Abstract
Background and aims: A subgroup of colorectal cancers (CRC) referred to as the CpG island methylator phenotype (CIMP+) shows simultaneous methylation of multiple CpG islands. The clinicopathological and molecular characteristics of this phenotype remain uncertain however.
Methods: We analysed methylation of CpG islands in the p16 and MDR1 genes and MINT-2 clone in 275 stage II/III CRCs.
Results: Concurrent methylation of two or more CpG islands was observed in 32% of cases and was considered to represent CIMP+. These were often poorly differentiated, had less TP53 mutations, and originated frequently in the proximal or higher stage CRC compared with CIMP− tumours (p<0.05 for each). CIMP+ had no prognostic significance in stage II or stage III CRC treated by surgery alone. hMLH1 methylated tumours comprised the majority (81%) of cases with microsatellite instability, were frequently observed in older female patients, were often poorly differentiated or CIMP+, and contained wild-type K-ras (p<0.05 for each). Females who were heterozygous or homozygous for the C677T MTHFR polymorphism were at increased risk of developing CIMP+ CRC (odds ratio 2.17, 95% confidence interval 1.03–4.57; p=0.037).
Conclusions: These observations made in a relatively large unselected series of CRC support the notion that CIMP+ characterises a subgroup of tumours with distinctive phenotypic features.
Keywords: colorectal cancer, CpG island methylator phenotype, hypermethylation
Transcriptional silencing of tumour suppressor genes by hypermethylation of CpG islands located in the promoter region has been proposed as an important mechanism for the development of a variety of cancer types.1,2 A CpG island methylator phenotype (CIMP+) has been described in colorectal cancer (CRC) and is characterised by simultaneous methylation of multiple genes including p16, THBS1, IGF2, HIC-1, and COX-2.3–5 In a small proportion of CRCs the DNA mismatch repair gene hMLH1 is also methylated. This specific epigenetic alteration gives rise to the majority of tumours with the microsatellite instability (MSI+) phenotype.6–8 The precise mechanism for the aberrant DNA methylation seen in CIMP+ tumours remains to be determined but two features of this phenotype that are consistently observed in CRC are origin in the proximal colon and poorly differentiated histology.4,9,10 Less clear cut are possible associations between CIMP+ and patient features such as age, sex, and survival.9,11–13 Contradictory reports have also been published on the relationship between CIMP+ and the tumour specific molecular features of TP53 mutation, K-ras mutation, and MSI+.11–15
One of the difficulties in attempts to characterise CIMP+ has been the use of selected CRC series, in particular enrichment with MSI+ cases. The majority of sporadic MSI+ tumours have a methylated hMLH1 gene6–8 and are therefore likely to be CIMP+ whereas familial MSI+ cases from the hereditary non-polyposis colon cancer syndrome show less hMLH1 methylation8 and are more likely to be CIMP−. Hence the methylation characteristics of MSI+ tumours may depend on whether they are sporadic or familial in origin. Another problem has been analysis of relatively small tumour cohorts, thereby limiting the statistical power to detect associations between CIMP+ and other molecular or clinical features. With the exception of a recent study in 119 CRC cases examined for p16 and p14 methylation,16 there have been no reports to our knowledge examining methylation of multiple CpG sites in a large number of unselected CRCs. Consequently, the biological and clinical properties of CIMP+ remain largely unknown. Possible involvement of either or both the MSI+ and CIMP+ phenotypes in response of CRC to 5-fluorouracil (5-FU) based adjuvant therapy17 highlights the need for further characterisation.
In the present study we have attempted to overcome the above limitations by examining the CIMP+ phenotype in a large (n=275) unselected CRC series. We chose to analyse hypermethylation of the p16 and MDR1 genes and of the MINT-2 clone because these sites are frequently (20–40%) and specifically methylated in CRC, but not in adjacent normal epithelium.3,4,15 Methylation of p1618,19 and MDR120 genes is known to be associated with transcriptional silencing of expression and may therefore have functional significance for the development of a malignant phenotype. The CRC series investigated in this study has previously been characterised for site of tumour origin, histological grade, patient outcome,17 TP53 mutation,21 K-ras mutation,22 MSI+,21 and the C677T polymorphism of the methylenetetrahydrofolate reductase (MTHFR) gene.23 This has allowed us to shed additional light on the characteristics of CIMP+ CRC.
