Abstract
Background: Bipotent liver progenitor (oval) cells with the ability to differentiate into hepatocytes and biliary epithelium have recently been identified in human subjects with hepatitis C. Animal studies suggest that members of the tumour necrosis factor family, including lymphotoxin β (LT-β), regulate oval cell proliferation in liver disease, but its role in human liver disease is unclear.
Aims: This study seeks to establish a role for LT-β in hepatitis C related liver injury and to provide evidence that its increased expression is related to the presence of oval cells.
Methods: Liver biopsy specimens were obtained from patients with chronic hepatitis C virus (HCV) infection (n=20). Control liver samples (n=5) were obtained from liver resection or transplant surgery. LT-β expression in liver biopsy specimens was studied using quantitative real time polymerase chain reaction and immunohistochemistry.
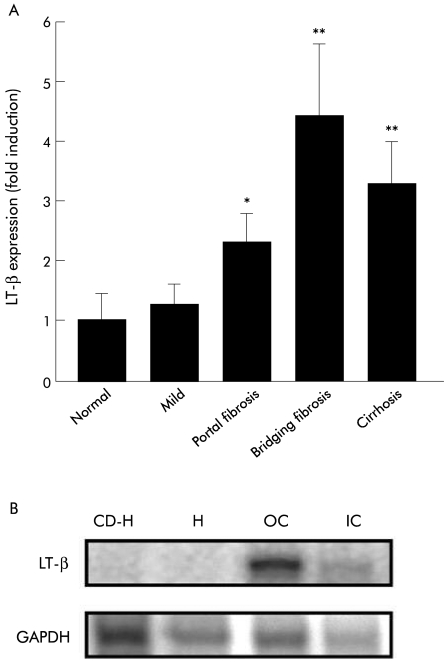
Results: LT-β mRNA levels were similar in control and HCV liver in the absence of fibrosis. In subjects with portal fibrosis, LT-β mRNA levels were elevated 2.2-fold over control liver levels (p=0.04). In subjects with bridging fibrosis, LT-β mRNA levels increased 4.4-fold over control liver levels (p=0.02). LT-β mRNA levels in subjects with established cirrhosis were increased 3.3-fold compared with controls and 2.6-fold compared with mild liver damage (p=0.02). Immunohistochemical analysis established that LT-β was expressed by oval cells, inflammatory cells, and small portal hepatocytes.
Conclusions: In chronic HCV infection, LT-β expression is observed in multiple hepatic cell types, including oval cells. LT-β expression is significantly increased when fibrosis or cirrhosis is present, suggesting a role for LT-β in the pathogenesis of chronic hepatitis C and a possible role in oval cell mediated liver regeneration.
Keywords: oval cell, liver, fibrosis, tumour necrosis factor
Hepatitis C virus (HCV) infection is associated with the development of chronic hepatitis, fibrosis, cirrhosis, and eventually hepatocellular carcinoma (HCC). Oval cells are a bipotent liver cell population of progenitor cells with the ability to differentiate into hepatocytes and biliary epithelium. They have been identified in human subjects with chronic HCV infection.1 Oval cells function as a facultative liver stem cell compartment and are induced to proliferate when the regenerative capacity of mature hepatocytes is compromised.2,3 There is a strong association between oval cell numbers and severity of liver injury that results from HCV infection.1 Furthermore, oval cell proliferation is associated with an increased risk of developing HCC in both rodents and humans, particularly when cirrhosis is present.2,3
In vivo studies have identified several potential factors that may mediate oval cell migration, proliferation, and/or differentiation during liver injury.4–8 The tumour necrosis factor (TNF) signalling pathway has an integral role in oval cell induction.4,9 Oval cell proliferation induced by 2-acetylaminofluorene administration followed by partial hepatectomy is inhibited by pretreatment with dexamethasone, an effect mediated by inhibition of nuclear factor κB (NFκB) activation downstream of TNF receptor 1 (TNFRI).9 The oval cell response induced by a choline deficient ethionine supplemented (CDE) feeding regimen is attenuated by 50% in mice lacking TNFRI, with a concurrent decrease in proliferation to a level of only 28% of wild-type mice.4 Thus it appears that TNF signalling through TNFRI is an integral mediator of liver regeneration via the oval cell compartment, although additional growth factors and/or signalling pathways must be involved as oval cell proliferation is not completely abrogated in TNFRI knockout mice.
