Abstract
Dystrophin forms part of a vital link between actin cytoskeleton and extracellular matrix via the transmembrane adhesion receptor dystroglycan. Dystrophin and its autosomal homologue utrophin interact with β-dystroglycan via their highly conserved C-terminal cysteine-rich regions, comprising the WW domain (protein–protein interaction domain containing two conserved tryptophan residues), EF hand and ZZ domains. The EF hand region stabilizes the WW domain providing the main interaction site between dystrophin or utrophin and dystroglycan. The ZZ domain, containing a predicted zinc finger motif, stabilizes the WW and EF hand domains and strengthens the overall interaction between dystrophin or utrophin and β-dystroglycan. Using bacterially expressed ZZ domain, we demonstrate a conformational effect of zinc binding to the ZZ domain, and identify two zinc-binding regions within the ZZ domain by SPOTs overlay assays. Epitope mapping of the dystrophin ZZ domain was carried out with new monoclonal antibodies by ELISA, overlay assay and immunohistochemistry. One monoclonal antibody defined a discrete region of the ZZ domain that interacts with β-dystroglycan. The epitope was localized to the conformationally sensitive second zinc-binding site in the ZZ domain. Our results suggest that residues 3326–3332 of dystrophin form a crucial part of the contact region between dystrophin and β-dystroglycan and provide new insight into ZZ domain organization and function.
Keywords: Duchenne muscular dystrophy, dystroglycan, dystrophin, epitope mapping, utrophin, zinc binding
Abbreviations: AAS, atomic absorption spectroscopy; BMD, Becker muscular dystrophy; CBP, CREB (cAMP-response-element-binding protein)-binding protein; DMD, Duchenne muscular dystrophy; KLH, keyhole-limpet (Diodora aspera) haemocyanin; LIM, Lin-11, Isl-1, Mec-3
INTRODUCTION
Dystrophin loss leads to the progressive muscle-wasting disease DMD (Duchenne muscular dystrophy). Following the identification of the DMD gene by Kunkel and co-workers [1] by positional cloning in the late 1980s, it was realized that the protein product dystrophin was a large cytoskeletal protein with homology to the spectrin family of proteins [2–4]. Like spectrin and α-actinin, dystrophin and utrophin can provide a link between the F-actin (filamentous actin) cytoskeleton and transmembrane proteins, and in the specific case of dystrophin through the transmembrane adhesion receptor protein dystroglycan, and so to the extracellular matrix [5]. The much rarer and often milder allelic form of DMD known as BMD (Becker muscular dystrophy) allows the expression of a variable amount of altered dystrophin protein [6]. Analysis of the genotype–phenotype relationship for deletion mutations in these patients has revealed valuable information about the functional importance of different regions of the dystrophin protein [7]. Dystrophin and its autosomal homologue utrophin [8] comprise four main regions. From N- to C-terminus: an actin-binding domain, a series of spectrin-like repeats, a cysteine-rich region and a dimeric coiled-coil region. The actin-binding domain and cysteine-rich region show highest overall sequence similarity between the two proteins, and the mutational spectrum from analysis of BMD patients points to a greater functional importance for these regions in dystrophin. Mutations in the actin-binding region give rise to severe BMD, while despite the presence of some dystrophin protein product, mutations in the cysteine-rich region give rise to a DMD phenotype [7]. These findings from human subjects were recapitulated in an elegant series of experiments using transgenic mice expressing dystrophin constructs containing a similar spectrum of deletion and mutations, reviewed in [9]. The human and mouse data demonstrate that from a functional point of view the physical links between dystrophin and the actin cytoskeleton via the actin-binding domain, and the link between cysteine-rich region and dystroglycan are crucial to dystrophin function. The spectrin repeats are to a large extent redundant, and only mild phenotypes are associated with loss of the C-terminal coiled-coil repeats. Within the cysteine-rich region, arguably the most important region of dystrophin, are three distinct domains that contribute to the interaction between dystrophin and the C-terminus of dystroglycan: the WW domain (protein–protein interaction domain containing two conserved tryptophan residues), EF hand and ZZ domains [10–12]. The WW domain is the primary site of interaction between dystrophin (or utrophin) and the last C-terminal 15 amino acids of β-dystroglycan [13,14]. The stability of the WW domain and its affinity for β-dystroglycan are greatly increased by the EF hand region which cradles the WW domain [15]. The overall complex is then further stabilized and supported by interactions between dystroglycan and the ZZ domain [13,16]. While the interactions between β-dystroglycan and the WW and WW–EF regions of dystrophin and utrophin have been studied in some detail, less attention has been paid to the ZZ domain. In the present study, we describe features of the ZZ domain with respect to its zinc-binding properties and, using a panel of new monoclonal antisera to the ZZ domain, define a binding site for dystroglycan on the ZZ domain.
MATERIALS AND METHODS
Peptides and constructs
All peptides used were obtained as previously described [17]. Overlapping seven-residue synthetic peptides were produced to span the dystrophin or utrophin ZZ domains, D3311–3342 and U3068–3099 respectively as represented in Figure 1. Dystrophin and utrophin differ by only two isoleucine residues in this region I3322 and I3336 (dystrophin numbering) replaced by two valine residues in utrophin sequences (V3079 and V3093). A ZZ domain construct comprising residues 3049–3114 of human utrophin was generated by PCR including NdeI and SalI restriction sites and cloned into the modified pET vector pSJW1 ([18], but see [18a]). Point mutations in cysteine residues were generated by overlap extension mutagenesis. The non-fusion protein was expressed in Escherichia coli BL21(DE3) and purified from inclusion bodies using CM-Sephadex and gel filtration chromatography in the presence of 6 M urea. Urea was removed by stepwise dialysis into 10 mM Pipes (pH 7.0) and 1 mM ascorbate with either 1 mM ZnCl2 or 1 mM phenanthroline. The cytoplasmic domain only of mouse β-dystroglycan, residues 775–895, was expressed in E. coli BL21(DE3) and purified as described previously [19].
Figure 1. Dystrophin and Utrophin ZZ domain alignment and peptides used in the present study.
Amino acid alignment of the ZZ domains of dystrophin (Dys) and utrophin (Utr), indicating identical residues (|) and highly conserved residues (:) and delimiting amino acid residue numbers (large boldface letters in the centre of the Figure). Above and below these sequences are the sequences of the series of overlapping seven-residue peptides used in the present study; all are numbered except the first four which could not be synthesized to high enough purity. Peptides above the alignment are common to dystrophin, including those that are underlined, which are common to both dystrophin and utrophin; those below the alignment are unique to utrophin.
Antibodies
The dystrophin ZZ domain sequence (3311–3342) was linked to KLH [keyhole-limpet (Diodora aspera) haemocyanin] and injected into mice according to a previously described protocol [20]. Spleen cells were fused to myeloma to obtain specific hybridomas and antibodies were selected by ELISA using the antigenic peptide. The dystrophin and utrophin C-terminal polyclonal antibodies (H4 and K7 respectively) were raised in New Zealand rabbits by repeated intradermal injections. Peptides of the last 16 and 11 C-terminal amino acids of dystrophin or utrophin respectively were conjugated via a cysteine residue to KLH and used as antigen. Antibodies were purified and characterized as previously described [21].
Tissues
All tissues, muscle, lung and sciatic nerve, were dissected immediately after death and rapidly frozen in 2-methylbutane, cooled in liquid nitrogen and stored at −80 °C until use. Crude muscle membrane homogenate was prepared using freshly dissected muscle according to a previous protocol [22].