MATERIALS AND METHODS
Patients
A series of 133 stage II or III CRCs that were surgically resected between 1991 and 1996 at the Sir Charles Gairdner Hospital, Nedlands, Australia, were prospectively included in a study designed to evaluate the prognostic significance of various molecular genetic alterations. Tumour samples were snap frozen immediately after surgery and stored at −80oC. An additional unselected series of 142 paraffin embedded stage III CRC cases that were surgically treated between 1991 and 1996 were also analysed. All tumours had negative resection margins. Adjuvant chemotherapy according to the Moertel regimen24 was administered to 39 patients. These cases were excluded from survival analyses. Ethics approval for the project was obtained from the Sir Charles Gairdner Hospital Human Research Ethics Committee. Information on disease specific survival was obtained from the West Australian Health Department death register and from hospital medical records. Survival data on patients who died from causes other than CRC were censored at the time of death. The splenic flexure was used as the anatomical boundary to define proximal and distal CRC.
Methylation specific PCR (MSP)
DNA suitable for polymerase chain reaction (PCR) analysis was extracted from frozen tissue according to standard methods and from paraffin embedded samples using previously described methods.17 Methylation specific PCR (MSP) for the detection of methylation in the promoter regions of p16, hMLH1, and the clone MINT-2 was carried out as described previously.9,25 This technique uses bisulphite modification to convert unmethylated, but not methylated, cytosine to uracil. MSP utilises this difference to specifically amplify either methylated or unmethylated DNA. Bisulphite (Sigma, St Louis, Missouri, USA) conversion of 1 μg of genomic DNA was carried out for 16 hours at 55°C according to a modified method26 but without prior digestion. Primers for the promoter region of MDR1 were designed to include six CpG dinucleotides that have been linked to regulation of MDR1 expression.20 Primer sequences for amplification of unmethylated MDR1 were:
forward 5′-GGGTGTGGGTTGAGTATAGTTGTTTT-3′
reverse 5′-CCAACTTTACATACCCCTACCTCACA-3′,
and for methylated MDR1 were:
forward 5′-GGGCGTGGGTTGAGTATAGTCGTTTC-3′
reverse 5′-CGCTCCTTAAAACAACCACCAAAACG-3′.
The annealing temperature was 56°C for unmethylated DNA and 60°C for methylated DNA, with a Mg2+ concentration of 1.6 mM. PCR reaction mixes contained 1×polymerisation buffer, 1×Q-solution (Qiagen, Melbourne, Australia), the optimal Mg2+ concentration for that primer pair, 0.4 μM of each primer, and 0.5 U Taq polymerase (Qiagen, Melbourne, Australia) in a total volume of 20 μl. PCR reactions were “hot started” at 94°C by addition of 100 ng of bisulphite converted DNA, followed by 35 cycles (30 seconds at 94°C, 30 seconds at the appropriate annealing temperature, 30 seconds at 72°C) and seven minutes of final extension at 72°C. PCR products were analysed on 2.5% agarose gels. Both methylated (HT-29 CRC cell line) and unmethylated DNA (peripheral blood lymphocytes) controls were included in every bisulphite conversion.
Screening for MSI+, TP53 mutation, K-ras mutation, and MTHFR genotype
The MSI status of each tumour was evaluated by fluorescent single strand conformation polymorphism (SSCP) analysis of deletions in the BAT-26 mononucleotide repeat, as described previously.27 Deletions in the quasimonomorphic BAT-26 allele establish the MSI status of tumours with more than 99% accuracy in Caucasian populations.28 Tumour samples were also screened for mutations in exons 4–10 of the TP53 gene21 and in codons 12 and 13 of the K-ras gene using fluorescent-SSCP,22 as described previously by our laboratory. All suspected TP53 and K-ras mutations were confirmed at least once by separate PCR-SSCP analyses as well as by DNA sequencing.29 Genotyping for the C677T polymorphism in the MTHFR gene was carried out using silver stain and fluorescent SSCP methods, as described recently by our laboratory, and confirmed by HinfI restriction enzyme digestion.23 The incidence of CT and TT MTHFR genotypes in CRC cases was compared to that observed in an age matched series of 467 healthy Australian individuals (range 60–92 years).23
Statistical analysis
Data were analysed using the SPSS 10.0 (Chicago, Illinois, USA) software package. Contingency tables were analysed using the χ2 test (Pearson statistic) or Fisher’s exact test when expected frequencies were lower than five. Kaplan-Meier analysis was used to assess five year cumulative survival probability and differences were evaluated using the log rank test. Cox’s proportional hazards univariate and multivariate analyses were used to calculate hazard ratios and 95% confidence intervals. Wald’s test was used to calculate 95% confidence intervals for odds ratios. All p values are derived from two tailed statistical tests.
