Abstract
Background: Mucins are large complex glycoproteins that protect intestinal mucosal surfaces by limiting access of environmental matter to their epithelial cells. Several mucin genes have been described, including MUC3 that is a membrane associated mucin of the small intestine. Increased MUC3 mRNA transcription is induced by incubation of intestinal epithelial cells with a Lactobacillus strain known to be adherent to them.
Aims: To determine whether increased epithelial cell MUC3 mucin expression in response to Lactobacillus strains results in increased extracellular secretion of MUC3 mucins and the importance of epithelial cell adherence in modulation of MUC3 mucin expression.
Methods: HT29 cells grown to enhance expression of MUC3 mucins were incubated with selected Lactobacillus strains. Spent cell culture medium was collected for detection of secreted MUC3 mucins using dot blot immunoassay with a generated MUC3 antibody. Post-incubation HT29 cell RNA was collected for analysis of MUC3 expression by northern blot analysis using a MUC3 cDNA probe. In vitro binding studies using Lactobacillus strains incubated alone or coincubated with enteropathogenic Escherichia coli strain E2348/69 were used for adherence and inhibition of adherence studies, respectively.
Results: Lactobacillus strains with minimal ability to adhere to HT29 cells failed to induce upregulation of mucin gene expression. There was a direct correlation between upregulation of MUC3 mucin mRNA expression and extracellular secretion of MUC3 mucin. The same Lactobacillus strains that increased extracellular secretion of MUC3 mucin led to reduced adherence of enteropathogen E coli E2348/69 during coincubation experiments.
Conclusion: Probiotic microbes induce MUC3 mucin transcription and translation with extracellular secretion of the MUC3 mucins. Epithelial cell adherence enhances the effects of probiotics on eukaryotic mucin expression.
Keywords: Lactobacillus, Escherichia coli, mucin, adherence
Mucosal surfaces employ a number of protective strategies to defend against noxious substances and pathogens found within the intestinal lumen. The intestinal epithelial cells lining the intestinal tract provide both a physical barrier and have evolved inducible innate protective strategies that offer rapid responses to pathogenic challenge. These include secretion of salt and water, and elaboration of antibacterial peptides or more complex molecules such as mucins.1 Mucins are complex glycoproteins synthesised and secreted by epithelial cells of a number of organ systems, including the mucosal epithelial cells of the intestinal tract. Mucins may protect epithelial cells from microbial pathogens by limiting their access through simple steric hindrance, by providing a physicochemical barrier, or through specific mucin-bacterial/viral interactions.2 Preventing epithelial cell adherence interrupts colonisation, toxin delivery, and invasion. Small intestinal mucins have also been reported to inhibit viral replication,3 which may limit disease and speed shedding of viral pathogens from the intestinal tract.
At least 12 mucin genes have been identified. MUC2 mucin exists only as a secreted form and is the major secreted mucin of the colon.4 MUC3 mucin is not highly expressed in the colon4 but is expressed in both goblet cells and enterocytes of the small intestine.5 It is a member of the membrane bound mucins6 with a 3′ terminus containing exons coding for two epithelial growth factor-like domains, a transmembrane domain and a cytoplasmic tail.7 However, by an alternative splicing mechanism, the MUC3 gene may encode variants in which some aforementioned domains are deleted,7,8 suggesting secreted forms of MUC3 mucins may also be produced.
We have previously shown that coincubation of Lactobacillus plantarum strain 299v (Lp299v) and L rhamnosus strain GG (LrGG) with an enteropathogenic Escherichia coli (EPEC) strain E2348/69 or enterohaemorrhagic E coli O157:H7 strain inhibited the adherence of either of the E coli strains to mucin producing intestinal epithelial cells.9 We also showed that incubation of Lp299v with HT29 cells could upregulate expression of MUC3 mRNA.9 One of the basic properties used to identify Lp299v and LrGG as potentially useful probiotics is their ability to adhere to intestinal epithelial cells.10,11
To determine whether adherence is important to the process of mucin gene upregulation, we studied adherence of different strains of Lactobacillus, including a mutant strain lacking an adhesin, and measured alteration in the expression of mucin gene mRNA expression. In addition, we evaluated whether the MUC3 mucin produced was a secreted form by antibody analysis of spent cell culture medium using an antibody generated for this purpose. Finally, to show that the mucin product secreted into the cell culture medium was biologically active, we employed a bioassay to study inhibition of adherence of an enteric pathogen to the mucin producing epithelial cells.
MATERIALS AND METHODS
Bacteria and growth conditions
A stock culture of EPEC strain E2348/69 (serotype O127: H6) was maintained at 4°C on trypticase soy agar slants (Becton-Dickenson Microbiology Systems, Cockeysville, Maryland, USA). Stocks of Lactobacillus strains were maintained at 4°C on MRS agar (Difco Laboratories, Detroit, Michigan, USA). Bacterial strains were kindly provided by Dr James Kaper (EPEC strain E2348/69; Center for Vaccine Development, Baltimore, Maryland, USA), and Dr Khem Shahani (L acidophilus strain DDS-1 (LaDDS); University of Nebraska, Lincoln, Nebraska, USA).