ELISA
ZZ domain sequence synthetic peptide (3311–3342) was coated on microtitre plates at 0.1 mg/ml, and incubated at 4 °C overnight in PBS. After washes, the plates were blocked with 0.05% Tween 20 in PBS buffer containing 1% (w/v) BSA for 20 min at 37 °C, then incubated with each monoclonal antibody for 2 h at 37 °C. Reaction was revealed by adding alkaline phosphatase-labelled anti-mouse IgG for 30 min at 37 °C and signal was detected at 405 nm using 1 mg/ml p-nitrophenylphosphate (Sigma) in 9.7% (v/v) diethanolamine buffer (pH 9.8). ELISA assay was performed to test epitope mapping for each monoclonal antibody using the seven-residue synthetic peptides corresponding to the overlapping sequences spanning the D3311–3342 or U3068–3099 ZZ domain. Freshly prepared peptide, 7.7 mg/ml in 0.05% Tween 20 PBS buffer, was coated on to microtitre plates in triplicate and then incubated with each monoclonal antibody to test their reactivity. For competitive ELISA experiments, microtitre plates were coated in triplicate with peptide 19. After blocking as above, plates were incubated at 4 °C overnight with either crude muscle membrane homogenate (0.1 mg/ml), purified β-dystroglycan (9 μg/ml) or with blocking buffer. Monoclonal antibody (13D2) and the commercial β-dystroglycan antibody (1:500, 43DAG/8D5; Novocastra) were added successively. Reactions were revealed as described above.
Immunofluorescence
Cryostat sections (10 μm) of unfixed muscle [rabbit and Torpedo marmorata (marbled electric ray)] were labelled with the antibodies described above. Immunoreactivity was detected with Cy3-conjugated sheep anti-mouse IgG (Euromedex).
Western blotting
Fresh extracts were prepared from 0.01 g of muscle tissue homogenized in 150 μl of 5% SDS buffer (50 mM Tris/HCl, pH 8.0, and 10 mM EDTA) supplemented with 1% trypsin inhibitor and 1% saponin. After centrifugation (10 min at 13000 g), supernatant protein concentrations were estimated using the BCA (bicinchoninic acid) protein assay (Pierce). Samples were separated in duplicate by SDS/PAGE (3–10 or 5–15% gel), transferred to nitrocellulose and developed with appropriate antibody, essentially as described previously [13]. All monoclonal antibodies were tested in competition with an excess of corresponding synthetic peptides (1 mg of peptide per μg/ml of monoclonal antibody [23]) on both cryostat sections and Western blots. All monoclonal antibodies were completely blocked by their specific peptides (see Figure 4).
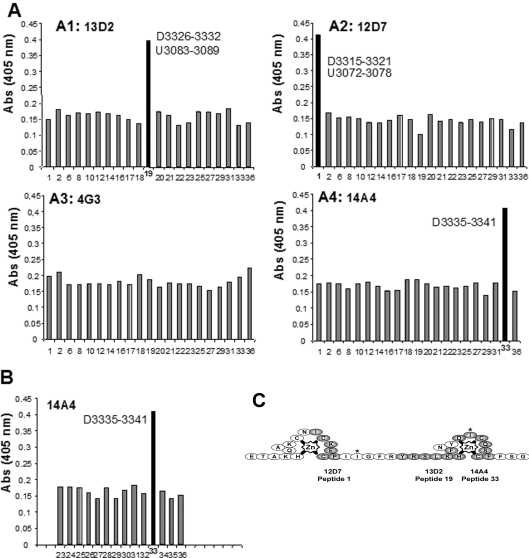
Figure 4. ZZ domain monoclonal antibody epitope mapping.
(A) Epitope determination of monoclonal antibody specificity by ELISA using synthetic peptides (number on x-axis; see Figure 1). Results are represented as histograms; 13D2, 12D7 and 14A4 antibodies all recognized a single specific dystrophin peptide, whereas antibody 4G3 did not recognize any peptide. 14A4 was reprobed against a series of overlapping dystrophin and utrophin peptides (B). Again the 14A4 antibody appeared specific for the dystrophin sequence D3335–3341 sequence, and did not recognize the Ile3336 to Val3039 substitution in the utrophin sequence (peptide 34). (C) The epitopes are represented on the topological map of the ZZ domain derived from Figure 3, individual epitopes are shaded grey and the non-conserved isoleucine residues (valine in utrophin) are indicated by asterisks.
Far-Western blotting overlay
Blotted nitrocellulose sheets (0.2 μm) containing total muscle protein extract were blocked for 1 h at room temperature with 10 mM triethanolamine (pH 7.6), 140 mM NaCl, 1 mM CaCl2 and 1 mM MgCl2 containing 5% BSA and then pre-incubated with peptide 19 (0.1 mg/ml) in 0.5% BSA containing 1 mM dithiothreitol or 0.5% BSA alone (control) for 24 h. Crude membrane homogenates were applied to the nitrocellulose membrane overnight at 4 °C with gentle agitation. Membranes were blocked in Tris buffer containing 0.1% Tween 20 and 3% BSA. After washing, proteins were revealed with dystrophin C-terminal antibody (H4).
ZZ domain zinc binding
CD was carried out on a Jobin Yvon CD6 spectrophotometer using 0.6 ml cuvettes. ZZ domain was analysed in the presence of 1 mM ZnCl2 or 1 mM 1,10-phenanthroline. Aliquots of similar samples were also subjected to SDS/PAGE in the absence or presence of zinc. AAS (atomic absorption spectroscopy) was performed on a Unicam Solaar 929 spectrometer using ZnCl2 solutions prepared in the same buffer as the ZZ domain as standards at a wavelength of 213.9 nm with a bandpass of 0.5 nm and taking the average of three readings at 1 s intervals. For metal ion competition assays, 10 μg of purified ZZ was spotted on to nitrocellulose and allowed to air dry. Membranes were probed with 65ZnCl2 as described previously [24] using 200 μM 65ZnCl2 (20 μCi/ml) and an excess of the indicated counter ions. Autoradiographs were quantified by densitometry and data expressed relative to 65ZnCl2 alone=1.0. A SPOTs peptide array comprising 46 peptides of 20 residues each differing by one amino acid covering the complete utrophin ZZ domain (3049–3114) was synthesized as described previously [13]. 65ZnCl2 binding was performed as described above; a separate control experiment was performed in the presence of excess 10 mM ZnCl2.