RESULTS
Methylation of p16, MDR1, and MINT-2 was analysed in 275 stage II/III CRC. To assess the sensitivity of MSP, DNA obtained from methylated and unmethylated control samples was mixed at different ratios prior to bisulphite treatment. MSP was able to detect as little as 3% methylated p16 allele content. The ratio of methylated/non-methylated allele for each sample was not evaluated because microdissection had not been carried out to remove contaminating normal tissue. Comparison of MSP results for p16, MDR1, and MINT-2 revealed perfect concordance between DNA obtained from 23 frozen tumours and from the corresponding paraffin embedded archival tissue blocks. This confirms previous observations on the suitability of DNA obtained from paraffin embedded tissue for methylation studies.30 Although the success rate for the MSP technique using archival DNA was approximately 70% in the current study, the intensity of bands was similar to that observed from frozen tissue DNA. Only samples showing clear results for all three CpG sites were included in the analysis. An example of MSP results obtained from archival DNA for the p16 gene are shown in fig 1 ▶.
Figure 1.
Methylation sensitive polymerase chain reaction (MSP) results for the p16 gene. DNA for MSP amplification was obtained from paraffin embedded tissue (lanes 1–4) and from frozen tissue (lanes 5–7). Samples 1, 2, 3, 5, and 6 were scored as unmethylated for the p16 gene whereas samples 4 and 7 were scored as methylated.
Methylation and clinicopathological features
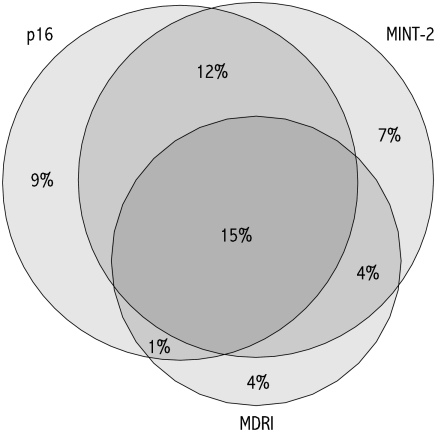
Methylation of p16, MDR1, and MINT-2 was detected in 36%, 24%, and 37% of tumours, respectively. The characteristics of tumours with individual or multiple methylated sites are summarised in table 1 ▶. When methylation of individual sites was analysed, no sex or age differences were apparent. Methylation of at least one or two CpG islands was more frequent in higher stage tumours. All three sites showed approximately twofold more frequent methylation in proximal compared with distal colon tumours, with even less methylation seen in rectal tumours (p<0.001 for each). Poorly differentiated tumours showed 2–3-fold higher frequency of p16, MDR1, and MINT-2 methylation compared with well/moderately differentiated tumours (p<0.005 for each). Methylation of CpG islands occurred in a non-random manner, with concordant methylation at two or more sites observed in 32% of cases (table 1 ▶, fig 2 ▶). These were defined arbitrarily in the present study as being CIMP+.
Table 1.
Associations between methylation and clinicopathological features of colorectal cancer
| Methylation (%) | ||||||
| Feature (n) | p16 | MDR1 | MINT-2 | One | Two | Three |
| Total (275) | 100 (36) | 67 (24) | 102 (37) | 141 (51) | 88 (32) | 40 (15) |
| Sex | ||||||
| Female (137) | 51 (37) | 35 (26) | 56 (41) | 70 (51) | 47 (34) | 25 (18) |
| Male (138) | 49 (36) | 32 (23) | 46 (33) | 71 (51) | 41 (30) | 15 (11)† |
| Age | ||||||
| <71 y (144) | 50 (35) | 35 (24) | 49 (34) | 69 (48) | 47 (33) | 18 (13) |
| ≥71 y (131) | 50 (38) | 32 (24) | 53 (40) | 72 (55) | 41 (31) | 22 (17) |
| Stage | ||||||
| II (72) | 21 (29) | 13 (18) | 20 (28) | 27 (38) | 16 (22) | 11 (15) |
| III (203) | 79 (39)* | 54 (27)* | 82 (40)† | 114 (56)‡ | 72 (36)‡ | 29 (14) |
| Site | ||||||
| Rectum (42) | 9 (21) | 2 (5) | 9 (21) | 15 (36) | 5 (12) | 0 (0) |
| Distal colon (77) | 20 (26) | 11 (14) | 18 (23) | 29 (38) | 16 (21) | 4 (5) |
| Proximal colon (152) | 71 (47)§ | 54 (36)§ | 75 (49)§ | 114 (64)§ | 67 (44)§ | 36 (24)§ |
| Grade | ||||||
| Well/moderate (211) | 62 (29) | 39 (19) | 65 (31) | 93 (44) | 55 (26) | 18 (9) |
| Poor (61) | 36 (59)§ | 27 (44)§ | 36 (59)§ | 46 (75)§ | 32 (53)§ | 21 (34)§ |
*p<0.2; †p<0.1; ‡p<0.05; §p<0.005.
Figure 2.
Venn diagram showing overlap in the methylation of p16, MDR1, and MINT-2 observed in 275 colorectal cancer cases. CpG island methylator phenotype tumours were defined as having two or more sites methylated. Percentages shown are absolute values.
