Abstract
Sterol 14α-demethylase encoded by CYP51 is a mixed-function oxidase involved in sterol synthesis in eukaryotic organisms. Completion of the Mycobacterium tuberculosis genome project revealed that a protein having homology to mammalian 14α-demethylases might be present in this bacterium. Using genomic DNA from mycobacterial strain H37Rv, we have established unambiguously that the CYP51-like gene encodes a bacterial sterol 14α-demethylase. Expression of the M. tuberculosis CYP51 gene in Escherichia coli yields a P450, which, when purified to homogeneity, has the predicted molecular mass, ca. 50 kDa on SDS/PAGE, and binds both sterol substrates and azole inhibitors of P450 14α-demethylases. It catalyzes 14α-demethylation of lanosterol, 24,25-dihydrolanosterol, and obtusifoliol to produce the 8,14-dienes stereoselectively as shown by GC/MS and 1H NMR analysis. Both flavodoxin and ferredoxin redox systems are able to support this enzymatic activity. Structural requirements of a 14α-methyl group and Δ8(9)-bond were established by comparing binding of pairs of sterol substrate that differed in a single molecular feature, e.g., cycloartenol paired with lanosterol. These substrate requirements are similar to those established for plant and animal P450 14α-demethylases. From the combination of results, the interrelationships of substrate functional groups within the active site show that oxidative portions of the sterol biosynthetic pathway are present in prokaryotes.
The origin and evolution of the sterol pathway continues to be an enigma. It has been proposed that it is very ancient, perhaps having arisen during the later stages of prokaryote evolution, after the introduction of molecular oxygen into the atmosphere (1, 2). For aerobic bacteria such as Mycobacterium tuberculosis (MT) (3), the architectural requirements of the cell membrane can be satisfied by either sterol surrogates, e.g., pentacyclic hopanoids (4, 5) synthesized directly from squalene by an anaerobic pathway, or by sterols synthesized from squalene by an aerobic pathway (1). However, based on our knowledge that sterols play a dual role in eukaryotic cell physiology at vastly different cellular concentrations, structurally as bulk inserts to affect permeability and hormonally to regulate growth, reproduction, and other processes (6–8), the level of cellular sterols in bacteria may be substantially less than even the hormonal level required in eukaryotes. In bacteria (1), sterol concentration was found to be two or more orders of magnitude less than the level in eukaryotic cells, which ranges from ca. 30 to 3,000 fg/cell depending on the size of the cell (9). Although most bacteria are reported not to contain sterols, chemical and biochemical studies have shown the occurrence and biosynthesis of distinct sterols in several nonphotosynthetic and photosynthetic bacteria (1, 10, 11). Only genes involved in the synthesis of isopentenyl pyrophosphate, a common precursor in isoprenoid and terpenoid biosynthesis, were identified in Escherichia coli (12, 13). Steps involved in the oxygen-requiring portion of sterol biosynthesis in eukaryotic organisms are still unknown in bacteria. Recently, Lamb et al. showed that Mycobacterium smegmatis, a closely related species to MT, is able to synthesize cholesterol from radiolabeled mevalonic acid (14), indicating the presence of genes encoding sterol biosynthetic enzymes in some oxygenic bacteria. Sequencing of genomic fragments from Mycobacterium leprae revealed a CYP (P450) gene fragment, and comparison between that and MT genomic sequences led to detection of one sterol 14α-demethylase cytochrome P450 (CYP51)-like P450 (15). Completion of the MT genome revealed the presence of 20 P450 genes (16), including one that had sequence similarity (29–39%) with known CYP51 genes in animals, plants, and fungi that encode the oxygen-requiring sterol 14α-demethylase (P45014DM) (17). Expressed in E. coli, this gene product, named CYP51-like, was shown to be a soluble P450 enzyme that binds ketoconazole, a known inhibitor of P450s including P45014DM (18).
We have characterized the enzymatic properties of the MT CYP51-like protein. Both a 3Fe-4S MT ferredoxin (Fdx) and E. coli flavodoxin (Fld) (19) are found to support the 14α-demethylase activity. Sterol specificity and mechanism of removal of the 14α-methyl group are unique to P450-dependent enzymes (Fig. 1B); P45014DM catalyzes the removal of the 14α-methyl group (C32) of sterols through three successive oxidations resulting in decarbonylation, releasing formic acid, and generation of an 8,14-diene sterol product (20). These steps appear to be common to all P45014DM transformations, although alternate double-bond introductions at 8,14-diene are known or suspected to occur (21, 22). The sterol substrate requirements of P45014DM from animal (20, 22), plant (23), and fungi (24) have been established; the C3-hydroxyl group, 14α-methyl group, and Δ8(9)-bond are obligatory for binding and catalysis. Purified MT P45014DM is found to act on 14α-methyl sterols specifically and to bind well known P45014DM azole inhibitors. Similar to the plant and fungal forms (23, 24), MT P45014DM binds preferentially C24-alkylated sterols, suggesting that the CYP51 enzyme from MT is more plant/fungal-like than animal-like.
Figure 1.