RESULTS
ZZ domain zinc binding
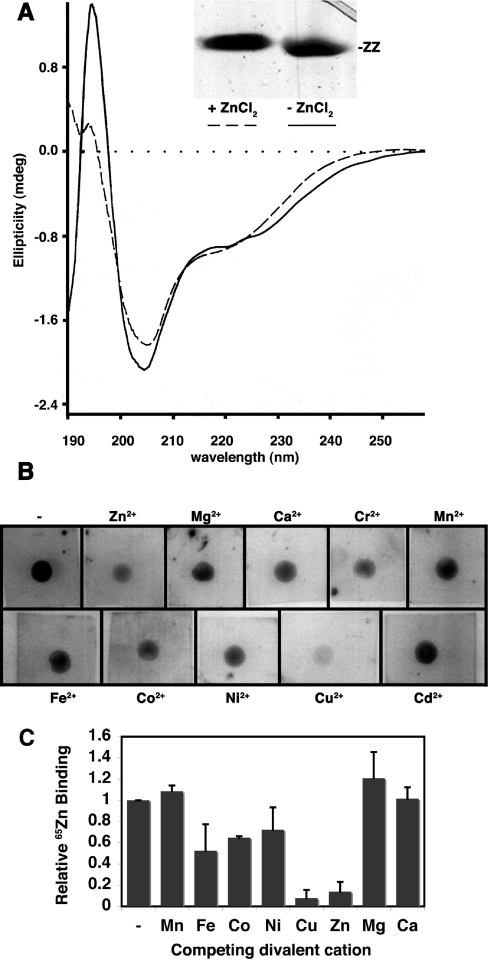
Like other proteins that bind bivalent cations, purified bacterially expressed utrophin ZZ domain showed a small upward mobility-shift on SDS/PAGE when the protein was prepared in the presence of zinc, whereas ZZ domain prepared in the presence of the chelator 1,10-phenanthroline ran slightly faster (Figure 2A, inset). This is indicative of a structural change induced by zinc binding altering the SDS/PAGE mobility, a phenomenon commonly seen in cation-binding proteins, such as calmodulin when subjected to SDS/PAGE in the presence or absence of calcium, see e.g. [25]. A conformational change upon zinc binding was also substantiated by CD. The CD spectrum of ZZ domain in the presence of zinc showed a slightly higher propensity towards helical structure compared with ZZ domain with phenanthroline (Figure 2A). Determination of bivalent cation specificity for the ZZ domain by 65Zn-overlay revealed most competition for zinc binding by excess zinc or copper (Figure 2B), which is in keeping with the complexing ability of bivalent cations of the transition metals which generally follow the Irving–Williams series Mn<Fe<Co<Ni<Cu>Zn, which in turn are all greater than the alkaline earth metals, Ca2+, Mg2+ etc. [26]. This trend is also evident on quantification of this and two other independent experiments (Figure 2C). These data suggest that the ZZ domain is a bona fide zinc-binding protein. The original description of the ZZ domain highlighted four conserved cysteine residues in the cysteine-rich regions of dystrophin and utrophin [12] leading to the suggestion that the ZZ domain might adopt a structure equivalent to half of a LIM (Lin-11, Isl-1, Mec-3) domain [27]. ZZ domains in most other proteins, however, contain six conserved cysteine residues in positions that could co-ordinate zinc with additional histidine and other potential liganding residues (see Figure 8) [12].
Figure 2. Zinc binding to the utrophin ZZ domain.
(A) CD spectrum of the utrophin ZZ domain in the presence of 1 mM ZnCl2 (dashed line) or absence of zinc (presence of phenanthroline; solid line) indicating a modest structural change on zinc binding. This is reflected in the slightly altered mobility of the ZZ domain on SDS/PAGE in the presence of ZnCl2 (inset). (B) Autoradiographs of individual 65ZnCl2 overlays of purified ZZ domain in the absence (top left square) or presence of an excess of the indicated competing bivalent cation. 65Zn binding is competed in keeping with the Irving–Williams series Mn<Fe<Co<Ni<Cu>Zn≫Ca/Mg. A subset of the data in (B) was published previously in brief [44]. (C) Densitometric analysis of the data in (B) and two additional independent experiments, with 65ZnCl2 binding in the presence of the indicated competing bivalent cations represented relative to 65ZnCl2 alone=1. Results shown are means±S.E.M. (n=3).
Figure 8. Alignment of ZZ domains of various proteins and positions of Zn-co-ordinating residues in ZZ domains.
(A) Alignment of ZZ domain sequences from dystrophin family proteins and other unrelated proteins from different phyla. Gene name, organism and residue numbers for the ZZ domain used in the alignment are shown. The alignment was generated using ClustalW and the 60% consensus sequence derived from the SMART database entry for the ZZ domain is shown at the bottom. All cysteines are coloured yellow, single amino acids that fit the consensus and conserved aromatic and basic residues that form the core of the ZZ domain are also coloured. Upper-case letters represent conserved amino acids (in the full alignment), and the lower-case letters and symbols denote conserved features; +, positive; a, aromatic; s, small; p, polar; b, big; and h, hydrophobic. A more extensive alignment of 47 ZZ domain sequences can be found at http://www.expasy.org/cgi-bin/aligner?psa=PS01357&color=1&maxinsert=10&linelen=0. (B) A scheme to demonstrate the positions of Zn-co-ordinating residues as proposed for the utrophin ZZ domain and as elucidated structurally for the CBP ZZ domain [36]. Numbers beneath each sequence refer to the positions of the liganding residues co-ordinating each individual zinc ion. For utrophin these are proposed to be consecutive in the linear sequence, whereas CBP adopts a cross-braced organization.
65Zn-overlay assays
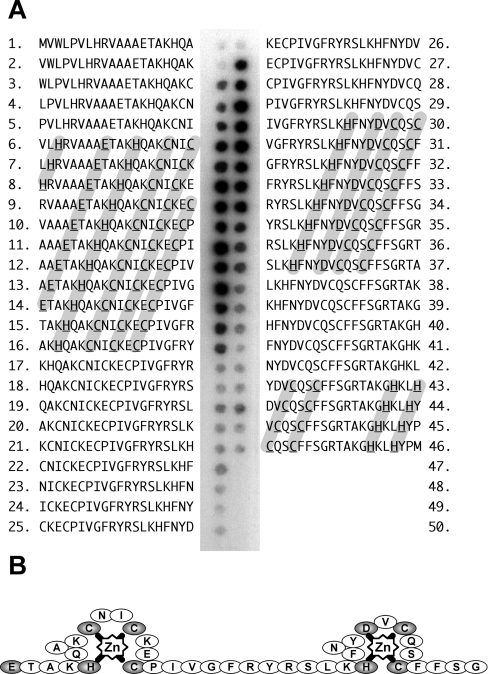
We therefore mutated each of the five cysteine residues in the utrophin ZZ domain and performed 65Zn-overlay assays. No qualitative difference was observed in zinc binding as determined by autoradiography, suggesting that the ZZ domain did not adopt a half LIM domain-like conformation, as removal of any one cysteine should prevent zinc co-ordination. For the constructs where enough soluble material could be produced, we further examined the ZZ domain zinc binding using AAS. The wild-type purified protein, refolded from urea, contained sub-stoichiometric quantities of zinc, 0.65±0.03 mol of Zn/mol of ZZ, suggesting that the protein was not all correctly folded. However, the zinc content of the utrophin ZZ domain mutant C3071A appeared stoichiometric at 2.14±0.1 mol of Zn/mol of ZZ: thus two, rather than one, zinc ions bound to the ZZ domain. We hypothesize that the mutation of Cys3071 to alanine removed a ‘free’ unco-ordinated cysteine residue that led to inappropriate zinc co-ordination, possible disulphide formation and aggregation of the domain leading to insolubility. To confirm the stoichiometry of zinc binding determined by AAS, we used a peptide SPOTs array of 20-amino-acid peptides each differing from the previous by one amino acid spanning the 66 residues of the ZZ domain to examine 65Zn binding. As shown in Figure 3(A), two distinct regions of the utrophin ZZ domain bound zinc, in part corroborating the AAS data. Zinc is typically co-ordinated by cysteine and histidine residues, but aspartic acid and glutamic acid residues are also able to act as ligands with water contributing in catalytic situations [28]. The first zinc-binding region, peptides 6–16, contained a glutamic acid residue, a histidine residue, two of the conserved cysteine residues and a further cysteine residue unique to dystrophin and utrophin. Each potential liganding residue was separated by two residues which could theoretically accommodate the co-ordination of a zinc ion (Figure 3A). The other region, peptides 30–37, contained the second pair of conserved cysteines, a histidine and an aspartic acid (Figure 3A). Rather than a zinc finger, the ZZ domain may therefore contain two zinc ‘knuckles’ as depicted in Figure 3(B), although other conformations are possible. The binding of zinc to the peptide regions described above must rely on some local sequence specificity and not the simple presence of potential co-ordinating ligands, as peptides 43–46, which all contain a pair of histidines and a pair of cysteines in a configuration one might expect to be able bind zinc, showed no labelling with 65Zn (Figure 3A). The synthesis of SPOTs peptides is not quantitative; therefore strength of binding interaction cannot necessarily be inferred from the intensity in any one SPOT as the quantities of peptide in each SPOT may differ. However, the presence of radioactivity of broadly similar intensity across several SPOTs is likely to indicate a similar affinity for the ligand over those SPOTs.
