Abstract
Mutations in the photoreceptor membrane guanylyl cyclase RetGC-1 have been linked to autosomal dominant cone–rod dystrophy. Three mutations were identified that alter strictly conserved residues within the RetGC-1 dimerization domain, a region predicted to form an amphipathic α-helical coil. Here we report on a biochemical characterization of one of the mutations, a substitution of cysteine for arginine at residue 838. We generated this mutation in vitro and measured its catalytic activity and sensitivity to guanylyl cyclase activating protein 1 (GCAP-1) and GCAP-2. The R838C substitution has several effects. It reduces the overall catalytic ability of RetGC-1 and dramatically reduces stimulation by GCAP-2, although GCAP-2 still appears to interact with the protein. The R838C substitution also increases the apparent affinity of RetGC-1 for GCAP-1 and alters the Ca2+ sensitivity of the GCAP-1 response, allowing the mutant to be stimulated by GCAP-1 at higher Ca2+ concentrations than wild type. The diminished response to GCAP-2, which we propose is not likely the cause of cone–rod degeneration in these patients, is interesting mechanistically because it separates the ability to bind a specific GCAP from the ability to be stimulated by it, and it also discriminates between the mechanisms of activation of GCAP-1 vs. GCAP-2. We suggest that the gain-of-function effects of R838C on RetGC-1 stimulated by GCAP-1, which are dominant in vitro and may cause an abnormal increase in cGMP synthesis in dark-adapted photoreceptors, may be the cause of the cone–rod degeneration.
Two membrane guanylyl cyclases, RetGC-1 and RetGC-2, synthesize cGMP in mammalian photoreceptor cells. cGMP gates cation channels, which control the membrane potential and signaling states of rods and cones. Light stimulates degradation of cGMP, causing the cGMP-gated channels to close. This reduces intracellular Na+ and Ca2+ concentrations, hyperpolarizes the cell, and slows neurotransmitter release. Lowered Ca2+ levels allow the Ca2+-binding proteins guanylyl cyclase activating protein 1 (GCAP-1) and GCAP-2 to stimulate RetGCs. Acceleration of cGMP synthesis reopens channels and restores photosensitivity to the photoreceptor cell.
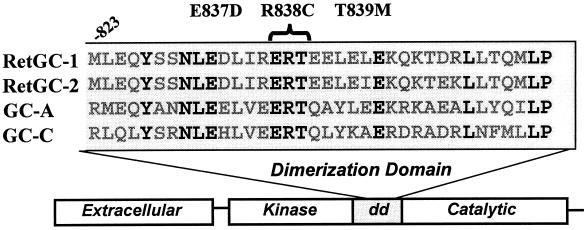
Recently, defects in the RetGC-1 gene were identified in patients with dominant cone–rod dystrophy, an autosomal-dominant disease causing initial degeneration of cones followed by loss of rod photoreceptors (1, 2). Cone degeneration causes an early loss of visual acuity and color vision that, as rods degenerate, leads to progressive night blindness and peripheral visual-field loss (3). Three mutations have been described: two heterozygous, single missense mutations, E837D and R838C (1), and a heterozygous, triple mutation, E837D; R838C; T839M (2). In all three cases, the substitutions are in conserved amino acids in the putative dimerization domain of RetGC-1 (Fig. 1).
Figure 1.
Location of the dominant cone–rod dystrophy amino acid changes in the dimerization domain (dd) of RetGC-1. A comparison of the sequences of several membrane GCs is also shown, including RetGC-2 (GC-F), the atrial natriuretic peptide receptor (GC-A), and the heat-stable enterotoxin receptor (GC-C). The amino acid residues in boldface are conserved among these membrane GCs.
To better understand the connection between mutations in the RetGC-1 dimerization domain and dominant cone–rod dystrophy, we generated and expressed the single R838C and E837D mutations in vitro. E837D is a conservative substitution and was predicted to have little effect on activity. Our preliminary work showing this mutant indeed has a mild phenotype prompted us to rescreen the original patient DNA for an additional mutation. This study, including the biochemical characterization of E837D, will be detailed in a separate paper. In this report, we characterize the biochemical effects of the R838C substitution, which dramatically alters the responses of RetGC-1 to GCAP-1 and GCAP-2.
MATERIALS AND METHODS
Mutagenesis.
The R838C point mutation was generated by using the Altered Sites Kit (Promega). A 1.6-kb KpnI/SphI cassette containing the codon to be mutated was subcloned from pBluescript-RetGC1 into the KpnI/SphI sites of pALTER1. Mutagenesis was performed with the 18-mer primer ATCCGGAGTGCACGGAGG. After verification by DNA sequencing, the plasmid was digested with AccI and AatII to yield a 586-bp fragment. This fragment was used to replace the equivalent fragment in the original pBluescript-RetGC1 plasmid. After sequencing to confirm the insertion of the mutagenized fragment, the mutant RetGC-1 cDNA (3.6 kb) was excised with HindIII/XbaI and subcloned into the eukaryotic expression vector pRC-CMV (Invitrogen).