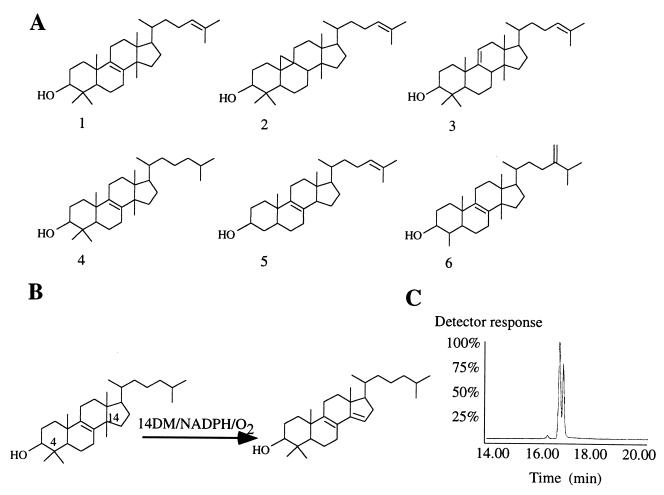
(A) Sterol molecule structures. 1, lanosterol; 2, cycloartenol; 3, parkeol; 4, DHL; 5, zymosterol; and 6, obtusifoliol. (B) DHL 14α-demethylation. Conversion of DHL to 4,4-dimethyl-5α-cholesta-8,14-diene-3β-ol in the presence of MT P45014DM, NADPH, and molecular oxygen. (C) GLC profile of overnight conversion of 2 mg DHL. E. coli Fld/Fdr system was used as P450 electron donor. The peaks at 16.38 and 17 min correspond to the DHL and metabolite retention times, respectively.
MATERIALS AND METHODS
General Methods.
Absolute spectra of purified MT P45014DM were recorded as described by Omura and Sato (25). Protein quantification was performed by using the Bradford method. DNA sequencing was carried out by using automated (Applied Biosystems PRISM Dye Terminator Cycle Sequencing Ready Reaction kit and Applied Biosystems PRISM 377 DNA sequencer) and manual sequencing (SequiTherm Cycle Sequencing; Epicentre Technologies, Madison, WI). Sterols were obtained from the Nes collection (26, 27) and purified by HPLC before assay with P45014DM. The six sterols assayed with MT P45014DM are shown in Fig. 1A.
MT P450 and Fdx Cloning and Expression.
Genomic DNA from MT strain H37Rv was provided by the TB Research Materials and Vaccine Testing Contract (NO1 AI-75320) at Colorado State University, and the MT CYP51-like gene was cloned by PCR using Vent polymerase (New England Biolabs, Beverly, MA). Primers were designed based on the sequence of cosmid MTCY369 from the MT genome, the upstream primer 5′-cgccatatgagcgctgttgcactaccc-3′, except the first 6 bases that were complementary to the sequence between bases 7495 and 7475, which is predicted to encode the N-terminal sequence of the MT CYP51-like protein. The downstream primer 5′- cgcaagcttcagtgatggtgatgaactcccgttcgccggcggtagc-3′, from bases 24 to 46, is identical to MTCY369 sequence between bases 6143 and 6165. For the Fdx gene, the upstream primer 5′-cgccatatgggctatcgagtcgaagcc-3′, except the first 6 bases, is complementary to MTCY369 sequence between bases 6137 and 6117 and the downstream primer 5′-cgcaagcttcagtgatggtgatgctctcccgtttctcggatggacagtgcctggg-3′ from bases 24 to 55 is identical to bases 5934–5965. The stop codon was removed in each gene, and four histidine codons followed by a new stop codon (bold characters) were inserted in the 3′ end of the coding sequences. The underlined bases are NdeI-cloning sites, including the initiator codon in the upstream primers and HindIII-cloning sites in the downstream primers. Amplification conditions were 94°C for 5 min and then 30 cycles of 94°C for 30 sec, 50°C for 30 sec, and 72°C for 45 sec. The PCR program ended by using one polymerization step at 72°C for 10 min, and the product was separated by electrophoresis on a 1% agarose gel. Bands of the expected sizes of MT P45014DM (1,377 bp) and Fdx (233 bp) were eluted from the gel by using Qiagen II kit (Qiagen, Valencia, CA). After digestion by NdeI and HindIII, the cDNAs were cloned into the E. coli expression vector pet17b (Novagen), giving MTP450/pet17b and MTFdx/pet17b. Those vectors were transformed separately into competent HMS174 (Novagen) cells. Single ampicillin-resistant colonies from each transformant were grown overnight at 37°C in 5 ml of Terrific Broth containing 100 μg/ml ampicillin. These precultures were used to inoculate (1:100) 500 ml of modified Terrific Broth medium (100 μg/ml ampicillin) (28). After 5-h growth at 37°C in a shaking incubator at 240 rpm, the culture was induced by using 1 mM isopropyl β-d-thiogalactopyranoside (Calbiochem). At the same time, δ-aminolevulinic acid (Sigma) was added to 2 mM final concentration for P450 expression. Growth was continued at 30°C with shaking at 190 rpm for 20 h.
P450 and Fdx Purification.
