Abstract
Background: Diffuse proliferation of interstitial cells of Cajal (ICCs) in the myenteric plexus layer of the intestine has been described in patients with familial and multiple gastrointestinal stromal tumours (GISTs). However, it is not fully understood whether proliferation is polyclonal or monoclonal.
Aims: To evaluate the clonal nature of diffuse ICC proliferation in familial and multiple GIST cases, we carried out clonal analysis using inactivation at the human androgen receptor (HUMARA) locus.
Materials and methods: Diffuse ICC proliferation tissues from three female patients were microdissected using a laser capture microdissection (LCM) system. Normal intestinal mucosal tissues were also microdissected for polyclonal controls and GIST tissues for monoclonal controls from the same patients, and genomic DNA was extracted. After digestion by restriction enzyme HhaI, the HUMARA locus was amplified by a fluorescent polymerase chain reaction (PCR) procedure and the PCR products were analysed.
Results: One case was uninformative because it was homozygous at the HUMARA locus. In the two other cases, PCR products from the diffuse ICC proliferation showed two alleles as well as those from normal intestinal mucosal tissues, indicating that ICC proliferation was polyclonal. In contrast, PCR products from associated GIST tissues showed only one allele, indicating that GISTs were monoclonal.
Conclusion: The results suggested that diffuse ICC proliferation in familial and multiple GIST cases was non-neoplastic hyperplasia.
Keywords: clonality, gastrointestinal stromal tumour, human androgen receptor gene, interstitial cells of Cajal, X chromosome inactivation, mosaicism
Gastrointestinal stromal tumours (GISTs) are the most common mesenchymal tumours in the human digestive tract. Most GISTs express a receptor tyrosine kinase, KIT.1,2 KIT is encoded by the c-KIT gene3,4 and its ligand is stem cell factor (SCF).5,6 GISTs frequently have a mutation of the c-KIT gene which results in constitutive activation of KIT without stimulation by SCF.1,7 As the development of interstitial cells of Cajal (ICCs) is dependent on SCF-KIT interaction and since ICCs and GISTs both express KIT and CD34,1,2 we considered that GISTs may originate from ICCs.
Recently, we and others found some families with multiple GISTs and a germline gain of function mutation of the c-KIT gene.7–13 In addition to the development of multiple GISTs, family members with the mutation showed diffuse proliferation of spindle shaped cells between the circular and longitudinal muscular layers—that is, the myenteric plexus of the small intestine.8–13 As the proliferative spindle shaped cells expressed both KIT and CD34, they were also considered to originate from ICCs. Although we regarded diffuse ICC proliferation as hyperplasia and the macroscopically apparent mass lesions as GISTs, the clonal nature of them remained unclear.
To examine the clonality of human samples, random inactivation of one of the two female X chromosomes (lyonisation) is most commonly used.14–16 Inactivation occurs by methylation during early embryogenesis in each somatic cell, and is stably transmitted to all progeny cells. It also persists during neoplastic transformation. Therefore, normal tissues or non-neoplastic hyperplastic lesions composed of a mosaic of cells showing either maternally or paternally derived inactivated X chromosomes display a polyclonal pattern, while true neoplastic lesions composed of uniform cells having either maternally or paternally derived inactivated X chromosomes demonstrate a monoclonal pattern. Currently, the polymorphism of the human androgen receptor (HUMARA) locus, which is located on the X chromosome, is most frequently used to examine clonality as it has a highly polymorphic tandem repeat.16–19 Approximately 90% of females are heterozygous for the number of CAG trinucleotide repeats.17 Moreover, methylation sensitive restriction enzyme sites of HpaII and HhaI are within the HUMARA locus.17
In the present study, we examined the clonality of diffuse ICC proliferation in familial and multiple GIST cases using polymorphism of the HUMARA locus. To obtain pure populations of cells from the diffusely proliferative lesions, the laser capture microdissection (LCM) method was used.20,21 GISTs showed a monoclonal pattern but diffuse ICC proliferation displayed a polyclonal pattern as did normal mucosal tissues.