EPEC strain E2348/69 is a member of the family of non-invasive, non-enterotoxin, diarrhoeagenic producing pathogens that produce a characteristic attachment/effacement lesion with epithelial cells.12 LrGG (American Type Culture Collection 53103, Rockville, Maryland, USA) was originally isolated in vitro from stool specimens of healthy humans.13 The LaDDS strain is from a dairy source and has been reported to stimulate murine macrophage production of interleukin 1α and tumour necrosis factor α.14 Lp299v is a member of a genetically well defined subgroup of L plantarum isolated from intestinal mucosa.15 Strains of this subgroup agglutinate Saccharomyces cerevisiae in a mannose sensitive manner and carry a mannose specific adhesin.16 The L plantarum strain adh− (Lp adh−) is a spontaneous mutant of Lp299v that no longer agglutinates S cerevisiae in a mannose sensitive manner. Restriction fragment length polymorphism analysis did not detect differences between Lp299v and its derivative Lp adh− (Siv Ahrne, personal communication).
E coli and Lactobacillus strains were grown in oxic conditions overnight at 37 °C in static, non-aerated Penassay (Difco), or MRS broth (Difco), harvested by centrifugation and quantified by determination of colony forming units (CFU), as previously described.9
Cell growth conditions
HT29 cells (American Type Culture Collection) were grown in McCoy’s 5a culture medium (Life Technologies, Gaithersburg, Maryland, USA) for enhanced MUC2 expression. Cells were progressively transferred from the usual glucose containing medium to a glucose free 5 mM galactose containing McCoy’s 5a culture medium to increase MUC3 mRNA expression and reduce MUC2 mRNA expression.17 Culture medium was supplemented with 10% heat inactivated qualified fetal bovine serum (Life Technologies). For bacteria free studies, an antibiotic/antimycotic mixture (100 U/ml penicillin G, 100 mg/ml streptomycin sulphate, and 0.25 mg/ml amphotericin B; Life Technologies) was added to cell culture medium. Cell cultures were grown at 37°C in a humidified atmosphere with 5% CO2 and were passaged after washing with Earle’s balanced salt solution (Life Technologies) using trypsin-EDTA (Life Technologies).
Microbial adherence assays
To evaluate binding of Lactobacillus strains to HT29 cells, 106 Lactobacillus CFU/well (5×105 CFU/ml) in 0.1 ml phosphate buffered saline (PBS), at pH 7.4, 25°C, were added to HT29 cells. After four hours of incubation at 37°C, cell culture medium was aspirated off the cells and cells were washed four times with Dulbecco’s PBS (pH 7.4, 37°C) to remove non-bound bacteria. Cells were released from polystyrene wells by adding 0.25 ml of trypsin-EDTA for 10 minutes. Then ice cold sterile PBS was added to each well followed by agitation to dissociate well contents. Serial dilutions of bacteria were plated on MRS agar and incubated overnight at 37°C for subsequent CFU quantification. All experiments were run in triplicate and results are based on at least five separate experiments. Viability of the microbes in cell culture medium without cells for the same incubation period and during the washing procedures was also determined.
The in vitro inhibition of EPEC adherence assay was similar to that previously described.9 Briefly, 109 Lactobacillus CFU/well in 0.1 ml PBS, pH 7.4, 25°C, were added to 1.9 ml of antibiotic free cell growth medium (5×108 CFU/ml) for an hour prior to addition of EPEC to maximise the effect. EPEC strain E2348/69 (106 CFU in 0.1 ml PBS, pH 7.4, 25°C) was added to wells (5×105 CFU/ml) of a 12 well polystyrene tray (Fisher) and incubated with the cells for three hours at 37°C. The inoculum of EPEC is a minimal infective dose in humans18 and does not increase mucin gene transcription.9 Quantification of adherent EPEC was performed by serial dilutions and enumeration of CFU on MacConkey agar. All experiments were run in triplicate.
Alteration in MUC2 and MUC3 mRNA expression
HT29 cells were grown to confluence in culture flasks (Falcon 3028; Becton-Dickinson, Franklin Lakes, New Jersey, USA). The same relative number of bacteria to cell contact area were added to culture flasks as were added to wells of 12 well plates used for adherence assays. Specifically, 4.5×1010 CFU/flask (1.5×109 CFU/ml) were added as the cell contact area of the culture flask is 45× greater than the cell contact area of a well of 12 well plates.
Total RNA was isolated from HT29 cells using the guanidine isothiocyanate-cesium chloride cushion ultracentrifugation technique with modifications to minimise mRNA shearing, as previously described.9,19 RNA was stored in 0.3 M sodium acetate with 2.5 volumes of ethanol at −70°C. Northern blots were formed and underwent probe hybridisation using either a cDNA probe to the tandem repeat region of MUC2 (clone SMUC41) or MUC3 (clone SIB 124; kindly provided by Drs James Gum and Young Kim), as previously described.9 Analysis of mucin gene signals using Phosphor screen autoradiography (Molecular Dynamics, Sunnyvale, California, USA) and area integration using ImageQuant software (version 3.3; Molecular Dynamics) relative to levels of 28s RNA were as previously described.9
Antibody to MUC3 peptide
To compare transcription levels of MUC3 mRNA to translation expression, an antibody to detect protein product was required. Polyvalent antiserum was produced by repeated intraperitoneal injections of Balb/C mice with a KLH conjugated peptide sequence (TSR RTT RIT). This 9 mer peptide sequence was deduced from the cDNA sequence of the human MUC3 gene at a position on the carboxyl side to the tandem repeat region.7 The methodology to generate antiserum in mice received prior approval from the Institutional Animal Care and Utilization Committee of the University of Nebraska Medical Center and was performed by the University of Nebraska Medical Center Antibody Facility.