Three liters of MT P450 culture was pelleted and resuspended in 200 ml of TES buffer (19). After addition of lysozyme (0.5 mg/ml) and stirring at 4°C for 15 min, 1 vol of ice-cold water containing 0.1 mM EDTA was added slowly, and stirring continued for 30 min. Spheroplasts were pelleted at 3,000 × g for 15 min. The supernatant (fraction A) was centrifuged at 225,000 × g for 30 min after addition of DNase I (1 μg/ml) and stirring at 4°C for 15 min. Spheroplasts were resuspended in 50 ml of 2-fold-diluted TES buffer and sonicated by using a Branson sonifier (Model 250) at duty cycle 30–40, 50% maximal output for 30 sec at room temperature followed by 1-min incubation on ice, repeated 10 times. After centrifugation at 225,000 × g for 30 min, the supernatant (fraction B) was combined with fraction A and the P450 was isolated by using a Ni2+-nitrilotriacetic acid (NTA) affinity column (Qiagen) equilibrated with 50 mM potassium phosphate, pH 7.4/20% glycerol. After washing with the same buffer containing 50 mM glycine and 500 mM NaCl, the P450 was eluted by using 40 mM l-histidine in place of glycine. The P450 eluate was dialyzed overnight against 50 mM potassium phosphate, pH 7.4/20% glycerol. One liter of MT Fdx culture grown as for MT P450 was pelleted and resuspended in 50 ml of 50 mM potassium phosphate, pH 7.4/0.1 mM EDTA/20% glycerol. After addition of lysozyme (0.5 mg/ml) and stirring at 4°C for 15 min, cells were sonicated as above. The cytosolic fraction after centrifugation at 225,000 × g was loaded on a Ni2+NTA affinity column. Washing and the elution conditions were the same as for MT P450.
Antibody Production.
Polyclonal antibodies against MT P45014DM purified by two passes over Ni2+NTA were raised in white New Zealand rabbits that were injected with 0.5 mg of MT P45014DM mixed with either complete Freund’s adjuvant (Sigma) or TiterMax@Gold (CytRx, Norcross, GA). Two weeks later, the rabbit injected with Freund’s adjuvant was boosted by using 0.5 mg of MT P45014DM in Freund’s incomplete adjuvant (Sigma), and the antiserum was collected after 4 weeks. From the rabbit injected with TiterMax@Gold, antiserum was collected after 19 days.
Electron Donor System Investigation.
Activities of P450 enzymes require support of a reductase, and a functional reductase system for MT P450s was unknown. The capacity of rat microsomal NADPH cytochrome P450 reductase, of bovine adrenodoxin/adrenodoxin reductase, and E. coli Fld/flavodoxin reductase (Fdr) (19) to reduce MT P45014DM was determined by formation of the reduced-CO spectrum. MT P45014DM (200 pmol) and rat P450 reductase (200 pmol) were incubated in 10 mM potassium phosphate buffer, pH 7.4, containing 20% glycerol and 200 μM final concentration of lanosterol with or without 100 μg/ml sonicated dilauroyl-l-a-phosphatidylcholine. After several cycles of degassing and bubbling with carbon monoxide, NADPH (Calbiochem) was added (final concentration, 1 mM) and the reduced-CO spectrum was recorded. In the control experiment, 40 μM final concentration of progesterone was added to 200 pmol bovine 17α-hydroxylase P450. To study the bovine adrenodoxin/adrenoxin reductase system, 20 mM Tris⋅HCl, pH 7.4, buffer containing 0.2% Tween 20/4 mM MgCl2/200 μM lanosterol (final concentrations) was used with 100 pmol MT P45014DM/1 nmol adrenodoxin/100 pmol adrenodoxin reductase with 1 mM NADPH. Bovine cholesterol side-chain cleavage P450 (100 pmol) with the substrate 25-hydroxycholesterol (30 μM final concentration) was used as positive control. Using the Fld/Fdr system, 500 pmol MT P45014DM, 2.5 nmol Fld, and 500 pmol Fdr were incubated on ice (10 min) in 3-(N-morpholino)propane sulfonic acid buffer containing 200 μM final concentration of lanosterol and 1 mM NADPH with or without 100 μg/ml dilauroyl-l-a-phosphatidylcholine. In the control experiment, 40 μM progesterone was added to 500 pmol P450 17α-hydroxylase.
Reconstituted Catalytic Activity and Sterol Analysis.