MATERIALS AND METHODS
Samples
Tissues were obtained from three female patients with multiple GISTs and a germline mutation of the c-KIT gene during operation. Case No 1 was a daughter of case No 2, and they had a c-KIT gene mutation at exon 11, as described previously.8,9 Case No 3 had no familial relationship with case Nos 1 and 2, and had a c-KIT gene mutation at exon 17, as described previously.13 In each case, some GIST tissues were resected, including macroscopically normal adjacent intestine, and were fixed in formalin and embedded in paraffin. Histological diagnosis was made by haematoxylin and eosin staining and immunohistochemistry of KIT and CD34. Immunohistochemistry was performed as described previously.13 In the present study, we regarded non-mass forming proliferation (<3 mm in diameter) of spindle shaped cells which were double positive for KIT and CD34 as diffuse ICC proliferation, while apparent mass forming lesions (>5 mm in diameter) which were double positive for KIT and CD34 as GISTs. The adjacent sections were used for DNA extraction.
DNA extraction
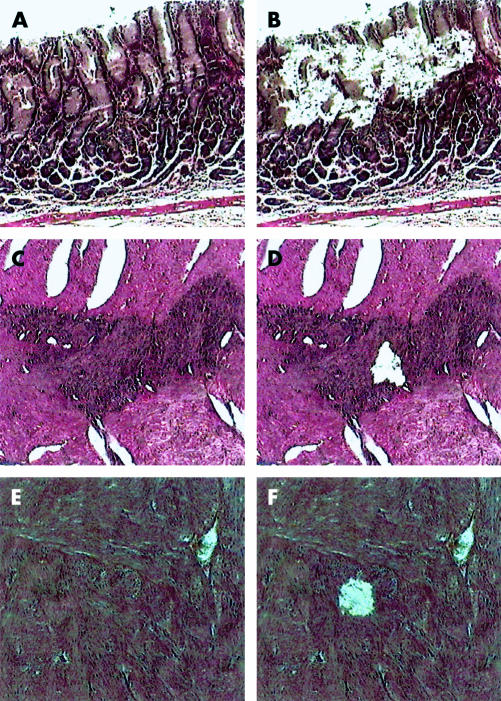
DNA samples were extracted from formalin fixed paraffin embedded tissues according to a previous method.22 Normal intestinal mucosa, diffuse ICC proliferation, and GISTs were microdissected from 4 μm sections stained with haematoxylin and eosin using an LCM system (LM200; Arcturus Engineering, Mountain View, California, USA). Representative histology before and after microdissection is shown in fig 1 ▶. In the section of large GISTs, three samples were extracted from three distinct sites, respectively. A total volume of 20 μl of PK buffer containing 1.0% proteinase K, 10 mM Tris HCl (pH 8.0), 1 mM ethylenediamine tetraacetic acid, and 1% Tween 20 was mounted on a microdissected specimen attached to a piece of LCM Transfer Film (Arcturus Engineering). Then, a 0.5 ml micro test tube (Eppendorf-Nethtler-Hinz-GmbH, Hamburg, Germany) was placed over it. After 16 hours of incubation at 37°C, proteinase K was inactivated by heating at 95°C for 10 minutes. The lysis mixture was centrifuged for five minutes to remove undigested tissue fragments.
Figure 1.
Tissues were microdissected using a laser capture microdissection system from normal intestinal mucosa ((A) before microdissection, (B) after microdissection), diffuse interstitial cells of Cajal proliferation ((C) before microdissection, (D) after microdissection), and gastrointestinal stromal tumours ((E) before microdissection, (F) after microdissection). Representative figures are shown.