The reactivity and specificity of the antibody in mouse immune serum was determined by dot blot immunoassay, as described by Towbin and Gordon.20 Briefly, spent culture media overlying HT29 cells grown to enhance MUC2 or MUC3 expression were collected separately and to purify the mucins, cell culture medium was subjected to density gradient ultracentrifugation, as described previously.9,21 Twenty μg (Lowry protein) from a mucin enriched fraction was applied as a spot onto nitrocellulose paper strips and allowed to air dry for one hour. Nitrocellulose strips were incubated with 3% bovine serum albumin (Sigma, St Louis, Missouri, USA) in 10 mM Trizma base (Sigma) with 0.9% saline at pH 7.4 for one hour at 37°C. Strips were then washed with Tris saline. Mouse immune serum was added in serial dilutions for incubation overnight at 4°C. Afterwards, the strips were washed with Tris saline at room temperature to remove the unbound antibody. A 1:5000 dilution of peroxidase conjugated goat antimouse IgG (Fcγ fragment specific; Jackson ImmunoResearch, West Grove, Pennsylvania, USA) was added to the nitrocellulose strips and incubated for one hour at room temperature. Horseradish peroxidase colour development reagent containing 4-chloro-1-naphthol (Bio-Rad, Richmond, California, USA) was added to develop any positive reaction.
Dot blot immunoassay was performed in a similar fashion for spent cell culture medium from HT29 cells grown in galactose containing medium incubated with Lp299v, LrGG, and LaDDS to quantify whether increased secreted MUC3 mucins were present. For this study, the cell culture media were changed to serum free medium prior to the addition of different Lactobacillus strains. After six hours of incubation, each cell culture medium was collected separately, and to minimise proteolytic degradation of native mucins22 protease inhibitors were added. The inhibitors included 5 mM N-ethylmaleimide (Sigma), 2 mM phenylmethylsulphonyl fluoride (Sigma), and 0.01% sodium azide (Sigma) in a 5 mM EDTA solution (Sigma). Spent culture medium was centrifuged to remove cellular and particulate debris and then passed through a 0.02 μm filter for sterilisation. The volume of soluble supernatant was reduced by vacuum centrifugation (SpeedVac SC200; Savant Instruments, Farmingdale, New York, USA). Equivalent amounts from the spent culture supernatants from cells incubating with different Lactobacillus strains (20 μg protein) were applied as a spot onto nitrocellulose paper and air dried, and the experimentation proceeded as above. For these experiments, mouse immune serum was added at a dilution of 1:100.
Indirect immunofluorescence
To further validate that the antibody generated was to MUC3 mucin, we performed indirect immunofluorescence on normal human tissue as tissue localisation in the small intestine is known.5 Biopsies of normal human duodenum were quick frozen in liquid nitrogen, fixed with OCT fixative, and stored at −70°C. Sections were cut 5 μm thick with a cold microtome and fixed onto microscope slides using acetate. After being warmed to room temperature and washed with PBS, the microscope slides were incubated for 30 minutes at room temperature in a 1:20 dilution of mouse anti-MUC3 mucin serum. The microscope slides were again washed with PBS, and a 1:20 dilution of fluorescein isothiocyanate conjugated to goat IgG/Fcγ fragment specific (Jackson ImmunoResearch) was added. The microscope slides were then incubated in darkness for 30 minutes at room temperature, washed with PBS, and examined by fluorescence microscopy. Control slides were stained with equal dilutions of PBS, fluorescein isothiocyanate alone, or preimmune mouse serum. The study protocol for obtaining a biopsy received prior approval from the Institutional Review Board of Children’s Hospital in Omaha.
Statistical analysis
Group data are expressed as means (SEM). Analyses between multiple groups were determined using one factor ANOVA with 95% confidence intervals. Post hoc ANOVA analyses were determined by Fisher’s protected least significant difference using the Statview software program (version 5.0.1; SAS Institute Inc., Cary, North Carolina, USA). Analysis between two groups were determined using the two tailed unpaired Student t test.
RESULTS
Adherence of Lactobacillus strains
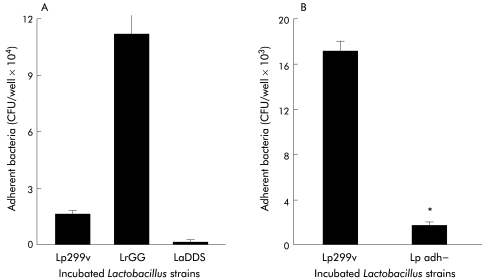
As shown in fig 1A▶, there were differences in adherence of the Lactobacillus strains to HT29 cells grown to enhance MUC3 expression. Adherence of LrGG (1.1×105 (1.1×104) CFU/well) was about 10× greater than Lp299v (1.6×104 (1.6×103) CFU/well) that was in turn about 10 times greater than LaDDS (1.3×103 (86) CFU/well). There was no loss of viability of any of these Lactobacillus strains in the galactose containing cell culture medium over a similar four hour incubation period (data not shown) or during the washing procedures. Similar results were determined for binding of the Lactobacillus strains to HT29 cells grown in glucose containing cell culture medium. That is, LrGG adherence to HT29 cells (1.1×105 (0.1×105) CFU/well; mean (SEM)) was greater than either Lp299v (1.6×104 (0.2×104) CFU/well; p<0.05) or LaDDS (1.3×103 (0.1×103) CFU/well; p<0.05).