Expression in Human Embryonic Kidney (HEK) 293 Cells.
Constructs were transiently transfected (calcium phosphate method) into HEK 293 cells. Cells were harvested after 48 hr, washed with PBS, and removed from dishes by agitation in PBS with 0.2% EGTA. Cells were pelleted gently by centrifugation, and pellets were swollen in homogenization buffer (10 mM Tris, pH 7.5/5 mM MgCl2/5 mM 2-mercaptoethanol) for 10 min. Four strokes of a 26-gauge needle then were used to lyse cells. Cell lysates were pelleted at 2,000 rpm at 4°C in a tabletop microcentrifuge to remove large debris. The supernatant from this spin was pelleted at 14,000 rpm, resuspended in homogenization buffer, and then frozen at –70°C in aliquots for use during assays.
Western Blotting.
Total membrane proteins from transfected HEK 293 cells were electrophoresed on a 7.5% SDS/PAGE gel and transferred to nitrocellulose membranes. Westerns were performed by using a polyclonal antibody that recognizes the kinase homology domain of RetGC-1 (4).
GC Assays.
Transiently transfected membranes containing equal amounts of total protein were resuspended in GC buffer (100 mM KCl/50 mM Mops/7 mM 2-mercaptoethanol/10 mM MgCl2/8 mM NaCl/1 mM EGTA). All reactions also contained 0.5 mM ATP. Measurement of guanylyl cyclase activity was carried out at 30°C for the indicated times essentially as described previously (5). Stimulated reactions contained recombinant myristoylated bovine GCAP-1 or GCAP-2 that was prepared as described by Olshevskaya et al. (6), except Fig. 5B, which used human GCAP-1 (provided by Krzysztof Palczewski, University of Washington). Assays measuring Mn2+/Triton X-100 activity contained 1% Triton X-100 and 10 mM MnCl2 instead of MgCl2. All experiments shown were repeated several times with similar results.
Figure 5.
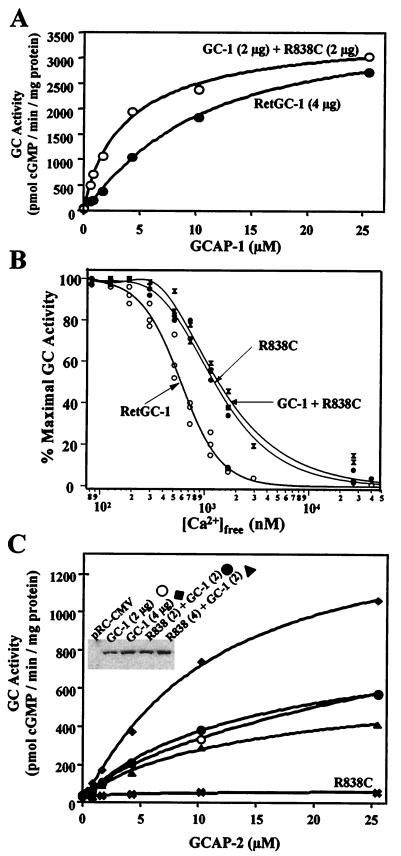
Coexpression of R838C with wild-type RetGC-1. HEK 293 cell membranes were transiently transfected with wild-type RetGC-1, R838C, or a mixture of both wild-type and R838C. (A) Response to GCAP-1 was measured from cell membranes transfected with 4 μg of wild-type RetGC-1 (●) or 2 μg of RetGC-1 and 2 μg of R838C (1:1 ratio) (○). Immunoblots of these membranes are shown in C Inset. Similar results were obtained from independent experiments with membranes transfected at the same ratios. (B) Ca2+ titration of wild-type RetGC-1 (○), R838C (●), and R838C coexpressed with wild-type RetGC-1 (X’s) stimulated by 8 μg of human GCAP-1. Wild-type and coexpressed membranes are the same as used in A. The half-maximal [Ca2+] for inhibition was 590 nM for wild-type RetGC-1, 1,230 nM for R838C, and 1,350 for coexpressed membranes. (C) Measurement of GCAP-2 responses in cotransfected membranes. Membranes from cells transfected with 2 μg of RetGC-1 (○), 4 μg of RetGC-1 (♦), 2 μg of RetGC-1 and 2 μg of R838C (●), or 2 μg of RetGC-1 and 4 μg of R838C (▴) were assayed for GCAP-2 stimulated activity. Inset shows an immunoblot of equivalent amounts of total membrane protein.
Ca2+ Buffers.
Ca2+-EGTA buffers were prepared from solutions of EGTA and EGTA saturated with CaCl2 by pH titration in accordance with the method of Tsien and Pozzan (7). Free Ca2+ concentrations were calculated by using a multifactor program (8).
Model for Regulation of RetGC by GCAP.