Clonality assessment
We used a quantitative fluorescent polymerase chain reaction (PCR) procedure that enables accurate measurement of peak intensities of each allele, as described previously.23 The HUMARA gene includes a polymorphic ((CAG)n) repeat located at 3′ of the methylation sensitive HhaI restriction enzyme sites.17 The PCR assay used primers, the product of which spanned both the HhaI sites and the ((CAG)n) polymorphism. Variations in length of the ((CAG)n) repeats on the paternal and maternal X chromosomes yield HUMARA alleles of different lengths. Methylation of the HhaI sites distinguishes the active (non-methylated) from the inactive (methylated) X chromosome. It is only the undigested inactive methylated allele that is subsequently amplified by PCR.17
Each DNA sample (7.5 μl) was digested overnight at 37°C in a 2.5 μl reaction mixture containing 0.5 μl of 16 units of HhaI (Toyobo, Osaka, Japan), 1 μl of concentrated (×10) TA buffer (330 mM Tris acetate, 660 mM KOAc, 100 mM MgOAc2, 5 mM dithiothreitol), and 1 μl of concentrated (×10) bovine serum albumin. For each case, a control sample without DNA was run simultaneously to rule out contamination of DNA in the reaction mixture. The restriction enzyme was then inactivated by heating at 95°C for 10 minutes.
For PCR, 1 μl of each digested DNA sample was added to 24 μl of a PCR reaction mixture containing 2.5 μl of concentrated (×10) PCR buffer,1.5 μl of deoxyribonucleoside triphosphate (200 μM), 0.2 μl of primers 1A and 1B (10 pM each), 0.1 μl of Taq polymerase (Roche Diagnostics GmbH, Mannheim, Germany), and 19.7μl of deionised H2O. The DNA samples were amplified using a sandwiched primer approach.24 The first step was performed using outer primers 1A (5′-GCT GTG AAG GTT GCT GTT CCT CAT-3′) and 1B (5′-CGT CCA AGA CCT ACC GAG GAG CTT-3′). The second step was performed with inner primers 2A (5′-TCC AGA ATC TGT TCC AGA GCG TGC-3′) and 2B (5′-ATG GGC TTG GGG AGA ACC ATC CTC-3′). Primer 2A was labelled at the 5′ end with 6-carboxyflurorescein. Initial denaturation was performed for 10 minutes at 95°C, followed by 35 cycles of one minute at 95°C, one minute at 60°C, and one minute at 72°C. In the final cycle, extension at 72°C was prolonged for 10 minutes. The second step PCR profile was the same as the first step PCR. Following the second step amplification, 5 μl of the PCR product was assessed using 2.0% agarose gel electrophoresis to confirm amplification of the HUMARA target. After amplification, 1 μl of the PCR products was mixed with 12 μl of a Template Suppression Reagent (Applied Biosystems, Foster City, California, USA) and 0.5 μl of internal size standards (GENESCAN-500 (TAMRA), Applied Biosystems). The mixture was denatured at 95°C for two minutes, and analysed through a DNA Sequencing Polymer (Applied Biosystems) with an ABI PRISM Genetic Analyser (Applied Biosystems), according to a previous method.25 Data were analysed using Genescan 310 Software (Applied Biosystems).
Data interpretation
Amplification of each HUMARA allele usually generated a set of multiple peaks, including one major peak and a few associated peaks of lesser intensity, as described previously.26 Clonality assessment was based on the major peak generated from each allele. Patients were considered heterozygous when PCR amplification of undigested DNA showed two major peaks of almost equal intensity. This suggested that maternal and paternal X chromosomes have HUMARA alleles of different molecular weights. PCR products showing a single major peak suggested that maternal and paternal X chromosomes have HUMARA alleles of the same molecular weight. Such patients were considered to be homozygous for the HUMARA gene, and thus uninformative for the analysis.
Samples were considered to be polyclonal when PCR amplification of digested DNA showed two major peaks similar to that of normal tissue from the same organ. PCR products showing only one of the two major peaks were considered to be monoclonal.
RESULTS
Specimens were obtained from three women. Immunohistochemistry of KIT and CD34 was carried out on sections with normal intestinal mucosa, diffuse ICC proliferation, and/or GIST tissues of each patient. Almost all cells of GIST or diffuse ICC proliferation tissues were double positive for KIT and CD34. Adjacent sections were stained with haematoxylin and eosin, then normal intestinal mucosa, diffuse ICC proliferation and GIST tissues of each patient were carefully removed by LCM (fig 1 ▶). In each case, two GISTs from different sites were studied. DNA was extracted from each sample, and the portion of the HUMARA locus containing the trinucleotide repeats was amplified by PCR and the length and intensity of the PCR products were analysed.
