunit cleavage-polyadenylation specificity factor from calf thymus. J. Biol. Chem. 1992;267:14804–14811. [PubMed] [Google Scholar]
- 22.Wilusz J., Shenk T., Takagaki Y., Manley J. L. A multicomponent complex is required for the AAUAAA-dependent cross-linking of a 64-kilodalton protein to polyadenylation substrates. Mol. Cell. Biol. 1990;10:1244–1248. doi: 10.1128/mcb.10.3.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takagaki Y., MacDonald C. C., Shenk T., Manley J. L. The human 64-kDa polyadenylation factor contains a ribonucleoprotein-type RNA binding domain and unusual auxiliary motifs. Proc. Natl. Acad. Sci. U.S.A. 1992;89:1403–1407. doi: 10.1073/pnas.89.4.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacDonald C. C., Wilusz J., Shenk T. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol. 1994;14:6647–6654. doi: 10.1128/mcb.14.10.6647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagai K., Oubridge C., Jessen T. H., Li J., Evans P. R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- 26.Varani G., Nagai K. RNA recognition by RNP proteins during RNA processing. Annu. Rev. Biophys. Biomol. Struct. 1998;27:407–445. doi: 10.1146/annurev.biophys.27.1.407. [DOI] [PubMed] [Google Scholar]
- 27.Pérez Cañadillas J. M., Varani G. Recognition of GU-rich polyadenylation regulatory elements by human CstF-64 protein. EMBO J. 2003;22:2821–2830. doi: 10.1093/emboj/cdg259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maris C., Dominguez C., Allain F. H. The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 2005;272:2118–2131. doi: 10.1111/j.1742-4658.2005.04653.x. [DOI] [PubMed] [Google Scholar]
- 29.Takagaki Y., Manley J. L. Complex protein interactions within the human polyadenylation machinery identify a novel component. Mol. Cell. Biol. 2000;20:1515–1525. doi: 10.1128/mcb.20.5.1515-1525.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richardson J. M., McMahon K. W., MacDonald C. C., Makhatadze G. I. MEARA sequence repeat of human CstF-64 polyadenylation factor is helical in solution: a spectroscopic and calorimetric study. Biochemistry. 1999;38:12869–12875. doi: 10.1021/bi990724r. [DOI] [PubMed] [Google Scholar]
- 31.Calvo O., Manley J. L. Evolutionarily conserved interaction between CstF-64 and PC4 links transcription, polyadenylation, and termination. Mol. Cell. 2001;7:1013–1023. doi: 10.1016/s1097-2765(01)00236-2. [DOI] [PubMed] [Google Scholar]
- 32.MacDonald C. C., Redondo J.-L. Reexamining the polyadenylation signal: were we wrong about AAUAAA? Mol. Cell. Endocrinol. 2002;190:1–8. doi: 10.1016/s0303-7207(02)00044-8. [DOI] [PubMed] [Google Scholar]
- 33.Liu D., Brockman J. M., Dass B., Hutchins L. N., McCarrey J. R., MacDonald C. C., Singh P., Graber J. H. Systematic variation in mRNA 3′-processing signals during mouse spermatogenesis. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkl919. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwalds-Gilbert G., Veraldi K. L., Milcarek C. Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res. 1997;25:2547–2561. doi: 10.1093/nar/25.13.2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H., Lee J. Y., Tian B. Biased alternative polyadenylation in human tissues. Genome Biol. 2005;6:R100. doi: 10.1186/gb-2005-6-12-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wallace A. M., Dass B., Ravnik S. E., Tonk V., Jenkins N. A., Gilbert D. J., Copeland N. G., MacDonald C. C. Two distinct forms of the 64,000 Mr protein of the cleavage stimulation factor are expressed in mouse male germ cells. Proc. Natl. Acad. Sci. U.S.A. 1999;96:6763–6768. doi: 10.1073/pnas.96.12.6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dass B., McMahon K. W., Jenkins N. A., Gilbert D. J., Copeland N. G., MacDonald C. C. The gene for a variant form of the polyadenylation protein CstF-64 is on chromosome 19 and is expressed in pachytene spermatocytes in mice. J. Biol. Chem. 2001;276:8044–8050. doi: 10.1074/jbc.M009091200. [DOI] [PubMed] [Google Scholar]