Figure 1 .
(A) Lactobacillus adherence to HT29 cells. Lactobacillus strains 106 colony forming unit (CFU)/well were incubated for four hours with HT29 cells grown in glucose free galactose containing cell culture medium to enhance MUC3 expression. Non-adherent bacteria were washed off (×4) and adherent bacteria were quantified by CFU determination on MRS agar plates. Lp299v, Lactobacillus plantarum strain 299v; LrGG, L rhamnosus strain GG; LaDDS, L acidophilus strain DDS-1. (B) Similar experiments were performed using the parent Lp299v strain and its adhesin negative genetic mutant Lp adh− strain. In both, five experiments were run in triplicate and results are expressed as mean (SEM).
In separate experiments, we compared adherence of Lp299v with the mutant Lp adh− strain which has lost the ability to agglutinate Saccharomyces due to loss of production of the mannose sensitive binding ligand required for HT29 cell attachment.16 As shown in fig 1B▶, adherence of Lp299v (1.7×104 (6.9×102) CFU/well) to MUC3 expressing HT29 cells was about 10 times greater than the non-agglutinating Lp adh− strain (1.9×103 (1.8×102)) which showed minimal binding similar to the LaDDS strain in the previous set of experiments (fig 1A▶).
Mucin mRNA expression
Levels of MUC3 mRNA expression of HT29 cells coincubated with Lactobacillus strains are shown in fig 2▶. Expression levels of MUC3 mRNA increased about threefold with addition of either Lp299v or LrGG compared with control cells. Both Lp299v and LrGG increased mucin gene expression levels to the same magnitude over control cells that were grown in sterile media. However, there was no difference in MUC3 mRNA expression for cells incubated with LaDDS compared with controls (p>0.05). For comparison, we also evaluated MUC2 mRNA expression as it is known to exist only as a secreted form. Similar to MUC3 mRNA expression levels, ANOVA analysis revealed that MUC2 mRNA expression levels of incubation of HT29 cells grown to express MUC2 for LrGG (304 (52)% of control) and Lp299v (227 (49)% of control) were similar and both greater than MUC2 mRNA expression by control cells incubated without bacteria or HT29 cells incubated with LaDDS (153 (27)% of control) (data not shown).
Figure 2 .
Mucin mRNA expression induced by Lactobacillus strains. Lactobacilli (4.5×1010 colony forming units) were added to flasks in an equivalent amount as cell surface contact area of 12 well plates. After one hour of incubation, northern blots were hybridised using random-primed 32P labelled cDNA probe to the MUC3 tandem repeat. Mucin mRNA levels were quantified by area integration of Phosphor screen autoradiography. Results were normalised to 28s RNA levels on agarose gels used for northern blots. Results are expressed as means (SEM) of five experiments for HT29 cells were grown in a glucose free galactose containing cell culture medium to enhance MUC3 expression. As shown in (A), MUC3 expression for cells incubated with Lactobacillus plantarum strain 299v (Lp299v) and L rhamnosus strain GG (LrGG) were increased compared with controls without bacteria added and HT29 cells with L acidophilus strain DDS-1 (LaDDS) added (*p<0.05). (B) MUC3 mRNA expression was increased for Lp299v compared with the ligand negative Lp adh− strain (*p<0.05).
In separate experiments, MUC3 mRNA expression levels were evaluated for the non-adhering Lp adh− compared with the parent Lp299v strain (fig 2B▶). The relative MUC3 mRNA expression following incubation of Lp299v (209 (31)% of control) was greater than incubation of similarly grown HT29 cells incubated with the Lp adh− strain (101 (8.5)% of control; p<0.05). There was no difference between expression levels of HT29 cells incubated with Lp adh− bacteria and cells grown under sterile conditions (p>0.05).
MUC3 mucin antiserum
To evaluate the specificity of the antisera generated to the 9 mer MUC3 mucin backbone peptide sequence, we performed a dot blot immunoassay with mucins purified from HT29 cell spent culture medium. In fig 3▶, a black dot is present identifying the application site of the spent culture medium for HT29 cells grown in glucose containing medium to enhance MUC2 expression (site A) and for HT29 cells grown in glucose free galactose containing medium to enhance MUC3 mRNA expression (site B).17 Surrounding the application dot of site B, there is a strong reactivity to the anti-MUC3 immune serum but only minimal reaction around application site A. A minimal reaction might be expected as the HT29 cells grown in glucose containing cell culture medium conditions enhance MUC2 mucin expression but we have previously shown that there is minimal MUC3 expression present.17
Figure 3 .