Figure 3. Utrophin ZZ domain peptide SPOTs 65Zn overlay.
(A) A 46-peptide, 20-residue SPOTs peptide array overlapping by one amino acid and covering the utrophin ZZ domain was overlaid with 65ZnCl2 and subjected to autoradiography. Individual peptides are numbered 1–46 with the corresponding sequence shown. Positions 47–50 in the array were blank. Two regions of 65Zn binding were observed spanning peptides 6–16 and 30–37 respectively, potential residues involved in the co-ordination of zinc are highlighted in grey and boldface underline. Another region with the potential to bind 65Zn (peptides 43–46: potential co-ordinating residues highlighted in grey) showed no 65Zn binding by this approach. The two zinc-binding regions are represented schematically in (B).
Monoclonal antibody selection and epitope mapping
Monoclonal antibodies directed towards the ZZ domain of dystrophin (3311–3342) were collected as hybrid supernatants, selected by ELISA using recombinant ZZ domain and named 13D2, 12D7, 14A4 and 4G3. Specificity and epitope mapping was performed for the four selected supernatants by ELISA against the series of seven-residue peptides from the dystrophin ZZ domain (Figure 1A) including those common to dystrophin and utrophin. A specific response is obtained with peptide 19 (D3326–3332/U3081–3089) for 13D2 supernatant, with peptide 1 (D3315–3331/U3072–3072) for 12D7 supernatant and with peptide 33 (D3335–3341) for 14A4 supernatant, while no specific response was obtained with supernatant 4G3 (Figure 4A). Peptide 33 is unique to dystrophin as it contains one of the isoleucines that is replaced by valine in the utrophin ZZ domain. We therefore re-screened serum 14A4 hybrid supernatant against all peptides that spanned the equivalent region in dystrophin and utrophin but, as shown in Figure 4(B), supernatant 14A4 was specific for peptide 33 and did not recognize the equivalent peptide from utrophin with an isoleucine to valine substitution (peptide 34). The respective epitopes of the selected antibodies are presented schematically in Figure 4(C) taking into account the topological information derived from Figure 3.
Immunodetection of the dystrophin ZZ domain
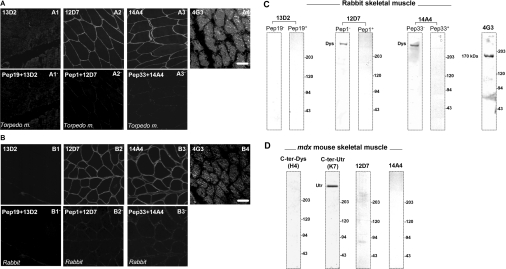
The four monoclonal antibodies were tested by immunofluorescence detection using both T. marmorata and rabbit muscle (Figures 5A and 5B). Serum 13D2 shows no specific sarcolemmal labelling, whereas both 12D7 and 14A4 antibodies produced the expected sarcolemmal labelling typical of dystrophin staining in skeletal muscle. Specific staining was blocked by pre-incubation of the hybrid supernatant with the respective peptide. The 4G3 antibody, which did not detect any specific dystrophin or utrophin ZZ domain peptide, produced a labelling pattern only in the cytoplasmic compartment of muscle fibres. These antibodies were further tested by Western blot on total protein homogenates from rabbit skeletal muscle (Figure 5C) or T. marmorata (results not shown). In keeping with the immunofluorescence detection pattern in muscle sections, no protein band was detected using the 13D2 antibody, while a 400 kDa protein band, corresponding to the expected molecular mass of dystrophin, was obtained by 12D7 and 14A4 antibodies (Figure 5). Specific staining was again blocked by pre-incubation of the serum with the respective peptide. Using protein extract from dystrophin-deficient mdx mouse muscle (Figure 5D), 12D7 and 14A4 antibodies fail to give a 400 kDa protein band, indicating that these antibodies recognize specifically the dystrophin ZZ domain sequence. The 4G3 monoclonal antibody revealed an unknown protein band with molecular mass of approx. 170 kDa. Monoclonal supernatant 13D2 specifically recognized peptide 19 common to the dystrophin/utrophin ZZ domain (Figure 4A). It is surprising therefore that it did not recognize dystrophin in tissue sections or on Western blots (Figures 5A and 5B). This lack of reactivity might imply that the epitope for 13D2 is masked in tissue sections or is dependent on conformation; the latter might also explain its lack of reactivity in Western blots.
Figure 5. Immunodetection of dystrophin in muscle.
Serial cryostat sections of T. marmorata (A) or rabbit muscle (B) were labelled using the four monoclonal antibodies. Antibodies 12D7 and 14A4 both produced a clear sarcolemmal staining pattern consistent with dystrophin staining (A2/B2, A3/B3), which in both cases was competed by an excess of the specific peptide to which the antibody was raised (A2′/B2′, A3′/B3′). Antibody 4G3, which did not detect a specific peptide by ELISA, exhibited extensive background staining in both T. marmorata and rabbit muscle (A4/B4). Surprisingly, antibody 13D2 produced only a very weak background staining pattern that was not further reduced in the presence of excess peptide, with no clear sarcolemmal staining in either muscle type (A1/A1′, B1/B1′). Scale bar, 50 μm. In keeping with the immunohistological observation, antibodies 12D7 and 14A4 recognized an approx. 400 kDa band in Western blots of rabbit muscle extracts, again consistent with dystrophin staining, which was competed away with the specific peptide, whereas antibody 13D2 did not detect anything (C). Antibody 4G3, which did not detect a specific peptide or label the sarcolemma, recognized an unknown 170 kDa protein in Western blots. The specificity of antibodies 12D7 and 14A4 was further tested on muscle extracts from mdx mice that lack dystrophin (D). A previously characterized antibody specific for the C-termini of dystrophin (H4, far left lane) confirmed the absence of dystrophin, whereas the presence of utrophin was revealed by antibody K7 (near left lane). Antibodies 12D7 and 14A4 detected no bands on Western blots of mdx skeletal muscle, confirming their specificity for dystrophin (right lanes). The protein bands detected are indicated by molecular mass markers (Bio-Rad).
ELISA competition assays
In order to further investigate the reasons behind this lack of reactivity, we performed competitive ELISA experiments in microtitre plates coated with peptide 19. In two separate experiments we examined the competition between purified β-dystroglycan or a skeletal-muscle membrane fraction enriched for β-dystroglycan, and the 13D2 ZZ domain antibody or 43DAG/8D5, a monoclonal hybrid supernatant against β-dystroglycan. All experiments, including controls, were carried out in the presence of 1% BSA. The two experiments produced qualitatively similar results (Figure 6). 13D2 incubation produced a consistent high signal across all wells (Figure 6), consistent with the specificity of this monoclonal antibody for peptide 19 (Figure 4). The monoclonal antibody 43DAG/8D5 against β-dystroglycan, however, gave a consistent low signal across all wells, demonstrating that, like the related antiserum MANDAG2 (mouse anti-β-dystroglycan 2), this hybridoma supernatant does not recognize peptide 19 and is specific for the WW domain interaction sequence and PPPYVP epitope at the C-terminus of β-dystroglycan (Supplementary Figure 1 at http://www.BiochemJ.org/bj/401/bj4010667add.htm) and as reported previously [13,29]. Increasing concentrations of either purified recombinant β-dystroglycan (Figure 6A) or dystroglycan-containing membrane fraction (Figure 6B), competed for 13D2 binding to peptide 19 reducing the signal from successive wells. Conversely the detection of β-dystroglycan with 43DAG/8D5 revealed an increase in β-dystroglycan binding to peptide 19 with increasing β-dystroglycan or dystroglycan-containing membrane fraction competition across successive wells. Thus peptide 19 appears to be able to bind β-dystroglycan which can compete specifically for the 13D2 monoclonal antibody, suggesting that the region of the dystrophin and utrophin ZZ domains, corresponding to peptide 19, is an additional binding site for β-dystroglycan on dystrophin and utrophin. Furthermore, the ability of 43DAG/8D5 to still recognize β-dystroglycan bound to peptide 19 suggests that the two binding events are independent in this context.