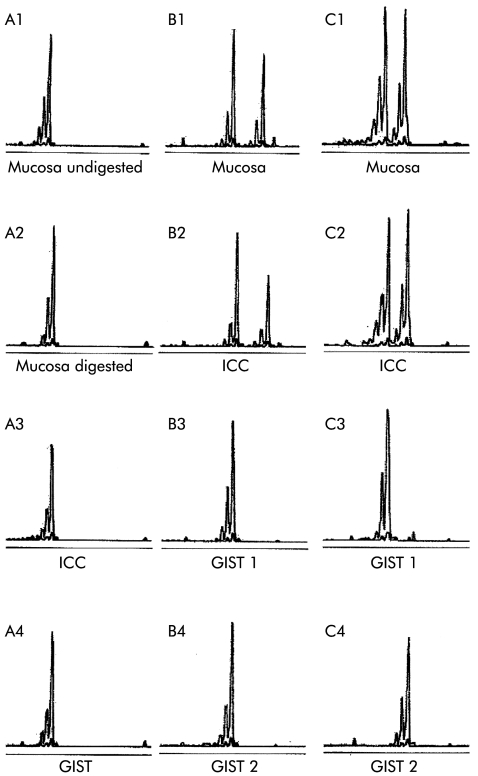
In case No 1, all of the PCR products using undigested and digested DNA from normal intestinal mucosa, diffuse ICC proliferation, and two GISTs showed only one major peak with an allelic size of 246 bases (fig 2A1–A4), indicating that the patient was homozygous for the number of trinucleotide repeats and uninformative for the analysis.
In case No 2, the mother of case No 1, the PCR products using undigested DNA from normal intestinal mucosa, diffuse ICC proliferation, and two GISTs showed two major peaks with allelic sizes of 231 and 246 bases, indicating that the patient was heterozygous for the number of trinucleotide repeats and informative for the analysis. After digestion by HhaI, the PCR product derived either from normal intestinal mucosa or from diffuse ICC proliferation demonstrated two major peaks (fig 2B1–B2). On the other hand, the PCR product derived either from a small GIST or from a large GIST showed only one allele with a lower molecular weight (allelic size of 231 bases) (fig 2 ▶ B3–B4). All of the PCR products from three distinct sites inside the same large GIST demonstrated disappearance of the same allele with higher molecular weight.
Figure 2.
Clonal analysis was carried out using undigested and digested DNA from normal mucosa, diffuse interstitial cells of Cajal (ICC) proliferation, and gastrointestinal stromal tumours (GISTs). In case No 1, all of the polymerase chain reaction (PCR) products of undigested (A1) and digested (A2) DNA from normal intestinal mucosa, digested DNA from diffuse ICC proliferation (A3), and GIST (A4) showed only one major peak with an allelic size of 246 bases, indicating that it was uninformative. In case No 2, the PCR product of digested DNA from normal intestinal mucosa (B1) and diffuse ICC proliferation (B2) showed two major peaks with allelic sizes of 231 and 246 bases, while that of digested DNA derived either from a small GIST (B3) or from a large GIST (B4) showed only one allele with a lower molecular weight (allelic size of 231 bases). In case No 3, the PCR product of digested DNA from normal intestinal mucosa (C1) and diffuse ICC proliferation (C2) showed two major peaks with allelic sizes of 240 and 252 bases, and that of digested DNA from a large GIST showed one allele with a lower molecular weight (allelic size of 240 bases) (C3), while that of digested DNA from a small GIST demonstrated the other allele with higher molecular weight (allelic size of 252 bases) (C4).
In case No 3, the PCR products using undigested DNA from normal intestinal mucosa, diffuse ICC proliferation, and two GISTs showed two major peaks with allelic sizes of 240 and 252 bases. After digestion by HhaI, the PCR product derived either from the normal intestinal mucosa or from diffuse ICC proliferation showed two major peaks (fig 2C1–C2). The PCR product derived from a large GIST showed only one allele with a lower molecular weight (allelic size of 240 bases) (fig 2C3). Again, all of the PCR products from three distinct sites inside the same large GIST demonstrated disappearance of the same allele with a higher molecular weight. On the other hand, the PCR product derived from a small GIST showed the other allele with a higher molecular weight (allelic size of 252 bases) (fig 2C4).