Immunoassay of HT29 spent cell culture medium. Immune serum generated against a 9 mer peptide (TSRRTTRIT) of deduced MUC3 sequence was incubated with MUC2 (A) and MUC3 mucins (B) isolated from HT29 cell culture medium. Mucin protein (20 μg) was blotted onto nitrocellulose. A 1:100 dilution of mouse antisera was added. After washings, bound antibody was detected by a horseradish peroxidase colorisation reaction. A black dot in each frame is pencil marking used to indicate where medium was blotted initially. High reactivity was observed for spent culture medium of cells grown in galactose containing medium to enhance MUC3 expression. In contrast, there was less reactivity to material from spent culture medium of HT29 cells grown in a glucose containing medium that is associated with high MUC2 mucin mRNA expression and low MUC3 mucin mRNA expression.17
Indirect immunofluorescence of human small intestinal biopsy incubated with the mouse immune serum is shown in fig 4▶. The cross sectional view of a villus tip showed fluorescence along the surface of the epithelial cells. These results are similar to those previously published by Chang and colleagues5 using an antibody to the MUC3 tandem repeat region. Taken together, these results demonstrate the expected location for antigenic material derived from mucin and that the polyclonal antiserum reacts strongly with material enriched with MUC3 mucins.
Figure 4 .
Indirect immunofluorescence of human duodenum. The photomicrograph using mouse antihuman MUC3 mucin antibody as the primary antibody is shown. The cross section view of a villus from a biopsy taken from the second part of the duodenum shows fluorescence of the epithelial cell cytoplasm with increased fluorescence at the surface of the epithelial cells (arrows). Controls using preimmune serum, FITC conjugate alone, and phosphate buffered saline were negative (not shown). Magnification 40×.
MUC3 mucin extracellular expression
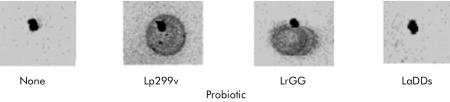
Anti-MUC3 serum was then used in a dot blot immunoassay using spent cell culture medium derived from HT29 cells grown in galactose containing medium incubated with different Lactobacillus strains. As shown in fig 5▶, a central black dot indicating the application site of the spent cell culture medium is visible in each panel. Surrounding the application spot of Lp299 and LrGG there was far greater reactivity than the blot of spent cell culture medium from HT29 cells without a probiotic added (none) or with LaDDS added.
Figure 5 .
Immunoassay for MUC3 mucins in spent cell culture medium from Lactobacillus strains. HT29 cells were grown in glucose free galactose containing medium to enhance MUC3 expression and incubated with equivalent colony forming units of different Lactobacillus strains. Cell culture medium (10 μg protein) was blotted onto nitrocellulose. A 1:100 dilution of mouse anti-MUC3 antisera was added. After washings, bound antibody was detected using a peroxidase colorisation reaction. A black dot in each frame is pencil marking used to indicate where medium was blotted initially. Greater immunoreactive MUC3 mucin was detected in cell culture medium incubated with Lactobacillus plantarum strain 299v (Lp299v) and L rhamnosus strain GG (LrGG) than with L acidophilus strain DDS-1 (LaDDS) and control cells without a probiotic added (none).
Inhibition of enteropathogenic Escherichia coli epithelial cell adherence
We correlated the biological significance of increased MUC3 gene transcription and secreted expression of MUC3 mucins in the presence of Lactobacillus strains differing in their ability to adhere to the HT29 cells by performing an in vitro inhibition of the adherence assay. For these experiments, controls were EPEC strain E2348/69 incubated with HT29 cells without Lactobacillus bacteria. As shown in fig 6A▶, recovery of EPEC bound to HT29 cells grown in galactose containing medium (for enhanced MUC3 expression) coincubated in the presence of Lp299v and LrGG was less than controls with EPEC alone. There was no difference in inhibition of EPEC coincubated with LaDDS compared with adherence of controls with EPEC alone for MUC3 expressing HT29 cells.
Figure 6 .
Comparative inhibition of enteropathogenic Escherichia coli (EPEC) adherence to MUC3 expressing HT29 cells. Each well received 109 colony forming units (CFU)/well Lactobacillus that were incubated at 37°C with the HT29 cells for an hour prior to the addition of 106 CFU/well of EPEC E2348/69. After a three hour incubation, EPEC adherent to cells were quantified by determining CFU on MacConkey agar. Results are expressed as means (SEM) of at least five independent experiments performed in triplicate. As shown in (A), both Lactobacillus plantarum strain 299v (Lp299v) and L rhamnosus strain GG (LrGG) decreased adherence of EPEC compared with controls of EPEC alone but L acidophilus strain DDS-1 (LaDDS) did not (*p<0.05). (B) The ligand negative strain Lp adh− showed less adherence than the parent ligand positive strain Lp299v (*p<0.05).
In fig 6B▶, the non-adherent Lp adh− strain was unable to impact the adherence of EPEC E2348/69 (8.8×105 CFU/well) to MUC3 producing HT29 cells. In contrast, Lp299v reduced the EPEC adherence (4.8×105 (3.8×1044)CFU/well) that is similar to the effect elicited by the adhering Lactobacillus strains depicted in fig 6A▶.