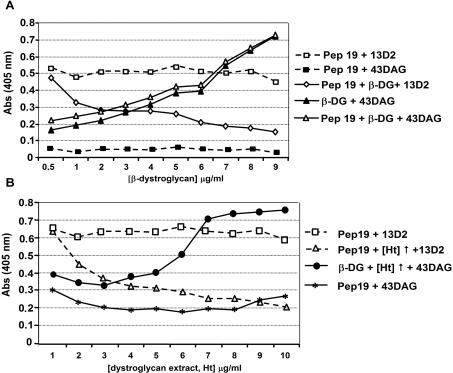
Figure 6. Competitive ELISA binding for peptide 19.
Microtitre plates coated with or without peptide 19 were detected with antibody 13D2 (raised against peptide 19) or 43DAG/8D5 (43DAG; raised against the β-dystroglycan cytoplasmic domain). In (A), peptide 19 was detected by 13D2 at similar levels in all wells (open squares) and not detected by 43DAG (closed squares). On incubation with increasing concentrations of purified β-dystroglycan cytoplasmic domain (β-DG; 0.5–9 μg/ml) 13D2 detection of peptide 19 was specifically reduced (open diamonds), whereas incubation of empty or peptide 19-coated wells with β-DG and detection with 43DAG showed a specific increase in signal. These results demonstrate a β-DG-specific competition for 13D2 binding to peptide 19, confirming peptide 19 as a β-dystroglycan-binding site. Qualitatively similar results were obtained in competitive ELISA binding for peptide 19 using a dystroglycan-enriched fraction of skeletal muscle (Ht; 1–10 μg/ml; B). Increasing concentrations of homogenate competed for 13D2 detection of peptide 19 (open triangle), whereas the homogenate had no effect on the detection of β-DG-coated wells by 43DAG (closed circles), other controls were as in (A).
Far-Western blotting of β-dystroglycan with dystrophin and utrophin ZZ domains
Total mouse muscle extract transferred to nitrocellulose was overlaid with a dystrophin-enriched crude membrane homogenate of mouse skeletal muscle, or with a utrophin-enriched crude membrane homogenate of mdx mouse skeletal muscle or sciatic nerve. The dystrophin in the enriched dystrophin homogenate bound to β-dystroglycan on the nitrocellulose membrane as revealed by blotting for the dystrophin C-terminus (Figure 7A3; Pep19–). When the blot was pre-incubated with peptide 19, however, the binding of dystrophin in the membrane homogenate to dystroglycan on the blot was strongly reduced (Figure 7A3; Pep19+). This result demonstrates that the peptide 19 can effectively block dystrophin binding to intact β-dystroglycan, again highlighting the importance of the ZZ domain for the dystrophin–dystroglycan interaction. In a similar experiment overlaying an enriched utrophin homogenate from dystrophin-deficient mdx muscle or sciatic nerve, utrophin detection using the C-terminal utrophin antibody (K7) showed that the fraction of utrophin that bound β-dystroglycan was less compared with that observed with dystrophin, suggesting that utrophin has a lower affinity for β-dystroglycan (Figure 7B3; Pep19−). However, when peptide 19 was applied, there was still a slight reduction in utrophin binding (Figure 7B3; Pep19+). The apparent difference in inhibition of binding to β-dystroglycan is not likely to be due to the slightly lower dystrophin+utrophin content seen in the mdx mice samples, compare quantifications in Figures 7(A4) and 7(B4). The difference in competition for β-dystroglycan binding probably reflects a real difference in the mode of binding between the dystrophin or utrophin ZZ domain, and β-dystroglycan. This may be due to other DGC (dystrophin–glycoprotein complex) components assembling differently in the mdx mouse in the presence of utrophin, which contributes to a slightly different mode of interaction with dystroglycan.
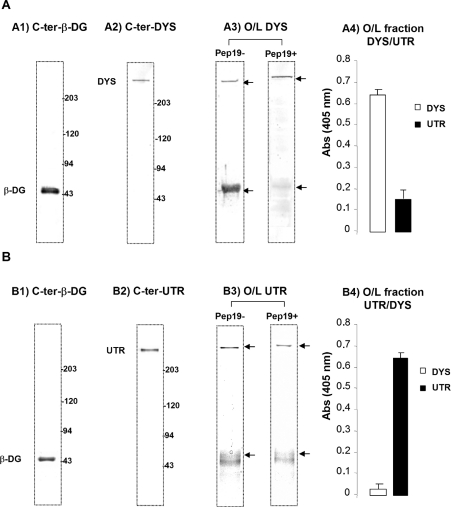
Figure 7. Overlay experiments using electroblotted total crude muscle membrane homogenate (Ht).
(A1) and (A2) correspond to the native nitrocellulose with total electrotransferred Ht revealed by LG5 and H4 polyclonal antibodies (anti-β-dystroglycan and anti-dystrophin respectively). Membranes were overlaid with enriched dystrophin homogenate comprising dystrophin and its associated proteins. Detection was performed with a dystrophin C-terminal antibody (H4). When peptide 19 was pre-incubated before overlay with the dystrophin-enriched homogenate, only a 400 kDa band was detected with H4 polyclonal antibody, suggesting that the binding site for β-dystroglycan was blocked (A3). Panel (B) represents the same overlay experiment performed with utrophin-enriched homogenate from mdx mouse comprising utrophin and its associated proteins detected by the C-terminal β-dystroglycan and utrophin antibodies (B1 and B2 respectively). Panel (B3) showed that the utrophin-binding properties for β-dystroglycan are slightly different from those of dystrophin and were not reduced to the same extent when peptide 19 was applied. Panels (A4) and (B4) represent densitometric quantification of the amounts of dystrophin (DYS) and utrophin (UTR) present in the two fractions.
DISCUSSION
The ZZ domain
ZZ domains are a highly conserved and widespread zinc-binding motif first identified in dystrophin/utrophin and the transcriptional co-activator CBP [CREB (cAMP-response-element-binding protein)-binding protein]/p300 [12] (Figure 8). In dystrophin, the first half of the C-terminal domain and ZZ- domain-containing cysteine-rich region has been implicated in binding to β-dystroglycan, and when mutated gives rise to a severe muscle-wasting phenotype [2,30,31]. This region contains several modular protein domains: a WW domain, two incomplete but putative calcium-binding sites (EF hands) [32] and a ZZ domain (zinc finger domain) [12]. An examination of the spectrum of point mutations in the human dystrophin gene in the ZZ domain region (Table 1) reveals that four of these mutations, where a full-length protein product may be produced, are in zinc-liganding residues implicated in our studies. The missense mutation C3313F produced a DMD phenotype, although no information is available as to whether this individual had any dystrophin protein expression [33]. D3335H on the other hand was identified in an individual with a normal level of correctly localized full-length dystrophin and dystrophin-associated proteins, but yet a severe DMD phenotype [34]. From our analysis of 65Zn peptide SPOTs overlay, D3335 is a potential liganding residue (Figure 3), but mutation of this residue to histidine in biochemical experiments had no effect on β-dystroglycan binding [35]. Although aspartic acid to histidine substitution is not normally considered a conservative substitution, in this context histidine could replace aspartic acid and still co-ordinate zinc, thus preserving protein conformation and, as suggested from the patient phenotype, presumably some function too. It may be that Asp3335 has another important functional role that so far has not been elucidated and that is not supported by a histidine substitution, but it is difficult to predict what that might be. Aspartic acid in this position is conserved in most ZZ domain sequences (Figure 8), suggesting a functional importance [12].