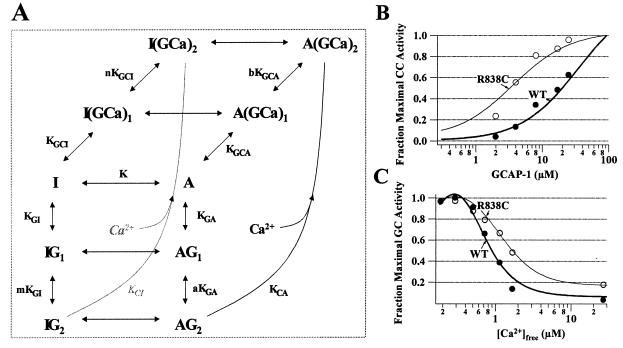
The fraction of RetGC in the active state was calculated based on equilibrium between the states shown in Fig. 8A. “A” represents the active state of the cyclase and “I” represents the inactive form. “G” represents GCAP, and “GCa” represents the fully Ca2+-liganded form of GCAP. The probability of being in the “A” state under any given set of conditions was calculated by using the formula: PA = [A]/[A] + [I] (Eq. 1), where [A] = 1 + 2[G]/KDA + (1/a) × (([G]/KDA)2) + 2((b/a)0.5)(([G][Ca])/(KDAKCA) + (1/a)(([G][Ca])/(KDAKCA))2) and [I] = K(1 + 2[G]/KDI + (1/m) × (([G]/KDI)2) + 2((n/m)0.5)(([G][Ca])/(KDIKCI) + (1/m)(([G][Ca])/(KDIKCI))2). The concentration of GCAP in the unliganded form was estimated to be: [G] = GT/(1 + (([Ca]/KGC)D), where GT is the total GCAP concentration, KGC is the dissociation constant for Ca2+ binding to GCAP, and D is a cooperativity factor.
Figure 8.
(A) Equilibrium model for regulation of RetGC by GCAP and Ca2+, as described in Discussion and Materials and Methods. For B and C, the probability of the cyclase being in the active “A” state was calculated by using Eq. 1 (described in Materials and Methods) as a function of GCAP concentration (B) and as a function of Ca2+ concentration (C). The data points shown are taken from the GCAP-1 titration in Fig. 3. The thick lines represent the calculated probability of being in the active state fit to the normal RetGC-1 data set. The values for the equilibrium constants and cooperativity factors used for that fit were: KDA = 1, KDI = .1, KCA = 3.6, KCI = 0.02, a = .025, b = 5 × 105, m = 5 × 105, n = 1, K = 70. The constants KGC and D defined in Materials and Methods were 0.044 and 2.6, respectively. The thin lines represent a fit to the R838C data made by keeping all the same values used for the normal RetGC-1 data except K, which was changed from 70 to 7 to simulate a mutation that makes the transition to the active state more favorable. The total GCAP concentration, GT, used in this calculation was 10 μM.
RESULTS
The cone–rod dystrophy mutations are in strictly conserved residues of the putative dimerization domain of RetGC-1. This domain, which corresponds to amino acids 817–857, lies between the kinase homology domain and the catalytic domain of RetGC-1. It is considered to promote dimerization because the corresponding domain of GC-A self-associates in a yeast two-hybrid system (9) and because it is predicted to form an amphipathic α-helical coil. We generated the R838C mutation, a substitution of Cys for the basic amino acid Arg, in the expression plasmid pRC-CMV-RetGC-1 and expressed the mutant and wild-type RetGC-1 proteins in HEK 293 cell membranes. The level of expression of R838C was equivalent to wild-type RetGC-1 (Fig. 2 Inset).
Figure 2.
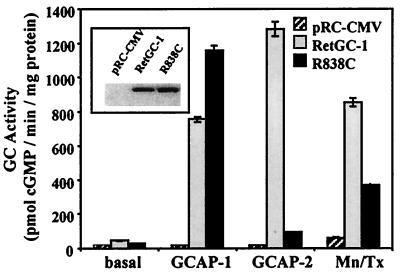
Measurement of R838C basal and stimulated activities. Constructs were transiently transfected in HEK 293 cells, and harvested membranes were immunoblotted with an antibody to the RetGC-1 kinase homology domain (Inset). The control lane labeled pRC-CMV contained membranes transfected with the plasmid pRC-CMV with no insert. Equal amounts of membrane protein were assayed for 20 min for basal-, GCAP-1-, GCAP-2-, or Mn2+/Triton X-100-stimulated GC activities. For GCAP-stimulated samples, 3.4 μM GCAP-1 or GCAP-2 and 1 mM EGTA were added. Mn/Tx-labeled samples contained 1% Triton X-100 and 10 mM MnCl2 instead of MgCl2.
The R838C Substitution Alters RetGC-1 Activity.