Recently, we reported that lymphotoxin β (LT-β) promoter activity is inducible by cytokines shown to have key roles in the regenerative response following chronic liver injury.10 LT-β is a member of the TNF superfamily of ligands and receptors. It exists predominantly as a membrane bound heterotrimer with lymphotoxin α (LT-α) in a stoichiometry of LT-α1β2.11 This complex signals exclusively through the LT-β receptor (LT-βR) and is essential for secondary lymphoid organogenesis, follicular dendritic cell development, compartmentalisation of splenic B and T cells, and natural killer cell development.12–16 In vitro studies have shown that LT-α1β2/LT-βR binding can transduce both apoptotic and antiapoptotic signalling pathways.17,18 Recent reports also show that activation of the LT-β receptor induces production of chemokines and adhesion molecules required for recruitment and trafficking of lymphocytes to sites of inflammation.15,19,20
The core protein of HCV associates with LT-βR and this protein-protein interaction modulates cytolytic activity triggered by LT-α1β2/LT-βR interaction.21 Association of HCV core protein with LT-βR potentiates NFκB activation in most cell types, which in turn may contribute to the chronic persistence of HCV infected cells.21 The association between members of the TNF superfamily and liver disease,22–25 and LT-α1β2/LT-βR signalling and HCV,21,26 prompted this study of LT-β protein and gene expression in liver biopsy specimens using quantitative real time polymerase chain reaction (PCR) and immunohistochemistry, respectively, in order to establish the pattern of LT-β expression in hepatitis C related liver injury.
MATERIALS AND METHODS
Liver specimens
Liver biopsy specimens were obtained from 20 patients with chronic HCV infection. Subjects were diagnosed with chronic HCV on the basis of HCV antibody positivity, HCV PCR positivity, and elevated alanine aminotransferase. Other liver diseases were excluded by standard pathological testing. Varying degrees of liver injury were scored using a modified Knodell system.27 Cases of non-fibrotic liver damage (n=5) scored 0; portal fibrosis (n=5) scored 1–2; bridging fibrosis (n=5) scored 3–5; and cirrhosis (n=5) scored 6. Control liver samples were normal tissue obtained from either (1) patients undergoing hepatic resection for secondary malignancy without underlying liver disease per se or (2) donor liver at the time of transplantation. Part of the biopsy (∼3 mm) was snap frozen in liquid nitrogen and stored at −80°C prior to RNA extraction. The remaining tissue was fixed in 10% buffered formalin and embedded in paraffin for pathological examination and immunohistochemistry. Approval for the study was obtained from the Fremantle Hospital Human Research Ethics Committee.
Quantitative real time PCR analysis
RNA was extracted from liver specimens using RNasol B (Bresatec, USA) according to the manufacturer’s instructions. cDNA was synthesised from 1 μg of total RNA using the Thermoscript Reverse Transcription System (Invitrogen, USA) with oligo-dT. PCR was performed using FastStart DNA Master SYBR Green I kit (Roche, USA) according to the manufacturer’s instructions. Each 20 μl PCR reaction contained 0.5 μM of each primer, 3 mM (LT-β) or 4 mM (glyceraldehyde-3-phosphate dehydrogenase (GAPDH)) MgCl2, and 2 μl of cDNA template. PCR primers used for amplification of LT-β and GAPDH (Invitrogen, USA) were: LT-β forward primer AAGCTGCCAGAGGAGGAGCC, LT-β reverse primer TCCCGCTCGTCAGAAACGC; GAPDH forward primer TGCCCCCTCTGCTGATGCC, GAPDH reverse primer CCTCCGACGCCTGCTTCACCAC. Quantitative real time PCR was performed using a LightCycler (Roche, USA). FastStart Taq polymerase was activated by preincubation at 95°C for two minutes, before cycling for 40 cycles of 95°C, 0 seconds; 60°C, five seconds; and 72°C, seven seconds. Fluorescence was measured at 86°C following the 72°C extension phase to distinguish between specific PCR product and primer-dimer. The level of LT-β mRNA expression was determined in arbitrary units by comparison with an external DNA standard (10-fold serial dilutions of purified PCR product amplified from a human LT-β genomic clone) that was amplified by the LT-β primers. The level of GAPDH mRNA expression was determined in arbitrary units by comparison with an external DNA standard (10-fold serial dilutions of purified PCR product amplified from human genomic DNA) that was amplified by the GAPDH primers. LT-β was normalised relative to GAPDH to control for variation in the reverse transcription step. Results were analysed according to the Mann-Whitney test. Data are expressed as mean (SEM). A p value of <0.05 was considered significant. PCR product specificity was confirmed by melting curve analysis and 2% agarose gel electrophoresis.