- 38.Handel M. A., Hunt P. A., Kot M. C., Park C., Shannon M. Role of sex chromosomes in the control of male germ-cell differentiation. Ann. N.Y. Acad. Sci. 1991;637:64–73. doi: 10.1111/j.1749-6632.1991.tb27301.x. [DOI] [PubMed] [Google Scholar]
- 39.Handel M. A., Park C., Kot M. Genetic control of sex-chromosome inactivation during male meiosis. Cytogenet. Cell Genet. 1994;66:83–88. doi: 10.1159/000133672. [DOI] [PubMed] [Google Scholar]
- 40.Wallace A. M., Denison T., Attaya E. N., MacDonald C. C. Developmental differences in expression of two forms of the CstF-64 polyadenylation protein in rat and mouse. Biol. Reprod. 2004;70:1080–1087. doi: 10.1095/biolreprod.103.022947. [DOI] [PubMed] [Google Scholar]
- 41.Wang P. J., McCarrey J. R., Yang F., Page D. C. An abundance of X-linked genes expressed in spermatogonia. Nat. Genet. 2001;27:422–426. doi: 10.1038/86927. [DOI] [PubMed] [Google Scholar]
- 42.Dass B., McDaniel L., Schultz R. A., Attaya E., MacDonald C. C. The gene CSTF2T encoding the human variant CstF-64 polyadenylation protein τCstF-64 is intronless and may be associated with male sterility. Genomics. 2002;80:509–514. [PubMed] [Google Scholar]
- 43.Dignam J. D., Lebovitz R. M., Roeder R. G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dixon M. The determination of enzyme inhibitor constants. Biochem. J. 1953;55:170–171. doi: 10.1042/bj0550170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Segel I. H. 2nd edn. New York: John Wiley and Sons; 1976. Biochemical Calculations: How to Solve Mathematical Problems in General Biochemistry. [Google Scholar]
- 46.Weeks K. M., Crothers D. M. RNA binding assays for Tat-derived peptides: implications for specificity. Biochemistry. 1992;31:10281–10287. doi: 10.1021/bi00157a015. [DOI] [PubMed] [Google Scholar]
- 47.Edwalds-Gilbert G., Milcarek C. Regulation of poly(A) site use during mouse B-cell development involves a change in the binding of a general polyadenylation factor in a B-cell stage-specific manner. Mol. Cell. Biol. 1995;15:6420–6429. doi: 10.1128/mcb.15.11.6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilusz J., Shenk T. A 64 kDa nuclear protein binds to RNA segments that include the AAUAAA polyadenylation motif. Cell. 1988;52:221–228. doi: 10.1016/0092-8674(88)90510-7. [DOI] [PubMed] [Google Scholar]
- 49.Deka P., Rajan P. K., Pérez-Cañadillas J. M., Varani G. Protein and RNA dynamics play key roles in determining the specific recognition of GU-rich polyadenylation regulatory elements by human Cstf-64 protein. J. Mol. Biol. 2005;347:719–733. doi: 10.1016/j.jmb.2005.01.046. [DOI] [PubMed] [Google Scholar]
- 50.Takagaki Y., Manley J. L. RNA recognition by the human polyadenylation factor CstF. Mol. Cell. Biol. 1997;17:3907–3914. doi: 10.1128/mcb.17.7.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Graber J. H., Cantor C. R., Mohr S. C., Smith T. F. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc. Natl. Acad. Sci. U.S.A. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Linden J. Calculating the dissociation constant of an unlabelled compound from the concentration required to displace radiolabel binding by 50% J. Cyclic Nucleotide Res. 1982;8:163–172. [PubMed] [Google Scholar]