Table 1. Missense mutations in the ZZ domain of dystrophin.
| Phenotype | Dystrophin ZZ domain | Source |
|---|---|---|
| Wild-type | A3311KCNICKECPIIGFRYRSLKHFNYDICQSCFF3343 | |
| DMD | AKFNICKECPIIGFRYRSLKHFNYDICQSCFF | [33] |
| BMD | AKCNICKERPIIGFRYRSLKHFNYDICQSCFF | [45] |
| DMD | AKCNICKECPIIGFRYRSLKHFNYHICQSCFF | [34] |
| DMD | AKCNICKECPIIGFRYRSLKHFNYDIYQSCFF | [43] |
| DMD* | AKCNICKECPIIGFRYRSLKHFNYDICLSCFF | [45] |
| DMD† | AKCNICKECPIIGFRYRSLKHFNYDICFSCFF | [46] |
*†These mutations produced frameshifting stop codons 34 (*) and 8 (†) codons respectively downstream of the point mutation. Consequently, the DMD phenotypes could be due to the frameshift and/or truncation rather than any specific point mutation. A rare single amino acid deletion of a conserved Glu3367 just C-terminal of the ZZ domain resulted in a DMD phenotype despite normal correctly localized protein [47].
Zinc binding
An NMR structure of the ZZ domain from CBP revealed a cross-brace zinc finger motif [36], where the individual zinc ions are co-ordinated by non-linear sequence elements, with the protein backbone crossing back on itself to form a zinc-binding site (Figure 8B). The SPOTs analysis carried out in Figure 3 could not reveal co-ordination of zinc through such a structure as the individual peptides were only 20 amino acids long, and from the CBP ZZ domain structure the liganding residues are 21 and 26 residues apart in the linear sequence [36]. Due to insertions in the dystrophin/utrophin sequences, this corresponds to 27 and 28 residues apart, too long to provide an unambiguous interpretation in the peptide array. But, as highlighted in Figure 3, it does not appear that just any combination of four residues that could co-ordinate zinc will bind zinc. The two pairs of cysteine and histidine residues in peptides 43–46 of the SPOTs are appropriately spaced to bind zinc (Figure 3A) but do not, suggesting a degree of specificity to the other sequences identified by this method that do bind zinc. Moreover, unlike almost all other ZZ domain sequences in the databases, vertebrate dystrophin and utrophin lack three of the conserved residues that form the second zinc-binding site in the CBP ZZ domain: the central pair of cysteines and one of the C-terminal histidines (Figure 8B) [12,36]. This would suggest that dystrophin and utrophin bind zinc in a different conformation and this conformation may be specific to the ability of these proteins to interact with dystroglycan through multiple modes of interaction as discussed below. Interestingly the vertebrate dystrobrevins and invertebrate dystrophins and dystrobrevins do have cysteine residues in these positions, suggesting some functional divergence with the emergence of distinct dystrophin and utrophin genes in vertebrates.
Dystroglycan binding: role of the WW domain
We and others have independently mapped the site for dystrophin and utrophin binding to dystroglycan to the last 15 amino acids at the C-terminus of β-dystroglycan [13,14,16,37–39]. The major determinant within these 15 residues is the sequence PPPY, a consensus for type I WW domain interactions [13,16,37,39]. Despite a necessary requirement for the PPPY motif in dystroglycan for dystrophin/utrophin binding via their WW domains [29], this is not sufficient. Where this requirement has been analysed in more detail, it has been found that constructs corresponding to the WW domain of dystrophin or utrophin alone cannot bind β-dystroglycan, the addition of the EF hand region allows some binding, but full binding potential is not realized until the ZZ domain is included [13,14,16,31,35,39,40]. Unlike most other WW domains that exhibit autonomous binding activity towards their cognate peptide ligands [29], the WW domains of dystrophin and utrophin as a minimum require the support of the EF hand region. The reasons for this became clear on elucidation of a crystal structure of the dystrophin WW–EF region in complex with a β-dystroglycan peptide [15]. This revealed that the WW domain was supported extensively by the EF hand region including a contribution to the β-dystroglycan binding interface and confirming earlier elegant biochemical studies by this group [16].
Dystroglycan binding: role of the ZZ domain
Detailed biochemical analyses have pointed to a role for the ZZ domain in further supporting and contributing to this binding interface [16,35]. Moreover, Rentschler et al. [16] even go so far as to suggest that there is an additional binding site between dystroglycan and non-WW domain regions in dystrophin, possibly the ZZ domain. Transgenic mdx mice expressing various dystrophin gene constructs showed that the dystrophic change in mdx muscle could be rescued [41,42]. Because the binding of dystrophin to β-dystroglycan is required to prevent the dystrophic phenotype, deletion models can be considered to exhibit effective and physiological binding activity in these mice. In this context, the full-length cDNA with deletion of exons 68–70 (ZZ domain) failed to rescue the phenotype [41] despite the presence of the WW–EF region. Another model with deletion of exons 64–67 (EF hands) also failed to rescue the phenotype [42]. This model, which includes the WW domain encoded by exons 62–63 but lacks the subsequent EF1 and EF2, suggests as in the biochemical experiments that the WW domain alone is not sufficient for effective binding to β-dystroglycan. On the other hand, a mouse with deletion of exons 71–78 could rescue the phenotype, suggesting that the C-terminal region encoded by these exons is not essential for the binding [42]. All these results are compatible with the fact that the C-terminal region of dystrophin spanning 3311–3342, the ZZ domain, is crucial for binding to β-dystroglycan. Our results suggest that we could limit the direct binding activity of this crucial region to the residues D3326–3332 or U3083–3089 of dystrophin or utrophin respectively (peptide 19). Taken together, these result are consistent with there being a second dystroglycan-binding site between dystrophin/utrophin ZZ domain and dystroglycan in addition to the WW domain-mediated interaction with the extreme C-terminus of β-dystroglycan. As shown in Figure 6, this region is likely to be distinct from the well-characterized PPPYVP site due to the inability of 43DAG/8D5 to compete for peptide 19 binding to purified β-dystroglycan. This additional binding site further stabilizes the β-dystroglycan–WW domain interaction, which itself is supported by the EF hand region, and explains why the complete WW–EF–ZZ region appears to be required for full binding activity between β-dystroglycan and dystrophin or utrophin.
ZZ domain mutations in humans
The first missense mutation reported in the C-terminal region was C3340Y from a patient with DMD and mental retardation, but with approx. 20% of normal levels of dystrophin immunoreactivity and some β-dystroglycan staining at the sarcolemma [43]. From our analyses, Cys3340 would be predicted to be essential for the formation of the second zinc knuckle (Figure 3), and mutation to tyrosine would ablate the zinc-binding site. Biochemical analysis of this mutation, and also an engineered C3340S substitution, in the context of the interaction between dystrophin and β-dystroglycan also demonstrates a loss of function [35] in support of the patient data and our own analysis of the zinc-binding site. From the biochemical analysis of mutations of all the cysteines in the dystrophin ZZ domain, only C3340Y prevents dystroglycan binding. Mutations C3313Y, C3316Y and C3319Y still allow dystroglycan binding [35], suggesting some plasticity in zinc binding in the first zinc knuckle. This is evident from the SPOTs overlay in Figure 3(A), where there are five potential liganding residues, but only four would be required to co-ordinate zinc. The situation for utrophin appears different however. Mutation of any of the three cysteine residues in the first zinc knuckle did not support dystroglycan binding, whereas mutation of either cysteine in the second zinc knuckle did support dystroglycan binding [35]. This observation is supported in part by our own data, where antibody 13D2 directed against the second zinc knuckle completely blocked dystroglycan binding to dystrophin but had a reduced ability to block utrophin binding (Figure 7), suggesting that the second zinc knuckle is less important for the utrophin–dystroglycan interaction.