Because methylation was more frequent in stage III cases (table 1 ▶), the prognostic significance of CpG island methylation was examined separately for patients with stage II or III disease. Only patients treated by surgery alone were included in these analyses because of a possible interaction between CIMP+ and chemotherapy.17 Kaplan-Meier and Cox regression analyses revealed no associations between methylation at individual or multiple sites and overall patient survival (results not shown).
Methylation and somatic alterations
Associations between CpG island methylation and several other molecular features that were previously characterised in this tumour series21,22,29 are shown in table 2 ▶. Tumours with wild-type TP53 showed more frequent methylation compared with those with mutant TP53, especially for MINT-2 and for multiple sites. Similarly, methylation was more frequent in tumours with wild-type K-ras, reaching significance for tumours with methylated MDR1, two or more, and with all three sites methylated. No differences between the type of TP53 or K-ras mutation and methylation were observed (results not shown). As expected, methylation was 2–3-fold more frequent in MSI+ compared with MSI− tumours.
Table 2.
Associations between methylation and molecular features of colorectal cancer
| Methylation (%) | ||||||
| Feature (n) | p16 | MDR1 | MINT-2 | One | Two | Three |
| Total (275) | 100 (36) | 67 (24) | 102 (37) | 141 (51) | 88 (32) | 40 (15) |
| TP53 | ||||||
| Wild-type (152) | 59 (39) | 40 (26) | 62 (41) | 79 (52) | 54 (36) | 28 (18) |
| Mutant (103) | 30 (29)* | 18 (18)† | 26 (25)‡ | 46 (44) | 21 (20)‡ | 7 (7)‡ |
| K-ras | ||||||
| Wild-type (138) | 51 (37) | 41 (30) | 50 (36) | 67 (46) | 49 (36) | 26 (19) |
| Mutant (65) | 17 (26)* | 7 (11)§ | 18 (28) | 28 (44) | 11 (17)‡ | 3 (5)‡ |
| MSI | ||||||
| − (228) | 70 (31) | 40 (18) | 70 (31) | 106 (45) | 58 (25) | 16 (7) |
| + (41) | 28 (68)§ | 27 (66)§ | 30 (73)§ | 32 (77)§ | 29 (71)§ | 24 (59)§ |
*p<0.2; †p<0.1; ‡p<0.05; §p<0.005.
MSI, microsatelite instability.
Differences between CIMP+ and MSI+ phenotypes
Recent studies suggest that the CIMP+ and MSI+ phenotypes are closely related.4,31 In order to characterise CIMP+ independently of MSI+, all 41 MSI+ cases were removed from the present series. The remaining 58 tumours with two or more methylated sites (CIMP+/MSI−) still showed significant associations with the features of proximal tumour origin, higher tumour stage, poor differentiation, and wild-type TP53 (p<0.05 for each), but not with the presence of wild-type K-ras. These features appear therefore to be characteristic of CIMP+ tumours independently of the MSI+ phenotype.
The majority of sporadic MSI+ tumours show methylation of the hMLH1 promoter6–8 and these are presumably CIMP+/MSI+. The remaining MSI+ cases are more likely to have germline or somatic mutations, or loss of heterozygosity of one or more of the DNA mismatch repair genes8 and might therefore be CIMP−/MSI+. In the present study, we examined hMLH1 methylation in the 41 MSI+ cases, as well as an additional 14 MSI+ cases from an earlier series.21 This allowed us to compare the features of MSI+ tumours with or without hMLH1 methylation (table 3 ▶). MSI+ tumours with methylated hMLH1 frequently were CIMP+ or poorly differentiated and tended to be more common in older (mean age 70 v 49 years for non-methylated cases) female patients. They also showed less frequent K-ras, but not TP53, mutations. Although the number of unmethylated MSI+ cases in this study was small (n=10), our findings suggest that in contrast with methylated tumours, they were mostly found in younger (9/10) male (7/10) patients and did not show a tendency to be poorly differentiated (1/10).
Table 3.
Characteristics of microsatellite instability (MSI+) colorectal cancer with or without hMLH1 methylation
| hMLH1 (%) | |||
| Feature (n) | Unmethylated | Methylated | p Value |
| Total (54) | 10 (19) | 44 (81) | |
| Sex | |||
| Female (33) | 3 (9) | 30 (91) | |
| Male (21) | 7 (33) | 14 (67) | 0.035† |
| Age | |||
| <68 y (27) | 9 (33) | 18 (67) | |
| ≥68 y (27) | 1 (4) | 26 (96) | 0.005‡ |
| Site | |||
| Distal (4) | 2 (50) | 2 (50) | |
| Proximal (49) | 7 (14) | 42 (86) | 0.13† |
| Grade | |||
| Well/moderate (31) | 9 (29) | 22 (71) | |
| Poor (22) | 1 (5) | 21 (95) | 0.033† |
| TP53 | |||
| Wild-type (42) | 9 (21) | 33 (79) | |
| Mutant (8) | 1 (13) | 7 (87) | NS |
| K-ras | |||
| Wild-type (41) | 5 (12) | 36 (88) | |
| Mutant (8) | 4 (50) | 4 (50) | 0.028† |
| CpG methylation* | |||
| <2 sites (15) | 10 (67) | 5 (33) | |
| ≥2 sites (34) | 0 (0) | 34 (100) | <0.001† |
*p16, MDR1, and MINT-2.