DISCUSSION
In this study, we examined the clonal nature of diffuse ICC proliferation observed in three female patients with a germline mutation of the c-KIT gene. One case was uninformative because the HUMARA locus was homozygous. In two informative cases, PCR products derived from diffuse ICC proliferation showed the two major peaks as those derived from the intestinal mucosa. In other words, diffuse ICC proliferation is polyclonal, suggesting that the lesion is hyperplastic but not neoplastic. In contrast, PCR products derived from apparent mass lesions (>5 mm in diameter) showed only one major peak. These mass lesions are monoclonal, suggesting that they are truly neoplastic—that is, GISTs.
In rare cases, X chromosome inactivation in normal tissue does not occur randomly, and is responsible for a false monoclonal pattern of inactivation in normal tissue.18,27 Therefore, interpretation of the clonal pattern of a lesion must always be performed with the knowledge of the X chromosome inactivation pattern of a normal tissue from the same organ.26 In the present study, we demonstrated that normal tissues from the intestine were apparently polyclonal, and therefore our clonality analysis can be considered reliable.
PCR products from three distinct sites inside the same large GIST showed disappearance of the same peak in case Nos 2 and 3. Although PCR products from a small GIST and a large GIST in case No 2 demonstrated disappearance of the same peak, those in case No 3 showed disappearance of different peaks. The results indicated that multiple GISTs observed in patients with a germline KIT mutation were not metastatic lesions from one GIST but developed independently.
Although monoclonality is considered to be a hallmark of neoplasia, the relationship between monoclonality/polyclonality and neoplasia/hyperplasia is currently controversial, especially in some familial syndromes. For example, in multiple endocrine neoplasia type 2A, each focus of C cell proliferation had been considered to be hyperplastic, but Diaz-Cano et al reported that the lesion was monoclonal intraepithelial neoplasia.28 Moreover, Merritt et al reported that Min (multiple intestinal neoplasia) mice frequently had intestinal adenomas showing a polyclonal structure.29 In the case of neurofibromatosis type 1, the clonality of each single neurofibroma is controversial. Some reports showed that each benign neurofibroma was polyclonal and neurofibrosarcoma probably developing from benign neurofibroma was monoclonal,30,31 while other reports demonstrated that each benign neurofibroma was monoclonal.32,33 In the present study, we showed that diffuse ICC proliferation was polyclonal and each GIST probably developing from diffuse ICC proliferation was monoclonal. Further studies are essential to clarify the relationship between monoclonality/polyclonality and neoplasia/hyperplasia.
Diffuse ICC proliferation in the myenteric plexus layer of the small intestine has been described in patients with familial and multiple GISTs.8–13 The present study suggested that diffuse ICC proliferation was a polyclonal hyperplasia while apparent mass lesions (>5 mm in diameter) were monoclonal neoplasia. A KIT mutation may contribute to the development of both diffuse ICC hyperplasia and GISTs but some other molecular mechanisms may be indispensable for the development of GISTs from diffuse ICC hyperplasia. Further research is needed for the elucidation of the relationship between diffuse ICC hyperplasia and the development of GISTs in patients with familial and multiple GISTs.
Acknowledgments
This study was supported by grants from the Ministry of Education, Science, Culture, and Sports of Japan, the Sumitomo Foundation, and Sasagawa Foundation. Hui Chen is a research fellow from Guangdong Provincial Hospital of TCM, China.