For comparison, we also evaluated inhibition of EPEC adherence to MUC2 producing HT29 cells coincubated with different Lactobacillus strains as MUC2 is the major mucin of the colon and exists only as a secreted mucin. For HT29 cells grown in glucose containing medium (for enhanced MUC2 expression), similar results to fig 6A▶ were determined. That is, there was decreased adherence of EPEC to HT29 cells with Lp299v (6.3×105 (0.7×105) EPEC CFU/well; p<0.005) and LrGG (1.0×105 (0.2×105) EPEC CFU/well; p<0.005) but not LaDDS (1.40×106 (2.5×105) EPEC CFU/well; p>0.05) compared with controls (1.60×106 (3.0×105) EPEC CFU/well (mean (SEM)).
DISCUSSION
In the current study we confirmed that incubation of Lp299v with HT29 cells upregulated expression of MUC3 mRNA. We also showed that other Lactobacillus strains may possess this same capability but it is not a universal property of probiotic strains (fig 2▶). To detect MUC3 mRNA expression, a cDNA probe to the central tandem repeat region was utilised. Recently, the existence of a second MUC3 gene has been proposed.23,24 Both human MUC3 genes, now called MUC3A and MUC3B, show such significant similarity that the cDNA probe that was used in these studies would recognise transcripts from both genes.23 MUC3 is a member of the membrane bound mucins along with MUC1, MUC4, and MUC12, and these various mucins can be expressed as membrane anchored, soluble, or secreted forms.6 Both membrane bound variants7 and splice variants coding for secreted forms7,8 have been described for MUC3 mucins. With upregulation of expression of MUC3 mRNA, the question then arises whether the mucins are cell anchored or secreted. This has biological consequences. If the upregulated mucin gene products were membrane bound, then in order for Lactobacillus strains to limit EPEC binding to intestinal epithelial cells all potential mucin binding sites for EPEC would have to be occupied by the Lactobacillus organisms. This would suggest there were common mucin binding sites for the gram positive Lactobacillus strains and the gram negative E coli. Alternatively, if the translated product of MUC3 mucin gene upregulation was a secreted mucin, then mucin EPEC interactions could occur away from the intestinal epithelial cell membrane surface and the trapped microbes could then be swept away with the peristaltic motions of the intestinal tract. As we demonstrated increased MUC3 mucins in the spent cell culture medium following probiotic incubation, the latter explanation is favoured. The rapidity of the response, which is within one hour, would match a biologically relevant time period and alteration of expression of other intestinal epithelial cell genes by non-virulent intestinal microbes has been demonstrated within a similar time frame.25 Rapidity of response would be expected to be an essential element for a member of the inducible innate protective response of the intestinal epithelial cell.
Traditionally, antibiotics have been administered for microbial related diseases of the intestinal tract. Their usage adds to the increasing concern regarding bacterial resistance or induction of other pathological processes such as haemolytic-uraemic syndrome,26 and this has led to a search for other interventions. Many enteric pathogens must first adhere to intestinal epithelial cells to initiate intestinal disease. Limiting access of pathogens to intestinal epithelial cells is one strategy to prevent disease that has been investigated previously. For example, the competitive inhibition of bacterial adherence by mimicry of receptors on the apical surface of enterocytes using oral administration of sialylated glycoproteins has been shown to protect newborn calves from an enterotoxigenic E coli strain K99.27 However, carbohydrate analogues or their synthetic imitators can be difficult and expensive to produce. Furthermore, identification of the causative organism for a given diarrhoeagenic to know which ligand should be administered takes time.
In contrast to specific glycomimetics, mucins are high molecular weight glycoproteins that express an array of glycan structures. There is a high content of carbohydrates with N-glycan oligosaccharides in the N and C terminal unique regions and O-glycosidic bonds between N-acetyl galactosamine and both serine and threonine in the central tandem repeat region of the mucin peptide backbone. In addition to N-acetyl galactosamine, other sugars found in O-glycans are fucose, N-acetyl glucosamine, galactose, and sialic acid. Terminal sugars that are attached to the core structures include galactose, N-acetyl glucosamine, fucose, and sialic acid.28 Thus this complex array of carbohydrates provides a broad range of potential sites for bacterial carbohydrate binding protein ligands of microbes. For EPEC, the adherence process is characterised by an initial relatively distant low affinity interaction mediated by the bundle forming pilus, followed by an essentially irreversible adherence to epithelial cells that is mediated via the protein intimin.12 We previously showed that exogenously added MUC3 mucins into an in vitro adherence assay system with EPEC incubating with non-mucin producing epithelial cells quantitatively inhibits EPEC adherence.9 Upregulation of intestinal mucins may provide both a rapid response and the potential to provide receptor mimicry against a broad range of organisms2 associated with diseases of the intestinal tract. Upregulation of mucin expression may explain the prevention of flare ups of chronic pouchitis following oral administration of probiotics reported by Gionchetti and colleagues29 as anaerobic bacterial concentrations correlate with the number of mucosal chronic inflammatory cells in pouchitis.30 In animal models, administration of Lactobacillus reduced the severity of mucosal inflammation of colitis in interleukin 10 gene deficient mice and methotrexate induced enterocolitis in rats. The diminished inflammation correlated with reduced adherent and translocated aerobic bacteria, and Enterobacteriaceae and gram negative anaerobes, respectively.31,32
In the current study, we have confirmed that preincubation of some probiotic strains of Lactobacillus with EPEC inhibit the adherence of the enteropathogen to intestinal epithelial cells. Not all Lactobacillus strains have this capacity as the mutant strain of Lp adh− and LaDDS failed to diminish the binding of EPEC E2348/69 compared with cells grown in a sterile cell culture medium. A difference between Lp adh− and LaDDS and the other two strains that we demonstrated was the ability to adhere to an intestinal epithelial cell. While LrGG had the greatest ability to bind to HT29 cells, Lp299v was demonstrated to have about 10 times greater ability to adhere to the cells compared with Lp adh− and LaDDS in our assay. The difference between LrGG and Lp299v may be a result of the vigour in which washings were performed—that is, LrGG may have a stronger binding strength than Lp299v. This would be in keeping with identification of LrGG in which ability to bind to epithelial cells was a major trait expected.13 Alternatively, upregulation of mucin mRNA may require a threshold value of adherent organisms per cell below which no effect is elicited. This suggests adherence is necessary but does not answer the question of whether it is sufficient, but irreversible adherence may not be as biologically important to define a useful probiotic as the ability to upregulate protective epithelial cell responses such as upregulation in mucin expression as we demonstrated mucin gene expression was similar between the Lp299v and LrGG strains.