MT P45014DM (365 pmol) was incubated on ice (10 min) with 18 nmol Fld and 2 nmol Fdr or 18 nmol MT Fdx and 2 nmol spinach ferredoxin reductase (Fnr). Because the electron donor to MT Fdx is unknown, Fnr (Sigma), shown to reduce ferredoxins from Streptomyces griseolus (29), was used. Substrate dispersed in Triton WR 1339 was resuspended in 3-(N-morpholino) propane sulfonic acid buffer (30). After mixing, the reaction was initiated (2 mM NADPH) in a final volume of 500 μl. After catalysis, sterols were extracted twice by using 5 vol of ethyl acetate for small-scale reactions or hexane for large-scale experiments. In the latter case, 1 vol of methanol containing 10% KOH was used to stop the reaction. One volume of DMSO then was added, and, after heating at 90°C and cooling to room temperature, sterols were extracted three times by using 3 vol of hexane and evaporated to dryness. Radiolabeled dihydrolanosterol ([24-3H]DHL) (20) and its tritiated 14-desmethyl sterol product were separated by HPLC on a Nova-Pak C18 column (30). Nonradioactive sterols were separated by HPLC on a 25-cm Zorbax C18 column (DuPont; 5 mm particle size, 4.6 mm i.d.) by elution with 100% methanol at room temperature (flow rate of 1 ml/min). TLC was performed on 250-μ silica gel G plates, which were developed twice with benzene/ether (85/15). GLC analysis was performed on a 3-foot spiral, 3% SE-30 packed column operated isothermally at 245°C. GLC-MS was performed on a Hewlett–Packard 5973 Mass Selective Detector interfaced with a 6890 GC system. The capillary column for GLC was a 30-m DB-5 column, 250 μM × 0.25 μM (from J&W Scientific, Folsom, CA). The temperature program was operated at: 170°C hold for 1 min; ramp at 20°C/min to 280°C; hold for 15 min. MS was performed by using MS transfer line at 280°C, with the inlet injector port kept at 250°C. The MS ion source temperature was maintained at 230°C. Helium gas, used as carrier, was maintained at a flow rate of 1.2 ml/min. 1H NMR spectroscopy was performed on samples dissolved in deuteriated chloroform at ambient temperature by using an AF-300 spectrometer (Bruker, Billeria, MA) with tetramethylsilane as internal standard (26, 27, 31, 32).
RESULTS AND DISCUSSION
Cloning, Expression, and Purification of MT P45014DM and MT Fdx.
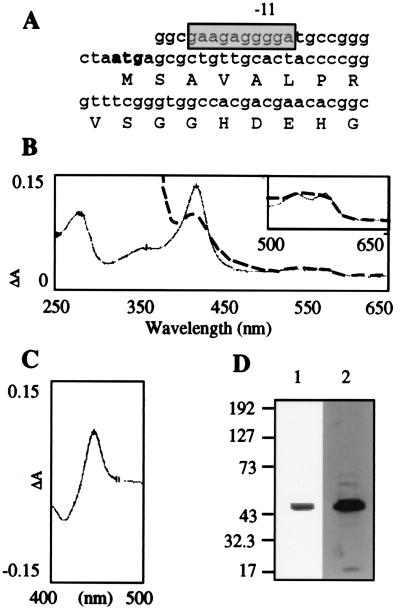
MTP450/pet17b and MTFdx/pet17b encode CYP51 and Fdx identical to the sequences deposited in GenBank (accession no. MTCY369). A purine-rich region, GAAGAGGGGA, located 10 bp upstream from the start codon is a potential Shine–Dalgarno sequence (Fig. 2A), and the length of the spacer between this and the start codon (10 bp) is similar to other mycobacterial genes (33). MT CYP51 produced in E. coli (2.5 μmol/liter) has a typical P450 reduced-CO spectrum (Fig. 2C), as observed previously (18). Cell fractionation reveals that MT P45014DM is soluble, with no P450 detected in the membranes as seen upon expression in JM109 E. coli strain (18). After two consecutive Ni2+ affinity column-purification steps, the specific content of the MT P45014DM is about 18 nmol/mg, and a single band is observed on SDS/PAGE at about 50 kDa, with the predicted molecular mass from the sequence being 51.4 kDa (Fig. 2D). The oxidized absolute spectrum of the purified enzyme, in the absence of substrate, showed a Soret band at 417 nm and α-, β-, and δ-bands at 569, 535, and 369 nm, which is typical for low-spin cytochrome P450 (Fig. 2B). Reduction by sodium hydrosulfite results in a Soret peak at 411 nm.
Figure 2.
(A) Potential Shine–Dalgarno sequence (shadowed box) of the MT CYP51 gene; the ATG is represented in boldface characters. (B) Absorbance of purified MT P45014DM (400 pmol), absolute oxidized form (regular trace), and sodium hydrosulfite reduced form (dashed trace). Inset shows the α- and β-bands for the oxidized and the reduced forms. (C) Differential CO-reduced P450 spectrum of purified MT P45014DM (400 pmol). (D) Silver staining (1) and immunoblot analysis (2) using 1 pmol and 0.4 pmol of purified MT P45014DM, respectively. MT P45014DM antibody prepared with TiterMax@Gold as adjuvant was used at a 1:5,000 dilution. Protein G-horseradish peroxidase conjugate (Bio-Rad) was used as a second antibody, and an enhanced chemiluminescence kit was used for detection.
Sterol 14α-Demethylase Reconstitution and Activity.
To investigate MT P45014DM enzymatic activity, it was first necessary to determine which electron donor can reduce the hemoprotein. In eukaryotes, P450s are localized in either the endoplasmic reticulum and reduced by ubiquitous NADPH cytochrome P450 reductase (34) or in the inner mitochondrial membrane and reduced by a two-component system of a flavoprotein reductase and a 2Fe-2S protein (35). Neither reducing system was capable of reducing MT P45014DM (Table 1). Nonetheless, we found that it can be reduced by a two-component system, Fld/Fdr from E. coli. Fld/Fdr reduces MT P45014DM at about 20% of the full reduction by sodium hydrosulfite when using the P450/Fld/Fdr ratio of 1:5:1, which reduces bovine P450c17 at 77% (Table 1).