†Fisher’s exact test.
‡χ2 test.
Methylation and MTHFR genotype
The C677T polymorphism in the MTHFR gene has only 30% the level of enzymatic activity when present as the homozygote (TT) compared with the wild-type.32 Compared with age matched controls, we recently found the TT genotype to be twice as frequent in CRC patients with MSI+ tumours.23 We hypothesised that aberrant methyl group metabolism in these individuals could predispose them to CIMP+ CRC. In the present study we investigated possible associations between MTHFR genotype and tumour specific methylation at CpG islands. Because of the relatively small numbers, individuals with the TT genotype were combined with CT genotype individuals. No associations between CIMP+ and the CT/TT genotype were observed for the entire group (table 4 ▶). However, a twofold increased risk of CIMP+ CRC was observed for CT/TT females compared with age matched controls. The CT/TT MTHFR genotype was also significantly associated with methylation of p16 (odds ratio 2.40 (95% confidence interval 1.19–4.85); p=0.01) and MINT-2 (1.97 (1.01–3.84); p=0.04) in females. When all MSI+ cases were excluded, significant associations between the MTHFR genotype and CIMP+, p16, or MINT-2 methylation were still observed. These results suggest that a CT/TT MTHFR genotype can predispose females to the development of CIMP+ tumours independently of MSI status.
Table 4.
Risk of CIMP+ and CIMP− colorectal cancer (CRC) according to the methylenetetrahydrofolate reductase (MTHFR ) genotype
| Females* (n=118) | Males* (n=109) | |
| Controls v all CRC | 1.13 (0.75–1.71) | 0.92 (0.60–1.39) |
| Controls v CIMP+† | 2.17 (1.03–4.57)‡ | 0.68 (0.32–1.42) |
| Controls v CIMP− | 0.84 (0.52–1.35)† | 1.03 (0.60–1.39) |
*Odds ratio and 95% confidence interval for frequency of the CT or TT genotype in CRC patients compared with age matched control group.
†Defined as having two or more CpG sites methylated.
‡p=0.037.
CIMP+, CpG island methylator phenotype.
DISCUSSION
In the present study, we examined methylation of p16, MDR1, and MINT-2 in a large unselected series of CRC. These cases were well characterised for clinicopathological features, including patient survival, as well as for various molecular alterations. The major findings were: (i) CIMP+ tumours were frequently poorly differentiated, often occurred in proximal or higher stage tumours, and had a lower frequency of TP53 mutations compared with CIMP− tumours; (ii) methylation of the CpG sites investigated had no prognostic value for patients treated by surgery alone; (iii) hMLH1 methylation status defined two groups of MSI+ tumours with different features; and (iv) females with the CT or TT MTHFR genotype were at increased risk of developing CIMP+ CRC.
Although CIMP+ was originally described using six CpG islands,4 no consensus panel currently exists for defining this phenotype. In a later publication the same authors stated that investigation of 2–4 CpG loci may be sufficient for identification of CIMP+ CRC.11 Previous studies have shown that CIMP+ tumours are characterised by concordant methylation of multiple type C CpG islands such as p16 and MINT-2.3–5 Our earlier work suggested that MDR1 was also a type C gene15 and hence in the current study we arbitrarily defined tumours with methylation at two or more of p16, MINT-2, and MDR1 as being CIMP+. Taking into account that most previous studies have been carried out on selected CRC series, the frequencies observed here for p16, MDR1, and MINT-2 methylation (table 1 ▶) are similar to those reported previously.4,9,13,15 These tumours comprised 32% of the current unselected stage II/III CRC series and were characterised by the features of poor histological grade and proximal tumour location. Previous studies on p164,9,10 and MINT-24 methylation have reported the same positive associations with tumour grade and site of origin. In agreement with a previous study,11 the present results show a lower frequency of TP53 mutation in CIMP+ tumours (table 2 ▶). The incidence of K-ras mutations in methylated tumours is less clear, with some studies reporting a higher incidence in CIMP+11 or p16 methylated14 tumours, but not others for p16 methylation alone.12,13 The present results show a trend for lower frequency of K-ras mutations in CIMP+ but this was due mainly to the low frequency of K-ras mutations in the closely related MSI+ group.22
To date, little is known of the prognostic significance of CpG island methylation in CRC. A study in 84 stage III CRC patients found an association between p16 methylation and shortened survival.12 However, the current work and that of Esteller and colleagues13 found no prognostic significance for p16 methylation alone. Methylation of MDR1, MINT-2, or multiple sites was also found to have no prognostic value for patients treated by surgery alone in the present study. We previously reported evidence for a greater extent of survival benefit from 5-FU based chemotherapy for CRC patients with proximal tumours compared with those with distal tumours, with the benefit appearing to be most pronounced for patients with MSI+ tumours.17 Interestingly, in the present study we found 44% of proximal (table 1 ▶) and 71% of MSI+ tumours (table 2 ▶) to be CIMP+. The degree of survival benefit from 5-FU based therapy appears therefore to correlate with the frequency of CIMP+. This phenotype is characterised by aberrant DNA methylation and we hypothesise that CIMP+ may be an important predictive factor for the response of CRC to antifolate therapies.