To examine the catalytic ability of R838C, we assayed basal activity and stimulation by Mn2+/Triton X-100, which constitutively activates membrane GCs and is used to quantitate general catalytic ability (10). Fig. 2 shows that R838C has reduced basal and Mn2+/Triton X-100 activities (2.3-fold reduction) compared with wild type.
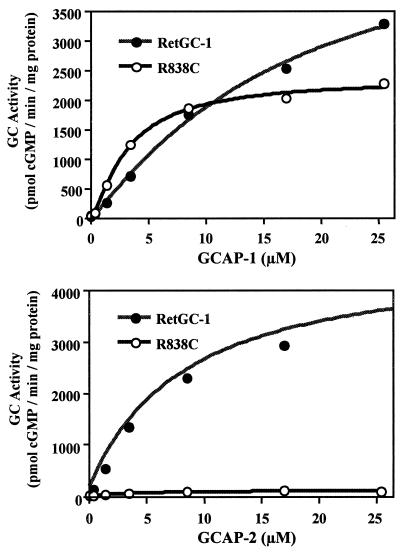
In the retina, RetGCs are activated by GCAPs, EF-hand Ca2+-binding proteins that stimulate RetGCs at Ca2+ concentrations below ≈300 nM (11–14) and inhibit them at micromolar Ca2+ concentrations (15, 16). Two GCAP isoforms, GCAP-1 and GCAP-2, have been identified. Both activate RetGCs, although they share only 40% identity in amino acid sequence. Because it is not yet clear whether RetGC-1 is activated physiologically by GCAP-1, GCAP-2, or both, we assayed the ability of R838C to respond to GCAP-1 as well as GCAP-2. As shown in Fig. 2, the behavior of R838C in response to the different GCAPs is striking. R838C has enhanced activity in response to 3.4 μM GCAP-1, but has a severely reduced response to 3.4 μM GCAP-2 (14-fold reduction from wild type).
Titrations (Fig. 3) reveal that R838C, compared with wild-type RetGC-1, has a higher apparent affinity for GCAP-1. The K1/2 [GCAP-1] values were 3.2 ± 0.31 μM for R838C, compared with 16.8 ± 6.7 μM for wild-type RetGC-1. R838C saturates earlier than wild type when stimulated by GCAP-1 or GCAP-2. When stimulated with GCAP-2, R838C has a very reduced response (a 25-fold reduction in activity with 25 μM GCAP-2).
Figure 3.
GCAP-1 and GCAP-2 titrations of R838C and wild-type RetGC-1. GC assays were done as in Fig. 1 with indicated concentrations of GCAP-1 or GCAP-2. A curve was fit to each plot by using the Hill equation v = [GCAP]n Vmax/K1/2n + [GCAP]n. K1/2 [GCAP-1] values derived from the curve fit were 3.2 ± 0.31 μM for R838C, and 16.8 ± 6.7 μM for wild-type RetGC-1.
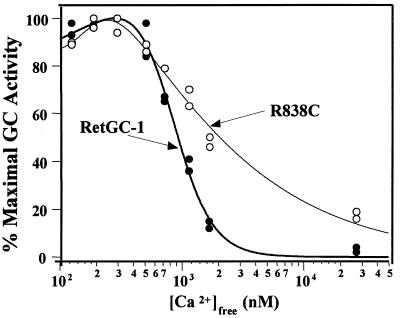
Recently, a GCAP-1 mutation, Y99C, was identified in patients with dominant cone dystrophy, a disease causing degeneration only in cones (17). The mutation alters the Ca2+ sensitivity of GCAP-1 so that it remains stimulatory at Ca2+ concentrations that would be inhibitory for wild type (18, 19). The consequent increase in cGMP in dark-adapted photoreceptors has been proposed as the cause of retinal degeneration in cone dystrophy patients. With the Y99C GCAP-1 mutation in mind, we examined the effect of the R838C mutation on the Ca2+ sensitivity of cGMP synthesis. We find that the R838C mutation alters the Ca2+ sensitivity of RetGC-1 activated by GCAP-1 (Fig. 4). Below 500 nM Ca2+, R838C behaves like wild-type RetGC-1. However, significantly higher than normal concentrations of Ca2+ are required for suppression of GCAP-1-catalyzed cGMP synthesis.
Figure 4.
Ca2+ sensitivity of R838C to GCAP-1 is altered. GC activity was measured in transiently transfected membranes expressing R838C (○) or wild-type RetGC-1 (●) stimulated with 8 μM bovine GCAP-1.
Coexpression of R838C with Wild-Type RetGC-1.