Table 1.
CO-based reduction of purified recombinant P450s by adrenodoxin/adrenodoxin reductase, rat P450 reductase, and E. coli Fld/Fdr
| Donor | SCC | bC17 | MT |
|---|---|---|---|
| Adx/Adr | 80% | — | NR |
| Rat P450 reductase | — | 73% | NR |
| Fld/Fdr | — | 77% | 20% |
Reduction is compared with 100%, determined by sodium hydrosulfite. Adx/Adr, adrenodoxin/adrenodoxin reductase; SCC, cholesterol side cleavage P450; bC17, bovine 17α-hydroxylase; MT, mycobacterial 14α-demethylase; NR, no reduction.
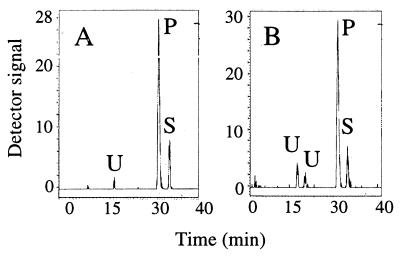
Preliminary studies on metabolism of [24-3H]DHL by MT P45014DM suggested the reconstituted enzyme by using Fld/Fdr-catalyzed 14α-demethylation. Cloning Fdx from MT showed that this 3Fe-4S ferredoxin was able to support the MT P45014DM activity to a level similar to E. coli Fld/Fdr when spinach Fnr was used as an Fdx electron donor (Fig. 3). To characterize the product, scaled-up batch enzyme experiments using Fld/Fdr as electron donor and three, different, nonradioactive sterol substrates—lanosterol, DHL, and obtusifoliol—were carried out overnight with 50 μM sterol and 365 pmol enzyme per assay. Total product recovered from the quenched reaction mixtures was 1% for lanosterol, 20% for DHL (Fig. 1C), and 98% for obtusifoliol. From incubation with lanosterol, a single sterol product was detected on TLC at Rf = 0.5 (the Rf value distinguishes whether C-demethylation occurs at C4, Rf of 0. 43, or C14, Rf of 0.50), by GC [3% Se-30: retention time relative to cholesterol (RRTc), 1.62], MS (M+, 410, and related diagnostic ions at m/z 395, 392, 377, 357, and 328), and UV (in ethanol) at λmax = 248 nm for a 8,14-diene (36). From incubation with DHL, a single sterol product was detected and identified: TLC, Rf = 0.5; GLC, RRTc = 1.53; MS, M+ = 412 (and related ions at 397, 394, 279, 351, 312, 285, 266, 245, 227, and 159); UV (in ethanol), λmax = 248 nm; 1H NMR analysis of the sample exhibited four singlets and three doublets in the methyl region of the spectrum between δ 0.76 and 1.01 ppm, consistent with loss of a methyl group from C14 and a single chemical shift at δ 5.34 ppm in the olefinic region corresponding to the Δ14(15)-bond. These structural assignments indicate a 4,4-dimethyl Δ8,14(15)-sterol (27). From incubation with obtusifoliol, a single sterol product was detected and identified: TLC, Rf = 0.43 (characteristic migration on TLC for a C4-monomethyl sterol); GLC, RRTc = 1.55; MS, M+ = 410 (and related ions at m/z 395, 392, 379, 357, 328, 267, 247, 227, and 189); and UV (in ethanol), λmax = 248 nm.
Figure 3.
Comparison of MT P45014DM activities supported by either Fld/Fdr or Fdx/Fnr. [24-3H]DHL was converted overnight at 30°C by 1 nmol MT P45014DM with either 20 nmol Fld and 2 nmol Fdr (A) or 20 nmol Fdx and 2 nmol Fnr (B) (30). Peaks S and P correspond to DHL and its 14α-demethylated product, respectively. Peaks U are unidentified products. MT P45014DM used in this experiment was purified further by HLPC (BIOCAD/Sprint, Perspective Biosystems, Framingham, MA) by using Poros HS and HQ columns (Perspective Biosystems). The HS flowthrough is loaded on an HQ column and eluted by using a NaCl gradient (150–500 mM).
Substrate Binding.
In the absence of substrates, most P450 enzymes are low-spin (37). Substrate addition shifts the heme to the high-spin state. For MT P45014DM, as for most P450s, the change in spin state leads to a peak at 390 nm and a trough at 420 nm in the substrate-induced difference spectrum (Fig. 4A). The amplitude of this difference is proportional to the P450-substrate complex. Addition of increasing amounts of substrate permits estimation of a binding constant similar to the Ks value. Binding constants were determined for obtusifoliol, lanosterol, and DHL. Obtusifoliol binds to the enzyme with a Ks value of 350 ± 150 nM, whereas DHL and lanosterol bind to the enzyme less effectively, ca. 1 ± 0.5 μM each (Fig. 4B). Neither parkeol, cycloartenol, nor zymosterol (Fig. 1A) were found to bind to the enzyme.
Figure 4.