Summary
The above data provide a conceptual advance on previously reported findings [16,35] and define the ZZ domain in concert with the WW domain as being a crucial structural and functional part of the dystrophin/utrophin–β-dystroglycan interaction that is involved in anchoring these proteins to the cell membrane. The identification of the short dystrophin peptide YRSLKHF defines a second β-dystroglycan-binding site in dystrophin and utrophin and raises the possibility of identifying further specific antibodies that could be used to distinguish between dystrophin and utrophin in this highly similar region. Finally, biochemical approaches, epitope mapping, ELISA and overlay provide new evidence for the zinc-binding properties and β-dystroglycan interaction site on the ZZ domain, and highlight distinct differences in the mode of interaction between β-dystroglycan and the dystrophin or utrophin ZZ domains.
Online data
Acknowledgments
K.H. was supported by the AFM (Association Française contre les Myopathies; Fellowship no. 10529), D.M. by Montpellier 1 University and CNRS (Centre National de la Recherche Scientifique), and S.J.W. by the BBSRC (Biotechnology and Biological Sciences Research Council; ICR07596), MRC (G0000104) and Wellcome Trust (042180). We are extremely grateful to Jürgen Wehland (Department of Cell Biology, Helmholtz Centre for Infection Research, Braunschweig, Germany) for synthesis of the ZZ domain peptide SPOT array and to Louise Anderson, who sadly and prematurely passed away in 2005, for generous supplies of 43DAG/8D5.
References
- 1.Koenig M., Hoffman E. P., Bertelson C. J., Monaco A. P., Feener C., Kunkel L. M. Complete cloning of the Duchenne muscular-dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman E. P., Brown R. H., Jr, Kunkel L. M. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 3.Koenig M., Monaco A. P., Kunkel L. M. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53:219–226. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- 4.Davison M. D., Critchley D. R. α-Actinins and the DMD protein contain spectrin-like repeats. Cell. 1988;52:159–160. doi: 10.1016/0092-8674(88)90503-x. [DOI] [PubMed] [Google Scholar]
- 5.Ervasti J. M., Campbell K. P. A role for the dystrophin glycoprotein complex as a transmembrane linker between laminin and actin. J. Cell Biol. 1993;112:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koenig M., Beggs A. H., Moyer M., Scherpf S., Heindrich K., Bettecken T., Meng G., Muller C. R., Lindlof M., Kaariainen H., et al. The molecular-basis for Duchenne versus Becker muscular-dystrophy – correlation of severity with type of deletion. Am. J. Hum. Genet. 1989;45:498–506. [PMC free article] [PubMed] [Google Scholar]
- 7.Beggs A. H., Hoffman E. P., Snyder J. R., Arahata K., Specht L., Shapiro F., Angelini C., Sugita H., Kunkel L. M. Exploring the molecular-basis for variability among patients with Becker muscular-dystrophy – dystrophin gene and protein studies. Am. J. Hum. Genet. 1991;49:54–67. [PMC free article] [PubMed] [Google Scholar]
- 8.Tinsley J. M., Blake D. J., Roche A., Fairbrother U., Riss J., Byth B. C., Knight A. E., Kendrick-Jones J., Suthers G. K., Love D. R., et al. Primary structure of dystrophin-related protein. Nature. 1992;360:591–593. doi: 10.1038/360591a0. [DOI] [PubMed] [Google Scholar]
- 9.Chamberlain J. The structure and function of dystrophin. In: Winder S. J., editor. Molecular Mechanisms of Muscular Dystrophies. Georgetown, TX: Landes Bioscience; 2006. pp. 14–34. [Google Scholar]
- 10.Andre B., Springael J. Y. WWP, a new amino acid motif present in single or multiple copies in various proteins including dystrophin and the SH3-binding Yes-associated protein YAP65. Biochem. Biophys. Res. Commun. 1994;205:1201–1205. doi: 10.1006/bbrc.1994.2793. [DOI] [PubMed] [Google Scholar]
- 11.Bork P., Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem. Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 12.Ponting C. P., Blake D. J., Davies K. E., Kendrick-Jones J., Winder S. J. ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem. Sci. 1996;21:11–13. [PubMed] [Google Scholar]
- 13.James M., Nuttall A., Ilsley J. L., Ottersbach K., Tinsley J. N., Sudol M., Winder S. J. Adhesion-dependent tyrosine phosphorylation of β-dystroglycan regulates its interaction with utrophin. J. Cell Sci. 2000;113:1717–1726. doi: 10.1242/jcs.113.10.1717. [DOI] [PubMed] [Google Scholar]
- 14.Jung D., Yang B., Meyer J., Chamberlain J. S., Campbell K. P. Identification and characterization of the dystrophin anchoring site on β-dystroglycan. J. Biol. Chem. 1995;270:27305–27310. doi: 10.1074/jbc.270.45.27305. [DOI] [PubMed] [Google Scholar]
- 15.Huang X., Poy F., Zhang R., Joachimiak A., Sudol M., Eck M. J. Structure of a WW domain containing fragment of dystrophin in complex with β-dystroglycan. Nat. Struct. Biol. 2000;7:634–638. doi: 10.1038/77923. [DOI] [PubMed] [Google Scholar]
- 16.Rentschler S., Linn H., Deininger K., Bedford M. T., Espanel X., Sudol M. The WW domain of dystrophin requires EF-hands region to interact with β-dystroglycan. Biol. Chem. 1999;380:431–442. doi: 10.1515/BC.1999.057. [DOI] [PubMed] [Google Scholar]
- 17.Pons F., Robert A., Fabbrizio E., Hugon G., Califano J.-C., Fehrentz J. A., Martinez J., Mornet D. Utrophin localisation in normal and dystrophin-deficient heart. Circulation. 1994;90:369–374. doi: 10.1161/01.cir.90.1.369. [DOI] [PubMed] [Google Scholar]
- 18.Winder S. J., Kendrick-Jones J. Protein production in 3 different expression vectors from a single PCR product. Anal. Biochem. 1995;231:271–273. doi: 10.1006/abio.1995.1534. [DOI] [PubMed] [Google Scholar]
- 18a.Erratum. Anal. Biochem. 1996;236:190. [Google Scholar]
- 19.Chen Y.-J., Spence H. J., Cameron J. M., Jess T., Ilsley J. L., Winder S. J. Direct interaction of β-dystroglycan with F-actin. Biochem. J. 2003;375:329–337. doi: 10.1042/BJ20030808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fabbrizio E., Leger J., Anoal M., Leger J. J., Mornet D. Monoclonal antibodies targetted against the C-terminal domain of dystrophin or utrophin. FEBS Lett. 1993;322:10–14. doi: 10.1016/0014-5793(93)81100-e. [DOI] [PubMed] [Google Scholar]
- 21.Chazalette D., Hnia K., Rivier F., Hugon G., Mornet D. a7B integrin changes in mdx mouse muscles after L-arginine administration. FEBS Lett. 2005;579:1079–1084. doi: 10.1016/j.febslet.2004.12.081. [DOI] [PubMed] [Google Scholar]