It was recently shown that hMLH1 methylation is more common in females compared with males.7 In both sexes, patients with hMLH1 methylated MSI+ tumours were on average 20 years older compared with those with no hMLH1 methylation.7 In the present study of 54 MSI+ tumours, similar sex and age differences were also noted between the two subgroups defined by hMLH1 methylation status (table 3 ▶). Tumours showing hMLH1 methylation were also more likely to be poorly differentiated, CIMP+, and to have wild-type K-ras in comparison with MSI+ tumours with unmethylated hMLH1. Kuismanen and colleagues8 reported similar tumour site differences between sporadic MSI+ cases depending on their hMLH1 methylation status. As shown in table 3 ▶, the large majority (34/39, 87%) of hMLH1 methylated tumours are CIMP+. Hence the above results suggest the existence of two groups of MSI+ tumours that can be distinguished by CIMP+ status and that are likely to evolve through different pathways. CIMP−/MSI+ tumours result from mutations and/or LOH of mismatch repair genes, whereas CIMP+/MSI+ tumours arise following widespread methylation of tumour DNA including hMLH1. The low number of CIMP−/MSI+ cases (n=10) prevents us from carrying out further characterisation of this subgroup. Our results (table 3 ▶) confirm those of previous studies6–8 showing that approximately 50–90% of sporadic MSI+ CRCs have methylated hMLH1. Since the frequency of MSI+ in population based CRC is approximately 10–15%, we therefore estimate the incidence of hMLH1 methylation to be about 7–12%. This is 2–3-fold lower than the frequencies of methylation observed for p16, MDR1, and MINT-2. The factors responsible for this difference have not been identified but may be gene, tumour, or host related.
Work by Slattery and colleagues33,34 has implicated lifestyle factors such as diet, smoking, oestrogen exposure, and alcohol consumption in the risk of developing MSI+ CRC. We recently reported that the TT MTHFR genotype could also predispose to MSI+ CRC.23 As the majority of sporadic MSI+ cases are CIMP+, it could be expected that these same lifestyle and genotypic factors are involved in the development of tumours with aberrant DNA methylation. Individuals with the TT MTHFR genotype have impaired remethylation of homocysteine to methionine,32 hence we speculated this could predispose them to the development of CIMP+ CRC. In the current study, we present evidence for a twofold increased risk of CIMP+ CRC in females, but not males, with the CT/TT MTHFR genotype (table 4 ▶). Although several previous studies have investigated associations between MTHFR genotype and the overall risk of CRC,35–37 this is the first study that has specifically examined the association between this genotype and a CRC subgroup characterised by DNA hypermethylation. Prospective studies that incorporate genotypic, dietary, and lifestyle factors are required to determine the relative contribution that each makes to the risk of developing CIMP+ tumours and may also shed light on the apparent sex difference observed here.
The site related distribution of CIMP+ observed in the current and previous studies4,9,10 add to the growing list of differences between proximal and distal CRC.38–40 We speculate that the higher incidence of CIMP+ tumours in the proximal compared with the distal colon may account for our previously reported site difference in the degree of survival benefit from antifolate based chemotherapy.17 Our results and those of others7,8 also reveal important differences between MSI+ tumours with or without hMLH1 methylation. The present work is the first to identify a genotypic risk factor for the development of CIMP+ CRC. Dietary and lifestyle factors are no doubt also involved in the aetiology of this CRC subgroup. A recent report suggesting that hyperplastic polyps and serrated adenomas may be the precursors of MSI+ CRCs with methylated hMLH141 should facilitate further investigations into the development of MSI+ and the closely related CIMP+ tumours.
Acknowledgments
This work was supported by grants from the Raine Foundation, the Cancer Foundation of Western Australia, and the Department of Radiation Oncology of the Sir Charles Gairdner Hospital, Nedlands, Australia.