(A) MT P45014DM type I binding spectrum for obtusifoliol (100 nM–5 μM). (B) Double reciprocal plot for obtusifoliol (●), DHL (▴), and lanosterol (■) binding with 10 μM MT P45014DM. (C) MT P45014DM type II binding spectrum in the presence of clotrimazole (500 nM–100 μM). (D) Double reciprocal plot for clotrimazole (○), ketoconazole (●), and fluconazole (▴) binding with 5 μM MT P45014DM.
Azole-Binding Spectra.
Binding of ketoconazole, clotrimazole, and fluconazole, known for their ability to inhibit 14α-demethylase activities (38, 39), was examined for MT P45014DM. These molecules produce type II binding spectra as a result of binding of the azole nitrogen to the sixth coordination position of the heme iron. The type II binding spectrum is characterized by a peak at 434 and a trough at 412 nm (Fig. 4C). Similar to the type I spectra, the P450-inhibitor complex can be titrated leading to an estimation of the inhibitor Ks (Fig. 4D). For ketoconazole and clotrimazole, these values are around 5 μM, whereas for fluconazole, the value is around 10 μM. Ketoconazole (20 μM) was found to inhibit the 14α-demethylation of DHL by MT P45014DM (not shown).
Expression of P45014DM in M. tuberculosis.
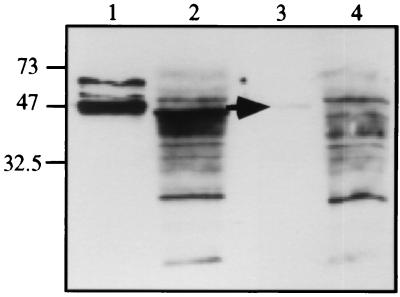
Immunoblot analysis was carried out on the cytosolic fraction of the MT virulent strain H37Rv (Fig. 5). A band near the expected molecular weight but lower than for purified recombinant MT P45014DM was obtained. Using antiserum depleted with an excess of purified recombinant MT P45014DM, this cytosolic band as well as that for purified P45014DM was reduced dramatically. This suggests that the MT P45014DM is expressed in MT. The difference in size between the recombinant and MT proteins might be explained by the presence of four additional histidines at the C terminus of the recombinant enzyme. Such modification can affect the mobility of proteins during SDS/PAGE. In fact, a single amino acid mutation in P4501B1 expressed in E. coli results in a shift to a greater size on the SDS-polyacrylamide gel (41).
Figure 5.
Western blot analysis of 0.4 pmol purified recombinant MT P45014DM (lanes 1 and 3) and 100 μg of MT cytosolic fraction (lanes 2 and 4) by using complete (lanes 1 and 2) and depleted (lanes 3 and 4) antisera. The antiserum raised by using Freund’s adjuvant was purified using an MT P45014DM Sepharose affinity column followed by batch chromatography with the same resin (40). The antiserum was depleted by overnight incubation with 6 nmol purified MT P45014DM at 4°C.
CONCLUSIONS
Results of this study demonstrate that MT contains a gene encoding an enzyme that catalyzes removal of the sterol 14α-methyl group stereoselectively, producing the 8,14-diene. The influence of substrate structure on MT P45014DM sterol binding has been determined by using a series of substrates that differ in a single molecular feature or in a combination of similar features. The tendency for preferential binding of obtusifoliol compared with the five other sterols tested indicates that the active site accommodates sterol side chains with a C24-alkyl group, suggesting the bacterial enzyme is plant/fungal-like in its active-site topology. Obtusifoliol also was found to be the best substrate for the MT P45014DM. The inability of parkeol or cycloartenol, structural isomers of lanosterol, to bind MT P45014DM indicates that the orientation of the substrate assumed upon binding requires a specific pseudoplanar conformation of the ring system and a specific, equatorially oriented tilt of the C3-hydroxyl group—analogous structural requirements as observed for the sterol methyl transferase enzyme from fungi and plants (26, 42). The lack of zymosterol binding (4,4,14-tridesmethyl lanosterol) indicates that one or both of the C4- and C14-methyl groups are important in sterol binding. The Δ8-bond is a critical stereoelectronic element of recognition; in each of the three sterols that were found to undergo 14α-demethylation by MT P45014DM, the product of the multistep reaction was a sterol with the conjugated Δ14(15)-bond system, suggesting the bacterial enzyme has evolved to bind and catalyze 14α-methyl sterols in a manner similar to P45014DM enzymes from higher species (17). Clearly, there is a conservation in sterol specificity for the P45014DM enzyme from primitive bacteria to advanced fungal and plant systems. The ketoconazole-binding constant estimated for maize microsomes is 10 μM (39), about the same as that for MT P45014DM, emphasizing similarities between bacterial and eukaryote enzymes.
A purine-rich region located 10 bp upstream of the ATG is associated with the MT CYP51 gene. Similar sequences are associated with other MT genes such as TB dnaj and TB 65 (33). The structure and the location of this putative MT CYP51 Shine–Dalgarno sequence are also in agreement with what is known in the most studied bacterium, E. coli, where the purine-rich region is separated from the ATG by 5–12 bases (43). No such sequence could be identified upstream of the Fdx gene in the P450 ORF, suggesting that the two genes that are separated by only 2 bp might be expressed as a polycistronic RNA.