The complete sequence of MUC3 and its promoter are unknown.6 In contrast, for the major secreted mucin of the colon, namely MUC2, both the full length cDNA sequence and regions important for its expression have been determined6 which has allowed for studies on its upregulation. For instance, ligation of ASGM1, a glycolipid that functions as a bacterial receptor, results in ATP release extracellularly, and by an autocrine mechanism mediated through activation of Erk 1/2 activates MUC2 mucin transcription.33 Whether similar pathways are involved in MUC3 activation remains to be determined.
In summary, selected probiotic Lactobacillus species that have the ability to adhere to intestinal epithelial cells rapidly induce eukaryotic MUC3 mucin expression. The upregulated MUC3 mucin gene product is a secreted mucin that has the ability to inhibit enteric pathogen epithelial cell adherence.
Acknowledgments
This work was supported by an unrestricted grant from Probi (DRM). The authors wish to thank Dr Deb Perry and Dr Tom Seemayer for their expertise in histology experiments.
Abbreviations
Lp299v
Lactobacillus plantarum strain 299v
LrGG, L rhamnosus strain GG
LaDDS, L acidophilus strain DDS-1
Lp adh−, L plantarum strain adh−
EPEC, enteropathogenic Escherichia coli
CFU, colony forming units
PBS, phosphate buffered saline
REFERENCES
- 1.Hecht G. Innate mechanisms of epithelial host defense: spotlight on intestine. Am J Physiol 1999;277:C351–8. [DOI] [PubMed] [Google Scholar]
- 2.Dai D, Nanthkumar N, Newburg DS, et al. Role of oligosaccharides and glycoconjugates in intestinal host defense. J Pediatr Gastroenterol Nutr 2000;30:S23–33. [PubMed] [Google Scholar]
- 3.Yolken RH, Ojeh C, Khatri IA, et al. Intestinal mucins inhibit rotavirus replication in an oligosaccharide-dependent manner. J Infect Dis 1994;169:1002–6. [DOI] [PubMed] [Google Scholar]
- 4.Ho SB, Niehans GA, Lyftogt C, et al. Heterogeneity of mucin gene expression in normal and neoplastic tissues. Cancer Res 1993;53:641–51. [PubMed] [Google Scholar]
- 5.Chang SK, Dohrman AF, Basbaum CB, et al. Localization of mucin (MUC2 and MUC3) messenger RNA and peptide expression in human normal intestine and colon cancer. Gastroenterology 1994;107:28–36. [DOI] [PubMed] [Google Scholar]
- 6.Moniaux N, Escande F, Porchet N, et al. Structural organization and classification of the human mucin genes. Front Biosci 2001;6:1192–206. [DOI] [PubMed] [Google Scholar]
- 7.Crawley SC, Gum JR, Hicks JW, et al. Genomic organization and structure of the 3′ region of human MUC3: alternative splicing predicts membrane-bound and soluble forms of the mucin. Biochem Biophys Res Comm 1999;263:728–36. [DOI] [PubMed] [Google Scholar]
- 8.Williams SJ, Munster DJ, Quin RJ, et al. The MUC3 gene encodes a transmembrane mucin and is alternatively spliced. Biochem Biophys Res Comm 1999;261:83–9. [DOI] [PubMed] [Google Scholar]
- 9.Mack DR, Michail S, Wei S, et al. Probiotics inhibit enteropathogenic E. coli adherence in vitro by inducing intestinal mucin gene expression. Am J Physiol 1999;276:G941–50. [DOI] [PubMed] [Google Scholar]
- 10.Johansson ML, Molin G, Jeppsson B, et al. Administration of different Lactobacillus strains in fermented oatmeal soap: in vivo colonization of human intestinal mucosa and effect on the indigenous flora. Appl Environ Microbiol 1993;59:15–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saxelin M, Elo S, Salminen S. Dose response colonization of feces after oral administration of Lactobacillus casei strain GG. Microbiol Ecol Health Dis 1991;4:209–14. [Google Scholar]
- 12.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clin Microbiol Rev 1998;11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorbach SL. The discovery of Lactobacillus GG. Nutr Today 1996;(suppl) 31:2–4S. [Google Scholar]
- 14.Rangavajhyala N, Shahani KM, Sridevi G, et al. Nonlipopolysaccharide component(s) of Lactobacillus acidophilus stimulate(s) the production of interleukin-1α and tumor necrosis factor-α by murine macrophages. Nutr Cancer 1997;28:130–4. [DOI] [PubMed] [Google Scholar]