Abbreviations
GISTs, gastrointestinal stromal tumours
HUMARA, human androgen receptor
ICCs, interstitial cells of Cajal
LCM, laser capture microdissection
PCR, polymerase chain reaction
SCF, stem cell factor
REFERENCES
- 1.Hirota S, Isozaki K, Moriyama Y, et al. Gain-of-function mutations of c-kit in human gastrointestinal stromal tumors. Science 1998;279:577–80. [DOI] [PubMed] [Google Scholar]
- 2.Kindblom L-G, Remotti HE, Aldenborg F, et al. Gastrointestinal pacemaker cell tumor (GIPACT): gastrointestinal stromal tumors show phenotypic characteristics of the intestinal cells of Cajal. Am J Pathol 1998;152:1259–69. [PMC free article] [PubMed] [Google Scholar]
- 3.Yarden Y, Kuang WJ, Yang-Feng T, et al. Human proto-oncogene c-kit: a new cell surface receptor tyrosine kinase for an unidentified ligand. EMBO J 1988;6:3341–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Besmer P, Murphy JE, George PC, et al. A new acute transforming feline retrovirus and relationship of its oncogene v-kit with the protein kinase gene family. Nature 1986;30:415–21. [DOI] [PubMed] [Google Scholar]
- 5.Flanagan JG, Leder P. The kit ligand: a cell surface molecule altered in steel mutant fibroblasts. Cell 1990;63:185–94. [DOI] [PubMed] [Google Scholar]
- 6.Zsebo KM, Williams DA, Geissler EN, et al. Stem cell factor is encoded at the Sl locus of the mouse and is the ligand for the c-kit tyrosine kinase receptor. Cell 1990;63:213–24. [DOI] [PubMed] [Google Scholar]
- 7.Nishida T, Hirota S, Taniguchi M, et al. Familial gastrointestinal stromal tumors with germline mutation of the KIT gene. Nat Genet 1998;19:323–4. [DOI] [PubMed] [Google Scholar]
- 8.O’Brien P, Kapusta L, Dardick I, et al. Multiple familial gastrointestinal autonomic nerve tumors and small intestinal neuronal dysplasia. Am J Surg Pathol 1999;23:198–204. [DOI] [PubMed] [Google Scholar]
- 9.Hirota S, Okazaki T, Kitamura Y, et al. Cause of familial and multiple gastrointestinal autonomic nerve tumors with hyperplasia of interstitial cells of Cajal is germline mutation of the c-Kit gene. Am J Surg Pathol 2000;24:326–7. [DOI] [PubMed] [Google Scholar]
- 10.Isozaki K, Terris B, Belghiti J, et al. Germline-activating mutation in the kinase domain of KIT gene in familial gastrointestinal stromal tumors. Am J Pathol 2000;157:1581–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maeyama H, Hidaka E, Ota H, et al. Familial gastrointestinal stromal tumor with hyperpigmentation: association with a germline mutation of the c-kit gene. Gastroenterology 2001;120:210–15. [DOI] [PubMed] [Google Scholar]
- 12.Beghini A, Tibiletti MG, Roversi G, et al. Germline mutation in the juxtamembrane domain of the kit gene in a family with gastrointestinal stromal tumors and urticaria pigmentosa. Cancer 2001;92:657–62. [DOI] [PubMed] [Google Scholar]
- 13.Hirota S, Nishida T, Isozaki K, et al. Familial gastrointestinal stromal tumors associated with dysphagia and novel type germline mutation of KIT gene. Gastroenterology 2002;122:1493–9. [DOI] [PubMed] [Google Scholar]
- 14.Vogelstein B, Fearon ER, Hamilton SR, et al. Use of restriction fragment length polymorphisms to determine the clonal origin of human tumors. Science 1985;227:642–5. [DOI] [PubMed] [Google Scholar]
- 15.Vogelstein B, Fearon ER, Hamilton SR, et al. Clonal analysis using recombinant DNA probes from the X-chromosome. Cancer Res 1987;47:4806–13. [PubMed] [Google Scholar]
- 16.Nakamura H, Hirota S, Adachi S, et al. Clonal nature of seborrheic keratosis demonstrated by using the polymorphism of the human androgen receptor locus as a marker. J Invest Dermatol 2001;116:506–10. [DOI] [PubMed] [Google Scholar]