Abbreviations
CRC, colorectal cancer
MSI, microsatellite instability
CIMP+, CpG island methylator phenotype
MTHFR, methylenetetrahydrofolate reductase
PCR, polymerase chain reaction
MSP, methylation sensitive PCR
SSCP, single strand conformation polymorphism
5-FU, 5-fluorouracil
REFERENCES
- 1.Baylin SB, Herman JG, Graff JR, et al. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res 1998;72:141–96. [PubMed] [Google Scholar]
- 2.Jones PA, Laird PW. Cancer epigenetics comes of age. Nat Genet 1999;21:163–7. [DOI] [PubMed] [Google Scholar]
- 3.Ahuja N, Li Q, Mohan AL, et al. Aging and DNA methylation in colorectal mucosa and cancer. Cancer Res 1998;58:5489–94. [PubMed] [Google Scholar]
- 4.Toyota M, Ahuja N, Ohe-Toyota M, et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 1999;96:8681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Toyota M, Shen L, Ohe-Toyota M, et al. Aberrant methylation of the cyclooxygenase 2 CpG island in colorectal tumours. Cancer Res 2000;60:4044–8. [PubMed] [Google Scholar]
- 6.Herman JG, Umar A, Polyak K, et al. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci USA 1998;95:6870–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malkhosyan SR, Yamamoto H, Piao Z, et al. Late onset and high incidence of colon cancer of the mutator phenotype with hypermethylated hMLH1 gene in women. Gastroenterology 2000;119:598. [DOI] [PubMed] [Google Scholar]
- 8.Kuismanen SA, Holmberg MT, Salovaara R, et al. Genetic and epigenetic modification of MLH1 accounts for a major share of microsatellite-unstable colorectal cancers. Am J Pathol 2000;156:1773–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiencke JK, Zheng S, Lafuente A, et al. Aberrant methylation of p16INK4a in anatomic and gender-specific subtypes of sporadic colorectal cancer. Cancer Epidemiol Biomarkers Prev 1999;8:501–6. [PubMed] [Google Scholar]
- 10.Burri N, Shaw P, Bouzourene H, et al. Methylation silencing and mutations of the p14ARF and p16INK4a genes in colon cancer. Lab Invest 2001;81:217–29. [DOI] [PubMed] [Google Scholar]
- 11.Toyota M, Ohe-Toyota M, Ahuja N, et al. Distinct genetic profiles in colorectal tumours with or without the CpG island methylator phenotype. Proc Natl Acad Sci U S A 2000;97:710–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liang JT, Chang KJ, Chen JC, et al. Hypermethylation of the p16 gene in sporadic T3N0M0 stage colorectal cancers: association with DNA replication error and shorter survival. Oncology 1999;57:149–56. [DOI] [PubMed] [Google Scholar]
- 13.Esteller M, Gonzalez S, Risques RA, et al. K-ras and p16 aberrations confer poor prognosis in human colorectal cancer. J Clin Oncol 2001;19:299–304. [DOI] [PubMed] [Google Scholar]
- 14.Guan RJ, Fu Y, Holt PR, et al. Association of K-ras mutations with p16 methylation in human colon cancer. Gastroenterology 1999;116:1063–71. [DOI] [PubMed] [Google Scholar]
- 15.Shannon BA, Iacopetta BJ. Methylation of the hMLH1, p16, and MDR1 genes in colorectal carcinoma: associations with clinicopathological features. Cancer Lett 2001;167:91–7. [DOI] [PubMed] [Google Scholar]
- 16.Esteller M, Tortola S, Toyota M, et al. Hypermethylation-associated inactivation of p14(ARF) is independent of p16(INK4a) methylation and p53 mutational status. Cancer Res 2000;60:129–33. [PubMed] [Google Scholar]
- 17.Elsaleh H, Joseph D, Grieu F, et al. Association of tumour site and sex with survival benefit from adjuvant chemotherapy in colorectal cancer. Lancet 2000;355:1745–50. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Zulueta M, Bender CM, Yang AS, et al. Methylation of the 5′ CpG island of the p16/CDKN2 tumour suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res 1995;55:4531–5. [PubMed] [Google Scholar]
- 19.Herman JG, Merlo A, Mao L, et al. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res 1995;55:4525–30. [PubMed] [Google Scholar]
- 20.Tada Y, Wada M, Kuroiwa K, et al. MDR1 gene overexpression and altered degree of methylation at the promoter region in bladder cancer during chemotherapeutic treatment. Clin Cancer Res 2000;6:4618–27. [PubMed] [Google Scholar]
- 21.Elsaleh H, Powell B, McCaul K, et al. P53 alteration and microsatellite instability have predictive value for survival benefit from chemotherapy in stage III colorectal carcinoma. Clin Cancer Res 2001;7:1343–9. [PubMed] [Google Scholar]