- 22.Fabbrizio E., Latouche J., Rivier F., Hugon G., Mornet D. Re-evaluation of the distributions of dystrophin and utrophin in sciatic nerve. Biochem. J. 1995;312:309–314. doi: 10.1042/bj3120309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khurana T. S., Watkins S. C., Kunkel L. M. The subcellular distribution of chromosome 6-encoded dystrophin-related protein in the brain. J. Cell Biol. 1992;119:357–366. doi: 10.1083/jcb.119.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schiff L. A., Nibert M. L., Fields B. N. Characterization of a zinc blotting technique: evidence that a retroviral gag protein binds zinc. Proc. Natl. Acad. Sci. U.S.A. 1988;85:4195–4199. doi: 10.1073/pnas.85.12.4195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chantler P. D. Calcium-dependent association of a protein complex with the lymphocyte plasma membrane: probable identity with calmodulin-calcineurin. J. Cell Biol. 1985;101:207–216. doi: 10.1083/jcb.101.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawson R. M. C., Elliott D. C., Elliott W. H., Jones K. M. Oxford: Clarendon Press; 1986. Data for Biochemical Research. [Google Scholar]
- 27.Winder S. J. Structure–function relationships in dystrophin and utrophin. Biochem. Soc. Trans. 1996;24:497–501. doi: 10.1042/bst0240497. [DOI] [PubMed] [Google Scholar]
- 28.Vallee B. L., Auld D. S. Zinc coordination, function, and structure of zinc enzymes and other proteins. Biochemistry. 1990;29:5647–5659. doi: 10.1021/bi00476a001. [DOI] [PubMed] [Google Scholar]
- 29.Ilsley J. L., Sudol M., Winder S. J. The WW domain: linking cell signalling to the membrane cytoskeleton. Cell. Signalling. 2002;14:183–189. doi: 10.1016/s0898-6568(01)00236-4. [DOI] [PubMed] [Google Scholar]
- 30.Campbell K. P., Kahl S. D. Association of dystrophin and an integral membrane glycoprotein. Nature. 1989;338:259–262. doi: 10.1038/338259a0. [DOI] [PubMed] [Google Scholar]
- 31.Suzuki A., Yoshida M., Yamamoto H., Ozawa E. Glycoprotein-binding site of dystrophin is confined to the cysteine-rich domain and the first half of the carboxy-terminal domain. FEBS Lett. 1992;308:154–160. doi: 10.1016/0014-5793(92)81265-n. [DOI] [PubMed] [Google Scholar]
- 32.Chung W., Campanelli J. T. WW and EF hand domains of dystrophin-family proteins mediate dystroglycan binding. Mol. Cell Biol. Res. Commun. 1999;2:162–171. doi: 10.1006/mcbr.1999.0168. [DOI] [PubMed] [Google Scholar]
- 33.Flanigan K., von Niederhausern A., Dunn D., Alder J., Mendell J., Weiss R. Rapid direct sequence analysis of the dystrophin gene. Am. J. Hum. Genet. 2003;72:931–919. doi: 10.1086/374176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goldberg L. R., Hausmanowa-Petrusewicz I., Fidzianska A., Duggan D. J., Steinberg L. S., Hoffman E. P. A dystrophin missense mutation showing persistence of dystrophin and dystrophin-associated proteins yet a severe phenotype. Ann. Neurol. 1998;44:971–976. doi: 10.1002/ana.410440619. [DOI] [PubMed] [Google Scholar]
- 35.Ishikawa-Sakurai M., Yoshida M., Imamura M., Davies K. E., Ozawa E. ZZ domain is essentially required for the physiological binding of dystrophin and utrophin to β-dystroglycan. Hum. Mol. Genet. 2004;13:693–702. doi: 10.1093/hmg/ddh087. [DOI] [PubMed] [Google Scholar]
- 36.Legge G. B., Martinez-Yamout M. A., Hambly D. M., Trinh T., Lee B. M., Dyson H. J., Wright P. E. ZZ domain of CBP: an unusual zinc finger fold in a protein interaction module. J. Mol. Biol. 2004;343:1081–1093. doi: 10.1016/j.jmb.2004.08.087. [DOI] [PubMed] [Google Scholar]
- 37.Ilsley J. L., Sudol M., Winder S. J. The interaction of dystrophin with β-dystroglycan is regulated by tyrosine phosphorylation. Cell. Signalling. 2001;13:625–632. doi: 10.1016/s0898-6568(01)00188-7. [DOI] [PubMed] [Google Scholar]
- 38.Rosa G., Ceccarini M., Cavaldesi M., Zini M., Petrucci T. C. Localisation of the dystrophin binding site at the carboxyl terminus of β-dystroglycan. Biochem. Biophys. Res. Commun. 1996;223:272–277. doi: 10.1006/bbrc.1996.0883. [DOI] [PubMed] [Google Scholar]
- 39.Tommasi di Vignano A., Di Zenzo G., Sudol M., Cesareni G., Dente L. Contribution of the different modules in the utrophin carboxy-terminal region to the formation and regulation of the DAP complex. FEBS Lett. 2000;471:229–234. doi: 10.1016/s0014-5793(00)01400-9. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki A., Yoshida M., Hayashi K., Mizuno Y., Hagiwara Y., Ozawa E. Molecular organisation at the glycoprotein-complex-binding site of dystrophin: three dystrophin-associated proteins bind directly to the carboxy-terminal portion of dystrophin. Eur. J. Biochem. 1994;220:283–292. doi: 10.1111/j.1432-1033.1994.tb18624.x. [DOI] [PubMed] [Google Scholar]
- 41.Rafael J. A., Cox G. A., Corrado K., Jung D., Campbell K. P., Chamberlain J. S. Forced expression of dystrophin deletion constructs reveals structure-function correlations. J. Cell Biol. 1996;134:93–102. doi: 10.1083/jcb.134.1.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crawford G. E., Faulkner J. A., Crosbie R. H., Campbell K. P., Froehner S. C., Chamberlain J. S. Assembly of the dystrophin-associated protein complex does not require the dystrophin COOH-terminal domain. J. Cell Biol. 2000;150:1399–1410. doi: 10.1083/jcb.150.6.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lenk U., Oxele K., Voit T., Ancker U., Hellner K.-A., Speer A., Hubner C. A cysteine 3340 substitution in the dystroglycan-binding domain of dystrophin associated with Duchenne muscular dystrophy, mental retardation and absence of the ERG b-wave. Hum. Mol. Genet. 1996;5:973–975. doi: 10.1093/hmg/5.7.973. [DOI] [PubMed] [Google Scholar]
- 44.Winder S. J. The membrane-cytoskeleton interface: the role of dystrophin and utrophin. J. Muscle Res. Cell Motil. 1997;18:617–629. doi: 10.1023/a:1018627705273. [DOI] [PubMed] [Google Scholar]
- 45.Aartsma-Rus A., Van Deutekom J. C. T. V., Fokkema I. F., Van Ommen G.-J. B., Dunnen J. T. D. Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 46.Hofstra R. M. W., Mulder I. M., Vossen R., de Koning-Gans P. A. M., Kraak M., Ginjaar I. B., van der Hout A. H., Bakker E., Buys C. H. C. M., van Ommen G.-J. B., et al. DGGE-based whole-gene mutation scanning of the dystrophin gene in Duchenne and Becker muscular dystrophy patients. Hum. Mutat. 2004;23:57–66. doi: 10.1002/humu.10283. [DOI] [PubMed] [Google Scholar]
- 47.Becker K., Robb S. A., Hatton Z., Yau S. C., Abbs S., Roberts R. G. Loss of a single amino acid from dystrophin resulting in Duchenne muscular dystrophy with retention of dystrophin protein. Hum. Mutat. 2003;21:651–656. doi: 10.1002/humu.9143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.