- 15.Johansson ML, Quendan M, Ahrne S, et al. Classification of Lactobacillus plantarum by restriction endonuclease analysis of total chromosomal DNA using conventional agarose gel electrophoresis. Int J Systemic Bacteriol 1995;45:670–5. [Google Scholar]
- 16.Adlerberth I, Ahrne S, Johansson ML, et al. A mannose-specific adherence mechanism in Lactobacillus plantarum conferring binding to the human colonic cell line HT-29. Appl Environ Microbiol 1996;62:2244–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mack DR, Hollingsworth MA. Alteration in expression of MUC2 and MUC3 mRNA levels in HT29 colonic carcinoma cells. Biochem Biophys Res Comm 1994;199:1012–18. [DOI] [PubMed] [Google Scholar]
- 18.Levine MM, Berquist EJ, Nalin DR, et al. Escherichia coli strains that cause diarrhoea but do not produce heat-labile or heat-stable enterotoxins and are non-invasive. Lancet 1978;8074:1119–22. [DOI] [PubMed] [Google Scholar]
- 19.Debailleul V, Laine A, Huet G, et al. Human mucin genes MUC2, MUC3, MUC4, MUC5AC, MUC5B, and MUC6 express stable and extremely large mRNAs and exhibit a variable length polymorphism. J Biol Chem 1998;273:881–90. [DOI] [PubMed] [Google Scholar]
- 20.Towbin H, Gordon J. Immunoblotting and dot blot immunoblotting-current status and outlook. J Immunol Methods 1984;72:313–40. [DOI] [PubMed] [Google Scholar]
- 21.Mack DR, Gaginella TS, Sherman PM. Effect of colonic inflammation on mucin inhibition of Escherichia coli RDEC-1 binding in vitro. Gastroenterology 1992;102:1199–21. [PubMed] [Google Scholar]
- 22.Mantle M, Forstner GG, Forstner JF. Antigenic and structural features of goblet cell mucin of human small intestine. Biochem J 1984;217:159–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pratt WS, Crawley S, Hicks J, et al. Multiple transcripts of MUC3: evidence for two genes, MUC3A and MUC3B. Biochem Biophys Res Comm 2000;275:916–23. [DOI] [PubMed] [Google Scholar]
- 24.Kyo K, Muto T, Nagawa H, et al. Associations of distinct variants of the intestinal mucin gene MUC3A with ulcerative colitis and Crohn’s disease. J Hum Genet 2001;46:5–20. [DOI] [PubMed] [Google Scholar]
- 25.Neish AS, Gewirtz AT, Zeng H, et al. Prokaryotic regulation of epithelial responses by inhibition of Iκβ-α ubiquitination. Science 2000;289:1560–3. [DOI] [PubMed] [Google Scholar]
- 26.Wong CS, Jelacic S, Habeeb RL, et al. The risk of the hemolytic-uremic syndrome after antibiotic treatment of Escherichia coli O157:H7 infections. N Engl J Med 2000;342:1930–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mouricout M, Petit JM, Carias JR, et al. Glycoprotein glycans that inhibit adhesion of Escherichia coli mediated by K99 fimbriae: treatment of experimental colibacillosis. Infect Immun 1990;58:98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Forstner JF, Forstner GG. Gastrointestinal mucus. In: Johnson LR, ed. Physiology of the Gastrointestinal Tract, 3rd edn. New York: Raven Press, 1994:1255–82.
- 29.Gionchetti P, Rizzello F, Venturi A, et al. Oral bacteriotherapy as maintenance treatment in patients with chronic pouchitis: a double-blind, placebo-controlled trial. Gastroenterology 2000;119:305–9. [DOI] [PubMed] [Google Scholar]
- 30.Sandborn WJ. Pouchitis following ileal-pouch anal anastomosis: definition, pathogenesis, and treatment. Gastroenterology 1994;107:1856–60. [DOI] [PubMed] [Google Scholar]
- 31.Madsen KL, Doyle JS, Jewell LD, et al. Lactobacillus species prevents colitis in interleukin 10 gene-deficient mice. Gastroenterology 1999;116:1107–14. [DOI] [PubMed] [Google Scholar]
- 32.Mao Y, Nobaek S, Kasravi B, et al. The effects of Lactobacillus strains and oat fiber on methotrexate-induced enterocolitis in rats. Gastroenterology 1996;111:334–4. [DOI] [PubMed] [Google Scholar]
- 33.McNamara N, Khong A, McKemy D, et al. ATP transduces signals from ASGM1, a glycolipid that functions as a bacterial receptor. Proc Natl Acad Sci U S A 2001;98:9086–91. [DOI] [PMC free article] [PubMed] [Google Scholar]