- 22.Gnanasampanthan G, Elsaleh H, McCaul K, et al. Ki-ras mutation type and the response of Dukes’ C colorectal caner to adjuvant chemotherapy. J Pathol 2001;195:543–8. [DOI] [PubMed] [Google Scholar]
- 23.Shannon B, Gnanasampanthan S, Beilby J, et al. A polymorphism in the methylenetetrahydrofolate reductase gene predisposes to colorectal cancers with microsatellite instability. Gut 2001;50:520–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moertel CG, Fleming TR, Macdonald JS, et al. Fluorouracil plus levamisole as effective adjuvant therapy after resection of stage III colon carcinoma: a final report. Ann Intern Med 1995;122:321–6. [DOI] [PubMed] [Google Scholar]
- 25.Ueki T, Toyota M, Sohn T, et al. Hypermethylation of multiple genes in pancreatic adenocarcinoma. Cancer Res 2000;60:1835–9. [PubMed] [Google Scholar]
- 26.Paulin R, Grigg GW, Davey MW, et al. Urea improves efficiency of bisulphite-mediated sequencing of 5′-methylcytosine in genomic DNA. Nucleic Acids Res 1998;26:5009–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iacopetta B, Grieu F. Routine detection of the replication error phenotype in clinical tumour specimens using fluorescence-SSCP. Biotechniques 2000;28:566–8, 70. [DOI] [PubMed] [Google Scholar]
- 28.Zhou XP, Hoang JM, Li YJ, et al. Determination of the replication error phenotype in human tumours without the requirement for matching normal DNA by analysis of mononucleotide repeat microsatellites. Genes Chromosomes Cancer 1998;21:101–7. [DOI] [PubMed] [Google Scholar]
- 29.Soong R, Powell B, Elsaleh H, et al. Prognostic significance of TP53 gene mutation in 995 cases of colorectal carcinoma. Influence of tumour site, stage, adjuvant chemotherapy and type of mutation. Eur J Cancer 2000;36:2053–60. [DOI] [PubMed] [Google Scholar]
- 30.Bian YS, Yan P, Osterheld MC, et al. Promoter methylation analysis on microdissected paraffin-embedded tissues using bisulfite treatment and PCR-SSCP. Biotechniques 2001;30:66–72. [DOI] [PubMed] [Google Scholar]
- 31.Ahuja N, Mohan AL, Li Q, et al. Association between CpG island methylation and microsatellite instability in colorectal cancer. Cancer Res 1997;57:3370–4. [PubMed] [Google Scholar]
- 32.Kim YI. Methylenetetrahydrofolate reductase polymorphisms, folate, and cancer risk: a paradigm of gene-nutrient interactions in carcinogenesis. Nutr Rev 2000;58:205–9. [DOI] [PubMed] [Google Scholar]
- 33.Slattery ML, Potter JD, Curtin K, et al. Estrogens reduce and withdrawal of estrogens increase risk of microsatellite instability-positive coloncancer. Cancer Res 2001;61:126–30. [PubMed] [Google Scholar]
- 34.Slattery ML, Anderson K, Curtin K, et al. Dietary intake and microsatellite instability in colon tumors. Int J Cancer 2001;93:601–7. [DOI] [PubMed] [Google Scholar]
- 35.Slattery ML, Potter JD, Samowitz W, et al. Methylenetetrahydrofolate reductase, diet, and risk of colon cancer. Cancer Epidemiol Biomarkers Prev 1999;8:513–18. [PubMed] [Google Scholar]
- 36.Ma J, Stampfer MJ, Giovannucci E, et al. Methylenetetrahydrofolate reductase polymorphism, dietary interactions, and risk of colorectal cancer. Cancer Res 1997;57:1098–102. [PubMed] [Google Scholar]
- 37.Chen J, Giovannucci E, Kelsey K, et al. A methylenetetrahydrofolate reductase polymorphism and the risk of colorectal cancer. Cancer Res 1996;56:4862–4. [PubMed] [Google Scholar]
- 38.Bufill JA. Colorectal cancer: evidence for distinct genetic categories based on proximal or distal tumour location. Ann Intern Med 1990;113:779–88. [DOI] [PubMed] [Google Scholar]
- 39.Breivik J, Lothe RA, Meling GI, et al. Different genetic pathways to proximal and distal colorectal cancer influenced by sex-related factors. Int J Cancer 1997;74:664–9. [DOI] [PubMed] [Google Scholar]
- 40.Lindblom A. Different mechanisms in the tumourigenesis of proximal and distal colon cancers. Curr Opin Oncol 2001;13:63–9. [DOI] [PubMed] [Google Scholar]
- 41.Hawkins NJ, Ward RL. Sporadic colorectal cancers with microsatellite instability and their possible origin in hyperplastic polyps and serrated adenomas. J Natl Cancer Inst 2001;93:1307–13. [DOI] [PubMed] [Google Scholar]