To determine how R838C could cause a dominant phenotype in patients who also have a normal RetGC-1 allele, we coexpressed R838C and wild-type RetGC-1 in HEK 293 cell membranes. The total amount of DNA transfected was kept constant by the addition of pRC-CMV to a total of 15 μg. Western blots show that the total amount of R838C or RetGC-1 DNA transfected correlates with the amount of protein expressed and used in the assays (Fig. 5 C Inset). We first examined the GCAP-1 sensitivity of R838C coexpressed with wild-type RetGC-1 at a 1:1 ratio (2 μg each) compared with RetGC-1 expressed alone (4 μg). We predicted that the increase in GCAP-1 affinity, a gain of function, would be dominant in our in vitro assay. Fig. 5A shows that, as predicted, the coexpressed membranes are more sensitive to GCAP-1 than wild type, which is similar to the results with R838C expressed alone.
We also analyzed the Ca2+ sensitivity of the GCAP-1 response in coexpressed membranes (Fig. 5B). The previous Ca2+ titrations shown in Fig. 4 were done by using bovine GCAP-1. To confirm that a similar change in Ca2+ sensitivity occurs when R838C is stimulated with human GCAP-1, we repeated the titrations shown in Fig. 4. We saw a similar change in Ca2+ sensitivity with human GCAP-1 as we had seen previously with bovine. We then tested the Ca2+ sensitivity of the coexpressed membranes. The coexpressed membranes gave a similar Ca2+ shift as R838C expressed alone, indicating that the Ca2+ shift induced by R838C is dominant in the presence of wild-type RetGC-1.
In contrast to the dominant effects of R838C observed with GCAP-1, we were unable to detect a dominant effect with regard to GCAP-2 stimulation. As shown in Fig. 5C, GC activity stimulated by GCAP-2 depends on the amount of wild-type RetGC-1 in the coexpressed membranes, but not on the amount of R838C. Membranes transfected with 2 μg of RetGC-1 and either 0, 2, or 4 μg of R838C gave similar responses to GCAP-2. In contrast, membranes transfected with 4 μg of RetGC-1 had twice the amount of GCAP-2-stimulated GC activity.
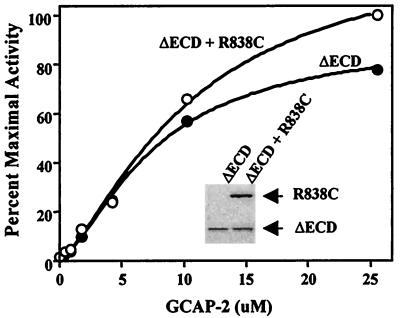
In the coexpression experiments reported in Fig. 5C, we were unable to determine the precise ratio of wild-type and mutant RetGC-1 expressed because the proteins colocalize on immunoblots. To verify the GCAP-2 results and visually discriminate between the two proteins, we repeated the coexpression experiments by using a mutant RetGC-1, ΔECD RetGC-1, which lacks the extracellular domain but behaves similarly to wild type in its responses to GCAPs (5). We transfected cells with ΔECD RetGC-1 or with ΔECD RetGC-1 and R838C and performed GC assays on membranes containing equivalent amounts of ΔECD RetGC-1 (see Fig. 6 Inset). Consistent with the results shown in Fig. 5C, the response to GCAP-2 depended on the amount of wild-type ΔECD RetGC-1, but not on the presence of R838C (Fig. 6). From these experiments, it appears that the R838C mutation does not have a dominant-negative effect with regard to GCAP-2 activation. We hypothesize that a person with one wild-type RetGC-1 allele and one R838C allele would have an approximately half-fold reduction in GCAP-2-stimulated GC activity.
Figure 6.
Coexpression of R838C and ΔECD RetGC-1. HEK 293 cells were transiently transfected with 7.5 μg of ΔECD RetGC-1 or R838C (5 μg) and ΔECD RetGC-1 (7.5 μg). Membrane proteins were immunoblotted with an antibody to the RetGC-1 kinase homology domain (Inset). Membranes containing equivalent amounts of ΔECD RetGC-1 were assayed for GC activity for 20 min with indicated concentrations of GCAP-2.
GCAP-2 Interacts with R838C but Cannot Activate It.
Evidence suggests that GCAPs bind to RetGCs independently of Ca2+ even though they activate RetGCs only at low Ca2+ concentrations. GCAP-1 and GCAP-2 inhibit RetGC (15, 16), protect RetGC from thermal inactivation (20), and protect RetGC from proteolysis (4), all at high Ca2+ conditions, where they cannot activate. Despite this evidence, no RetGC mutant has been reported that is known to bind but not be activated by a GCAP. Because the R838C mutant appeared to respond slightly (though not significantly) to GCAP-2, we considered that the defect would be in activation and not binding. Direct binding experiments of GCAP to RetGC have not yet been possible, so we used an indirect method: we investigated whether GCAP-2 can compete with and prevent activation by GCAP-1.