- 53.Chou Z. F., Chen F., Wilusz J. Sequence and position requirements for uridylate-rich downstream elements of polyadenylation signals. Nucleic Acids Res. 1994;22:2525–2531. doi: 10.1093/nar/22.13.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Legendre M., Gautheret D. Sequence determinants in human polyadenylation site selection. BMC Genomics. 2003;4:7. doi: 10.1186/1471-2164-4-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilusz J., Shenk T. A uridylate tract mediates efficient heterogeneous nuclear ribonucleoprotein C protein–RNA cross-linking and functionally substitutes for the downstream element of the polyadenylation signal. Mol. Cell. Biol. 1990;10:6397–6407. doi: 10.1128/mcb.10.12.6397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weiss E. A., Gilmartin G. M., Nevins J. R. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J. 1991;10:215–219. doi: 10.1002/j.1460-2075.1991.tb07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Beyer K., Dandekar T., Keller W. RNA ligands selected by cleavage stimulation factor contain distinct sequence motifs that function as downstream elements in 3′-end processing of pre-mRNA. J. Biol. Chem. 1997;272:26769–26779. doi: 10.1074/jbc.272.42.26769. [DOI] [PubMed] [Google Scholar]
- 58.McLauchlan J., Gaffney D., Whitton J. L., Clements J. B. The consensus sequence YGTGTTYY located downstream from the AAUAAA signal is required for efficient formation of mRNA 3′ termini. Nucleic Acids Res. 1985;13:1347–1368. doi: 10.1093/nar/13.4.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nussinov R. TGTG, G clustering and other signals near non-mammalian vertebrate mRNA 3′ termini: some implications. J. Biomol. Struct. Dynan. 1986;3:1145–1153. doi: 10.1080/07391102.1986.10508491. [DOI] [PubMed] [Google Scholar]
- 60.Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr. Opin. Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 61.Minvielle-Sebastia L., Keller W. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr. Opin. Cell Biol. 1999;11:352–357. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- 62.Meijer D., Hermans A., von Lindern M., van Agthoven T., de Klein A., Mackenbach P., Grootegoed A., Talarico D., Valle G. D., Grosveld G. Molecular characterization of the testis specific c-abl mRNA in mouse. EMBO J. 1987;6:4041–4048. doi: 10.1002/j.1460-2075.1987.tb02749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Oppi C., Shore S. K., Reddy E. P. Nucleotide sequence of testis-derived c-abl cDNAs: implications for testis-specific transcription and abl oncogene activation. Proc. Natl. Acad. Sci. U.S.A. 1987;84:8200–8204. doi: 10.1073/pnas.84.23.8200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Øyen O., Myklebust F., Scott J. D., Cadd G. G., McKnight G. S., Hansson V., Jahnsen T. Subunits of cyclic adenosine 3′,5′-monophosphate-dependent protein kinase show differential and distinct expression patterns during germ cell differentiation: alternative polyadenylation in germ cells gives rise to unique smaller-sized mRNA species. Biol. Reprod. 1990;43:46–54. doi: 10.1095/biolreprod43.1.46. [DOI] [PubMed] [Google Scholar]
- 65.Wingett D., Reeves R., Magnuson N. S. Characterization of the testes-specific pim-1 transcript in rat. Nucleic Acids Res. 1992;20:3183–3189. doi: 10.1093/nar/20.12.3183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foulkes N. S., Schlotter F., Pévet P., Sassone-Corsi P. Pituitary hormone FSH directs the CREM functional switch during spermatogenesis. Nature. 1993;362:264–267. doi: 10.1038/362264a0. [DOI] [PubMed] [Google Scholar]
- 67.Ravnik S. E., Wolgemuth D. J. The developmentally restricted pattern of expression in the male germ line of a murine cyclin A, cyclin A2, suggests roles in both mitotic and meiotic cell cycles. Dev. Biol. 1996;173:69–78. doi: 10.1006/dbio.1996.0007. [DOI] [PubMed] [Google Scholar]
- 68.Stefl R., Skrisovska L., Allain F. H. RNA sequence- and shape-dependent recognition by proteins in the ribonucleoprotein particle. EMBO Rep. 2005;6:33–38. doi: 10.1038/sj.embor.7400325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huber Z., Monarez R. R., Dass B., MacDonald C. C. The mRNA encoding τCstF-64 is expressed ubiquitously in mouse tissues. Ann. N.Y. Acad. Sci. 2005;1061:163–172. doi: 10.1196/annals.1336.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.