MT P45014DM is an endogenous P450 that has been found to accept electrons from both an iron–sulfur protein (Fdx) and an FMN-containing protein (Fld). Perhaps this reflects a transition in the P450 evolution between prokaryotic electron transfer (iron–sulfur protein) and the eukaryotic type (FMN-containing protein for microsomal P450s). The endogenous MT reductase remains to be established, although the 3Fe-4S ferredoxin seems a good candidate. The formulation of binding topology from studies with sterol substrates and the sensitivity of the P45014DM to azole inhibitors is consistent with MT having a functional sterol pathway and further refines the general picture of sterol evolution that has emerged from classical natural product chemistry approaches to identify sterol biosynthetic pathways. The identification of P45014DM in MT and its possible role in sterol biosynthesis support the recent demonstration that cholesterol biosynthesis occurs in M. smegmatis via a mevalonic pathway (14). Targeting MT P45014DM opens a possibility in drug design for new treatments of tuberculosis, a disease infecting one-third of the world population (44).
Acknowledgments
We thank Dr. Chris Jenkins for supplying purified E. coli Fld, Fdr, rat NADPH cytochrome P450 reductase, and bovine P45017α-hydroxylase; Dr. Irina Pikuleva for bovine adrenodoxin, adrenodoxin reductase, and P450scc; Dr. Norio Kagawa for helping with the antibody preparation; Dr. Heather DeHart for helping with the Fdx gene cloning; Dr. John Biliesle from Colorado State University for providing genomic DNA and cytosolic fractions of MT strain H37Rv; and Dr. James Trzaskos for providing tritiated DHL. This work was supported by Grants GM37942 and ES 00267-32 from the National Institutes of Health to M.R.W. and Grant D1276 from the Welch Foundation to W.D.N.
ABBREVIATIONS
- MT
Mycobacterium tuberculosis
- CYP51
sterol 14α-demethylase cytochrome P450 gene
- P45014DM
14α-demethylase cytochrome P450
- Fld
E. coli flavodoxin
- Fdr
E. coli flavodoxin reductase
- Fdx
MT ferredoxin
- Fnr
spinach ferredoxin reductase
- DHL
24,25-dihydrolanosterol
References
- 1.Nes W R, Nes W D. Lipids in Evolution. New York: Plenum; 1980. [Google Scholar]
- 2.Nes W D, Norton R A, Crumley F G, Madigan S J, Katz E R. Proc Natl Acad Sci USA. 1990;87:7565–7569. doi: 10.1073/pnas.87.19.7565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burrow W, Moudler J W, Lewert R M. Text Book of Microbiology. 18th Ed. Philadelphia: Sanders; 1966. p. 748. [Google Scholar]
- 4.Nes W R. Lipids. 1974;9:596–612. doi: 10.1007/BF02532509. [DOI] [PubMed] [Google Scholar]
- 5.Ourisson G, Rohmer M, Poralla K. Annu Rev Microbiol. 1987;42:301–333. doi: 10.1146/annurev.mi.41.100187.001505. [DOI] [PubMed] [Google Scholar]
- 6.Ramgopal M, Bloch K. Proc Natl Acad Sci USA. 1983;80:712–715. doi: 10.1073/pnas.80.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pinto W J, Lozano R, Sekula B C, Nes W R. Biochem Biophys Res Commun. 1983;112:47–54. doi: 10.1016/0006-291x(83)91795-3. [DOI] [PubMed] [Google Scholar]
- 8.Nes W D. ACS Symp Ser. 1987;325:304–328. [Google Scholar]
- 9.Nes W D. Recent Adv Phytochem. 1990;24:283–327. [Google Scholar]
- 10.Bird C W, Lynch J M, Pirt F J, Reid W W. Nature (London) 1971;230:473–474. doi: 10.1038/230473a0. [DOI] [PubMed] [Google Scholar]
- 11.Kohl W, Gloe A, Reichembach H. J Gen Microbiol. 1983;129:1629–1636. [Google Scholar]
- 12.Lois L M, Campos N, Surya R P, Danielsen K, Rohmer M, Boronat A. Proc Natl Acad Sci USA. 1998;95:2105–2110. doi: 10.1073/pnas.95.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takahashi S, Kuzuyama T, Watanabe H, Seto H. Proc Natl Acad Sci USA. 1998;95:9879–9884. doi: 10.1073/pnas.95.17.9879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lamb D C, Kelly D E, Manning N J, Kelly S L. FEBS Lett. 1998;437:142–144. doi: 10.1016/s0014-5793(98)01218-6. [DOI] [PubMed] [Google Scholar]