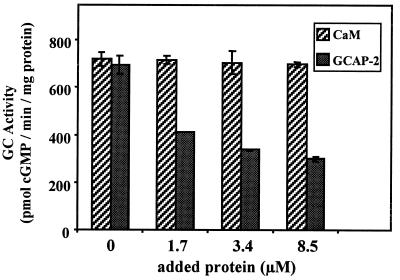
GCAP-2 was shown previously to compete with a constitutively active form of GCAP-1 at high Ca2+ concentrations (16), suggesting either that GCAP-1 and GCAP-2 bind to the same or overlapping sites on RetGC-1 or that activation of GCAP-1 and GCAP-2 are mutually exclusive. We tested whether GCAP-1-dependent activation of R838C could be blocked by increasing concentrations of GCAP-2 (Fig. 7). If GCAP-2 competes with GCAP-1, then a decrease in total activity would be observed because GCAP-2 does not activate R838C significantly itself. We stimulated R838C with 2.5 μM GCAP-1 and 0, 1.7, 3.4, or 8.5 μM of either calmodulin or GCAP-2. Addition of GCAP-2, but not calmodulin, reduced GCAP-1-dependent activation of R838C by as much as 2.5-fold, indicating that it competes effectively. The simplest interpretation of this result is that R838C is competent in binding GCAP-2, but is defective in being stimulated by the bound GCAP-2.
Figure 7.
GCAP-2 competes with GCAP-1 for activation of R838C. GC activity was measured in transiently transfected membranes expressing R838C stimulated with 2.5 μM GCAP-1 and indicated concentrations of calmodulin (CaM) or GCAP-2.
DISCUSSION
In this report, we characterize a mutation of RetGC-1 that was identified first in patients with dominant cone–rod dystrophy. The mutation, a substitution of Cys for Arg at residue 838 (R838C), is in a conserved amino acid in the putative RetGC-1 dimerization domain. We expressed this mutant protein in a heterologous system (HEK 293 cells) and assayed its abilities to synthesize cGMP and to be regulated. We find that R838C has reduced catalytic ability and responds poorly to GCAP-2, but its sensitivity to GCAP-1 is enhanced. We also find that R838C also has an altered response to GCAP-1/Ca2+, such that Ca2+-dependent turnoff of R838C is impaired.
How might the altered response of the R838C mutant cause dominant retinal degeneration? Because null alleles of RetGC-1 in several reported cases of Leber’s congenital amaurosis are deleterious only in homozygotes (21), it appears that even a half-fold reduction in the amount of functional RetGC-1 does not cause a significant visual defect, perhaps because of the presence of protein encoded by the wild-type allele or the other membrane cyclase, RetGC-2. The severely reduced response of R838C to GCAP-2, which is not dominant in the presence of wild-type RetGC-1, therefore should not, in itself, cause a dominant disease phenotype. On the other hand, we identified other properties of this mutant that cause a gain of function and are dominant in our in vitro system even in the presence of wild-type RetGC-1. R838C is less sensitive than normal to suppression of its activity by Ca2+/GCAP-1 and shows an increased sensitivity to activation by GCAP-1. If the localized concentration of GCAP-1 in the outer segments is limiting, the increase in sensitivity could accelerate synthesis of cGMP. The altered Ca2+ sensitivity of R838C could cause higher than normal rates of cGMP synthesis in dark-adapted photoreceptors. The consequent alteration in the balance between Ca2+ concentration, enzyme activity, and cGMP-gated channel activity may be the cause of the cone and rod degeneration. Although a GCAP-1-mediated cause of degeneration is plausible, other possible causes of the disease, such as a dominant effect of R838C on transport of RetGC-1 to photoreceptor outer segments, cannot be dismissed based only on our findings here. A R838C transgenic mouse model would be especially useful in understanding further the cause and progression of the disease.
The similarities between the R838C RetGC-1 mutation and the Y99C mutation of GCAP-1 associated with dominant cone dystrophy (18, 19) are intriguing. Both mutations cause a shift in GCAP-1 Ca2+ sensitivity so that Ca2+-dependent turnoff is hampered, which suggests the two diseases may have a similar molecular etiology. Both diseases prominently affect cones, which suggests that RetGC-1 and GCAP-1 may function more prominently in cones. Although the precise localization of RetGCs and GCAPs has been debated, a recent immunocytochemical study of GCAPs in human retinas reported that the relative ratios of GCAP-1 to GCAP-2 indeed were significantly higher in cones than rods (22).
All three of the identified RetGC-1 mutations associated with dominant cone–rod dystrophy are in conserved residues in the dimerization domain of RetGC-1. The role of this domain in membrane GC regulation is not fully understood. The R838C mutation does not appear to simply block dimerization, as originally suggested (1), because the catalytic domain of RetGC-1 appears to function as a homodimer (23, 24) and R838C is catalytically active. Furthermore, it is unlikely that abolishing dimerization in itself would produce a dominant effect. The mutation does alter the Ca2+ sensitivity and affinity of the GCAP-1/GC interaction. Essentially, R838C is easier to activate (via GCAP-1) and harder to turn off, consistent with a stabilization of the active conformation of RetGC-1.