- 17.Allen RC, Zoghbi HY, Moseley AB, et al. Methylation of HpaII and HhaI sites near the polymorohic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Am J Hum Genet 1992;51:1229–39. [PMC free article] [PubMed] [Google Scholar]
- 18.Busque L, Gilliland DG. Clonal evolution in acute myeloid leukemia. Blood 1993;82:337–42. [PubMed] [Google Scholar]
- 19.Busque L, Zhu J, Dehart D, et al. An expression based clonality assay at the human receptor locus (HUMARA) on chromosome X. Nucleic Acids Res 1994;22:697–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emmert-Buck MR, Bonner RF, Smith PD, et al. Laser capture microdissection. Science 1996;274:998–1001. [DOI] [PubMed] [Google Scholar]
- 21.Simone NL, Bonner RF, Gillespie JW, et al. Laser capture microdissection: opening the microscopic frontier to molecular analysis. Trends Genet 1998;14:272–6. [DOI] [PubMed] [Google Scholar]
- 22.Mashal RD, Lester SC, Sklar J. Clonal analysis by study of X chromosome inactivation in formalin-fixed paraffin-embedded tissue. Cancer Res 1993;53:4676–9. [PubMed] [Google Scholar]
- 23.Wu CD, Wickert RS, Williamson JE, et al. Using fluorescence-based human androgen receptor gene assay to analyze the clonality of microdissected dendritic cell tumors. Am J Clin Pathol 1999;111:105–10. [DOI] [PubMed] [Google Scholar]
- 24.Fujita M, Enomoto T, Wada H, et al. Application of clonal analysis: differential diagnosis for synchronous primary ovarian and endometrial cancers and metastatic cancer. Am J Clin Pathol 1996;105:350–9. [DOI] [PubMed] [Google Scholar]
- 25.George KS, Zhao X, Gallahan D, et al. Capillary electrophoresis methodology for identification of cancer related gene expression patterns of fluorescent differential display polymerase chain reaction. J Chromatogr B 1997;695:93–102. [DOI] [PubMed] [Google Scholar]
- 26.Paradis V, Laurent A, Flejou J-F, et al. Evidence for the polyclonal nature of focal nodular hyperplasia of the liver by the study of X-chromosome inactivation. Hepatology 1997;26:891–5. [DOI] [PubMed] [Google Scholar]
- 27.Gale RE, Wheadon H, Linch DC. X-chromosome inactivation patterns using HPRT and PGK polymorphisms in haemotomogically normal and post-chemotherapy females. Br J Haematol 1991;79:193–7. [DOI] [PubMed] [Google Scholar]
- 28.Diaz-Cano SJ, de Miguel M, Blanes A, et al. Germline RET 634 mutation positive MEN 2A- related C-cell hyperplasia have genetic features consistent with intraepithelial neoplasia. J Clin Endocrinol Metab 2001;86:3948–57. [DOI] [PubMed] [Google Scholar]
- 29.Merritt AJ, Gould KA, Dove WF. Polyclonal structure of intestinal adenomas in ApcMin/+ mice with concomitant loss of Apc+ from all tumor lineages. Proc Natl Acad Sci USA 1997;94:13927–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Friedman JM, Fialkow PJ, Greene CL, et al. Probable clonal origin of neurofibrosarcoma in a patient with hereditary neurofibromatosis. J Natl Cancer Inst 1982;69:1289–91. [PubMed] [Google Scholar]
- 31.Rey JA, Bello MJ, de Campos JM, et al. Cytogenetic clones in a recurrent neurofibroma. Cancer Genet Cytogenet 1987;26:157–63. [DOI] [PubMed] [Google Scholar]
- 32.Skuse GR, Kosciolek BA, Rowley PT. The neurofibroma in von Recklinghausen neurofibromatosis has a unicellular origin. Am J Hum Genet 1991;49:600–7. [PMC free article] [PubMed] [Google Scholar]
- 33.Daschner K, Assum G, Eisenbarth I, et al. Clonal origin of tumor cells in a plexiform neurofibroma with LOH in NF1 intron 38 and in dermal neurofibromas without LOH of the NF1 gene. Biochem Biophys Res Commun 1997;234:346–50. [DOI] [PubMed] [Google Scholar]