- 15.Philipp W, J, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom B R, Jacobs W R, Jr, Cole S T. Proc Natl Acad Sci USA. 1996;93:3132–3137. doi: 10.1073/pnas.93.7.3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, III, et al. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida Y, Noshiro M, Aoyama Y, Kawamoto T, Horiuchi T, Gotoh O. J Biochem (Tokyo) 1997;122:1122–1128. doi: 10.1093/oxfordjournals.jbchem.a021870. [DOI] [PubMed] [Google Scholar]
- 18.Aoyama Y, Horiuchi T, Gotoh O, Noshiro M, Yoshida Y. J Biochem (Tokyo) 1998;124:694–696. doi: 10.1093/oxfordjournals.jbchem.a022167. [DOI] [PubMed] [Google Scholar]
- 19.Jenkins C M, Waterman M R. J Biol Chem. 1994;269:27401–27408. [PubMed] [Google Scholar]
- 20.Fischer R T, Stam S H, Johnson P R, Ko S S, Magolda R L, Gaylor J L, Trzaskos J M. J Lipid Res. 1989;30:1621–1632. [PubMed] [Google Scholar]
- 21.Pascal R A, Cheng P, Schroepfer G F. J Am Chem Soc. 1980;102:6599–6601. [Google Scholar]
- 22.Venkatramesh M, Nes W D. Arch Biochem Biophys. 1995;324:189–199. doi: 10.1006/abbi.1995.9912. [DOI] [PubMed] [Google Scholar]
- 23.Rahier A, Taton M. Biochem Soc Trans. 1990;18:52–56. doi: 10.1042/bst0180052. [DOI] [PubMed] [Google Scholar]
- 24.Yoshida Y, Aoyama Y. Biochem Soc Trans. 1991;19:778–782. doi: 10.1042/bst0190778. [DOI] [PubMed] [Google Scholar]
- 25.Omura T, Sato R. J Biol Chem. 1964;239:2379–2385. [PubMed] [Google Scholar]
- 26.Venkatramesh M, Guo D A, Jia Z, Nes W D. Biochim Biophys Acta. 1996;1299:313–324. doi: 10.1016/0005-2760(95)00218-9. [DOI] [PubMed] [Google Scholar]
- 27.Nes W D, Janssen G G, Crumley F G, Kalinowska M, Akihisa T. Arch Biochem Biophys. 1993;300:724–733. doi: 10.1006/abbi.1993.1100. [DOI] [PubMed] [Google Scholar]
- 28.O’Keefe D P, Gibson K J, Emptage M H, Lenstra R, Romesser J A, Litle P J, Omer C A. Biochemistry. 1991;30:447–455. doi: 10.1021/bi00216a021. [DOI] [PubMed] [Google Scholar]
- 29.Bauer S, Shiloach J. Biotechnol Bioeng. 1974;16:933–941. doi: 10.1002/bit.260160707. [DOI] [PubMed] [Google Scholar]
- 30.Stromstedt M, Rozman D, Waterman M R. Arch Biochem Biophys. 1996;329:73–81. doi: 10.1006/abbi.1996.0193. [DOI] [PubMed] [Google Scholar]
- 31.Xu S H, Norton R A, Crumley F G, Nes W D. J Chromatogr. 1988;452:377–398. doi: 10.1016/s0021-9673(01)81462-x. [DOI] [PubMed] [Google Scholar]
- 32.Nes W D, Koike K, Jia Z, Sakamoto Y, Satou T, Nikaido T, Griffin J F. J Am Chem Soc. 1998;120:5970–5980. [Google Scholar]
- 33.Dale J W, Patki A. In: Molecular Biology of the Mycobacteria. McFadden J, editor. San Diego: Academic; 1990. pp. 173–198. [Google Scholar]
- 34.Vermilion J L, Coon M J. J Biol Chem. 1978;253:8812–8819. [PubMed] [Google Scholar]
- 35.Coghlan V M, Vickery L E. J Biol Chem. 1991;266:18606–18612. [PubMed] [Google Scholar]
- 36.Goad L J, Akhisa T. Analysis of Sterols. New York: Blakie; 1997. [Google Scholar]
- 37.Guengerich F P. Biochemistry. 1983;22:2811–2820. doi: 10.1021/bi00281a007. [DOI] [PubMed] [Google Scholar]
- 38.Yoshida Y, Aoyama Y. Biochem Pharmacol. 1987;36:229–235. doi: 10.1016/0006-2952(87)90694-0. [DOI] [PubMed] [Google Scholar]
- 39.Salmon F, Taton M, Benveniste P, Rahier A. Arch Biochem Biophys. 1992;297:123–131. doi: 10.1016/0003-9861(92)90649-h. [DOI] [PubMed] [Google Scholar]
- 40.Gough N M, Adams J M. Biochemistry. 1978;17:5560–5566. doi: 10.1021/bi00618a036. [DOI] [PubMed] [Google Scholar]
- 41.Shimada T, Wunsch R M, Hanna I, Sutter T R, Guengerich F P, Gillam E M J. Arch Biochem Biophys. 1998;357:111–120. doi: 10.1006/abbi.1998.0808. [DOI] [PubMed] [Google Scholar]
- 42.Nes W D, Janssen G G, Bergenstrahle A. J Biol Chem. 1991;266:15202–15212. [PubMed] [Google Scholar]
- 43.De Boer H A, Hui A. In: Gene Expression Technology. Goeddel D V, editor. Vol. 185. San Diego: Academic; 1991. pp. 103–114. [Google Scholar]
- 44.Fenton M J, Vermeulen M W. Infect Immunol. 1996;64:683–690. doi: 10.1128/iai.64.3.683-690.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]