To more fully dissect the molecular basis of the R838C substitution, we generated the model shown in Fig. 8A of the regulation of RetGC. “A” represents the catalytically active state of the cyclase, and “I” represents the inactive state. The fraction of cyclase molecules in the active state is determined by the dissociation constants, KDA, KDI, KCA, and KCI; the equilibrium constant K, which is equal to [I]/[A]; the cooperativity factors a, b, m, and n; and the concentrations of Ca2+ and GCAP-1 as described in Eq. 1 (see Materials and Methods). The darker lines in Fig. 8 B and C were calculated by using Eq. 1 and fit to our Ca2+ and GCAP titration data for wild-type RetGC-1 (solid circles). We used the model to investigate the effect of shifting the equilibrium to favor the active state of the molecule on the Ca2+ sensitivity and sensitivity to GCAP-1. The lighter lines in Fig. 8 B and C show the effects of shifting the equilibrium constant “K” by a factor of 10 to favor the active “A” state. The results fit well with the experimental data from the R838C mutation (open circles). A similar result also can be produced by decreasing the cooperativity factor, a, which represents the degree of cooperativity of GCAP binding. Both of these events, stabilization of the active state and enhanced cooperativity of GCAP binding, seem reasonable based on the nature and location of the R838C mutation in the dimerization domain of RetGC-1.
The R838C substitution is especially interesting mechanistically because it demonstrates differential activation by GCAPs and separates GCAP binding from activation. The two retinal GCAPs are 40% identical overall, but most of the homology resides in the central region of the molecules containing the Ca2+-binding EF-hands. The carboxyl and amino termini of GCAP-1 and GCAP-2 are quite different. Several groups have reported functional differences between GCAP-1 and GCAP-2. Myristoylation is important for GCAP-1 function, but not for GCAP-2 (6, 25). The Y99C mutation of GCAP-1 causes a shift in Ca2+ sensitivity (18, 19), but the corresponding mutation in GCAP-2 does not (19). The functional roles of the individual EF-hands of the GCAPs also appear to be different (15, 16). Recent chimeric analyses of GCAP-1 (26) and GCAP-2 (27) have pointed to the N and C termini as important for activation and portions of the core as important for inhibition/binding. Because the central region is somewhat conserved between GCAPs, it seems plausible that binding occurs at the same or overlapping sites on RetGC.
The only other reported RetGC-1 mutants showing differential activation by GCAPs were mutants of bovine RetGC-1 (ROS-GC) that were missing the kinase homology domain or that were substituted with the GC-A kinase homology domain (28). These responded poorly to GCAP-1, but retained GCAP-2 stimulation. It was not clear from that study whether the defect was in binding or activation. The R838C mutant should provide a useful tool for further analysis and dissection of the mechanisms of RetGC activation, the differences between activation by GCAP-1 or GCAP-2, and the different requirements for binding and activation.
Acknowledgments
We thank Bill Zagotta (University of Washington) for assistance in generating the model shown in Fig. 8. We also thank Krzysztof Palczewski (University of Washington, Department of Ophthalmology) for a gift of human GCAP-1. These studies were supported by grants from the National Institutes of Health (EY06641) to J.B.H. and the Wellcome Trust to D.M.H.
ABBREVIATIONS
- GC
guanylyl cyclase
- GCAP
GC activating protein
- HEK
human embryonic kidney
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Kelsell R E, Gregory-Evans K, Payne A M, Perrault I, Kaplan J, Yang R, Garbers D L, Bird A C, Moore A T, Hunt D M. Hum Mol Genet. 1998;7:1179–1184. doi: 10.1093/hmg/7.7.1179. [DOI] [PubMed] [Google Scholar]
- 2.Perrault I, Rozet J-M, Gerber S, Kelsell R E, Souied E, Cabot A, Hunt D M, Munnich A, Kaplan J. Am J Hum Genet. 1998;63:651–654. doi: 10.1086/301985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moore A T. J Med Genet. 1970;29:289–290. doi: 10.1136/jmg.29.5.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Laura R P, Hurley J B. Biochemistry. 1998;37:11264–11271. doi: 10.1021/bi9809674. [DOI] [PubMed] [Google Scholar]
- 5.Laura R P, Dizhoor A M, Hurley J B. J Biol Chem. 1996;271:11646–11651. doi: 10.1074/jbc.271.20.11646. [DOI] [PubMed] [Google Scholar]
- 6.Olshevskaya E V, Hughes R E, Hurley J B, Dizhoor A M. J Biol Chem. 1997;272:14327–14333. doi: 10.1074/jbc.272.22.14327. [DOI] [PubMed] [Google Scholar]
- 7.Tsien R, Pozzan T. Methods Enzymol. 1989;172:230–262. doi: 10.1016/s0076-6879(89)72017-6. [DOI] [PubMed] [Google Scholar]
- 8.Marks P W, Maxfield F R. Anal Biochem. 1991;193:61–71. doi: 10.1016/0003-2697(91)90044-t. [DOI] [PubMed] [Google Scholar]
- 9.Wilson E M, Chinkers M. Biochemistry. 1995;34:4696–4701. doi: 10.1021/bi00014a025. [DOI] [PubMed] [Google Scholar]
- 10.Potter L R, Garbers D L. J Biol Chem. 1992;267:14531–14534. [PubMed] [Google Scholar]
- 11.Gorczyca W A, Gray-Keller M P, Detwiler P B, Palczewski K. Proc Natl Acad Sci USA. 1994;91:4014–4018. doi: 10.1073/pnas.91.9.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palczewski K, Subbaraya I, Gorczyca W A, Helekar B S, Ruiz C C, Ohguro H, Huang J, Zhao X, Crabb J W, Johnson R S, et al. Neuron. 1994;13:395–404. doi: 10.1016/0896-6273(94)90355-7. [DOI] [PubMed] [Google Scholar]
- 13.Dizhoor A M, Lowe D G, Olshevskaya E V, Laura R P, Hurley J B. Neuron. 1994;12:1345–1352. doi: 10.1016/0896-6273(94)90449-9. [DOI] [PubMed] [Google Scholar]
- 14.Dizhoor A M, Olshevskaya E V, Henzel W J, Wong S C, Stults J T, Ankoudinova I, Hurley J B. J Biol Chem. 1995;270:25200–25206. doi: 10.1074/jbc.270.42.25200. [DOI] [PubMed] [Google Scholar]
- 15.Dizhoor A M, Hurley J B. J Biol Chem. 1996;271:19346–19350. doi: 10.1074/jbc.271.32.19346. [DOI] [PubMed] [Google Scholar]
- 16.Rudnicka-Nawrot M, Surgucheva I, Hulmes J D, Haeseleer F, Sokal I, Crabb J W, Baehr W, Palczewski K. Biochemistry. 1998;37:248–257. doi: 10.1021/bi972306x. [DOI] [PubMed] [Google Scholar]
- 17.Payne A M, Downes S M, Bessant D A R, Taylor R, Holder G E, Warren M J, Bird A C, Bhattachraya S S. Hum Mol Genet. 1998;7:273–277. doi: 10.1093/hmg/7.2.273. [DOI] [PubMed] [Google Scholar]
- 18.Sokal I, Li N, Surgucheva I, Warren M J, Payne A M, Bhattacharya S S, Baehr W, Palczewski K. Mol Cell. 1998;2:129–133. doi: 10.1016/s1097-2765(00)80121-5. [DOI] [PubMed] [Google Scholar]
- 19.Dizhoor A M, Boikov S G, Olshevskaya E V. J Biol Chem. 1998;273:17311–17314. doi: 10.1074/jbc.273.28.17311. [DOI] [PubMed] [Google Scholar]
- 20.Tucker C L, Laura R P, Hurley J B. Biochemistry. 1997;36:11995–12000. doi: 10.1021/bi971212k. [DOI] [PubMed] [Google Scholar]
- 21.Perrault I, Rozet J M, Calvas P, Gerber S, Camuzat A, Dollfus H, Chatelin S, Souied E, Ghazi I, Leowski C, et al. Nat Genet. 1996;14:461–464. doi: 10.1038/ng1296-461. [DOI] [PubMed] [Google Scholar]
- 22.Kachi S, Nishizawa Y, Olshevskaya E, Yamazaki A, Miyake Y, Wakabayashi T, Dizhoor A, Usukura J. Exp Eye Res. 1999;68:465–473. doi: 10.1006/exer.1998.0629. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Ruoho A E, Rao V D, Hurley J H. Proc Natl Acad Sci USA. 1997;94:13414–13419. doi: 10.1073/pnas.94.25.13414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tucker C L, Hurley J H, Miller T R, Hurley J B. Proc Natl Acad Sci USA. 1998;95:5993–5997. doi: 10.1073/pnas.95.11.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otto-Bruc A, Buczylko J, Surgucheva I, Subbaraya I, Rudnicka-Nawrot M, Crabb J W, Arendt A, Hargrave P A, Baehr W, Palczewski K. Biochemistry. 1997;36:4295–4302. doi: 10.1021/bi963000d. [DOI] [PubMed] [Google Scholar]
- 26.Krylov D M, Niemi G A, Dizhoor A M, Hurley J B. J Biol Chem. 1999;274:10833–10839. doi: 10.1074/jbc.274.16.10833. [DOI] [PubMed] [Google Scholar]
- 27.Olshevskaya E V, Boikov S, Hurley J B, Dizhoor A M. J Biol Chem. 1999;274:10823–10832. doi: 10.1074/jbc.274.16.10823. [DOI] [PubMed] [Google Scholar]
- 28.Krishnan A, Goraczniak R M, Duda T, Sharma R K. Mol Cell Biochem. 1998;178:251–259. doi: 10.1023/a:1006860018300. [DOI] [PubMed] [Google Scholar]