Detection of murine LT-β mRNA expression in different liver cell types
To specifically demonstrate expression of LT-β in different liver cell types, BDF-1 mice were administered a CDE diet for four weeks to induce oval cell proliferation.4 Liver cell isolations were carried out by centrifugal elutriation as previously described to generate oval cell, CDE treated hepatocyte, normal hepatocyte, and inflammatory cell samples.4 RNA was isolated as previously described.4 The Riboquant Multi-Probe RNase Protection Assay System and mCK-3b Multi-Probe Template were used to detect and quantitate expression of LT-β and GAPDH in 20 μg of target RNA, according to the manufacturer’s instructions. Protected products were visualised using a FujiImager (Fuji, Japan). All animals received humane care according to guidelines set out by the National Health and Medical Research Council of Australia.
Immunohistochemistry
Sections (4 μm) were prepared from paraffin embedded tissue specimens. Microwave antigen retrieval (one minute at 700 W, 14 minutes at 70 W, and 15 minutes at room temperature in 1 mM EDTA, pH 8.0) was performed on sections for M2-pyruvate kinase (M2-PK). Sections for LT-β staining were digested with protease (type VIII; Sigma, USA) for four minutes at 37°C.1 Immunohistochemistry was performed using a three step indirect method and detected with Liquid DAB (Dako, USA). Cells expressing LT-β were identified using mouse antihuman LT-β (B27 clone, diluted 1:50, a kind gift from Dr Jeffrey Browning, Biogen, USA). Oval cells were detected by expression of M2-PK (diluted 1:50; ScheboTech, Germany) and π-glutathione S-transferase (GST, diluted 1:400; NCL, USA). These antibodies have been previously shown to stain oval cells in human biopsies.1 Infiltrating CD45 positive inflammatory cells were identified by expression of leucocyte common antigen (LCA, diluted 1:50; Dako, USA). Proliferating cells were identified by expression of proliferating cell nuclear antigen (PCNA, diluted 1:100; Dako).
RESULTS
Hepatic LT-β mRNA levels are elevated in chronic HCV infection
LT-β mRNA was detected in all of the biopsy specimens studied (fig 1A ▶). Low levels of LT-β mRNA expression were observed in control liver samples. There was no significant difference between LT-β mRNA levels in control subjects and subjects with non-fibrotic liver damage. In subjects with portal fibrosis, LT-β mRNA levels were elevated 2.2-fold over control liver levels (p=0.04). In subjects with bridging fibrosis, LT-β mRNA levels increased 4.4-fold over control liver levels (p=0.02). LT-β mRNA levels in subjects who had established cirrhosis were increased 3.3-fold compared with controls and 2.6-fold compared with mild liver damage (p=0.02). These data show that LT-β mRNA levels are increased when fibrosis or cirrhosis is present.
Figure 1.
(A) Expression of lymphotoxin β (LT-β) mRNA in liver in chronic hepatitis C. LT-β mRNA levels detected in liver biopsies from chronic hepatitis C patients increased significantly with fibrosis and cirrhosis. No significant difference was observed between control liver (n=5) and subjects with non-fibrotic liver damage (n=5). LT-β mRNA levels were 2.3-fold and 4.4-fold higher than control liver in subjects with portal fibrosis (n=5) and bridging fibrosis (n=5), respectively. LT-β mRNA levels were increased 3.3-fold in cirrhotic liver (n=5) compared with control liver. Results are expressed as mean (SEM) (*p=0.04, **p=0.02). (B) Ribonuclease protection analysis performed on RNA extracted from hepatocytes, oval cells, and inflammatory cells isolated from mice that received a choline deficient ethionine supplemented (CDE) diet for four weeks. LT-β mRNA is expressed by oval cells (O) and inflammatory cells (IC). LT-β mRNA was undetectable in normal hepatocytes (H) and CDE-treated hepatocytes (CD-H). GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Oval cells express LT-β
As LT-β expression was found to increase with fibrosis and cirrhosis, we undertook to identify the hepatic cell types capable of expressing LT-β to ascertain whether oval cells were responsible for the increased expression. As sufficient oval cells cannot be isolated from small human liver biopsies, a mouse model of oval cell mediated liver regeneration was utilised. Ribonuclease protection analysis was performed on RNA extracted from hepatocytes, oval cells, and inflammatory cells isolated from mice that received a CDE diet for four weeks. Cells were separated by centrifugal elutriation and inflammatory cell and oval cell fractions were collected. The oval cell fraction was further purified to remove any contaminating inflammatory cells.4 After purification, inflammatory cell contamination in the oval cell fraction was reduced to 0.96 (0.25)%. Oval cells were not present in the inflammatory cell fraction. Strong expression of LT-β mRNA was detected in oval cells, with low level expression in inflammatory cells (fig 1B ▶). LT-β mRNA was undetectable in normal and CDE treated hepatocytes (fig 1B ▶).
Cellular localisation of LT-β expression in HCV infected liver
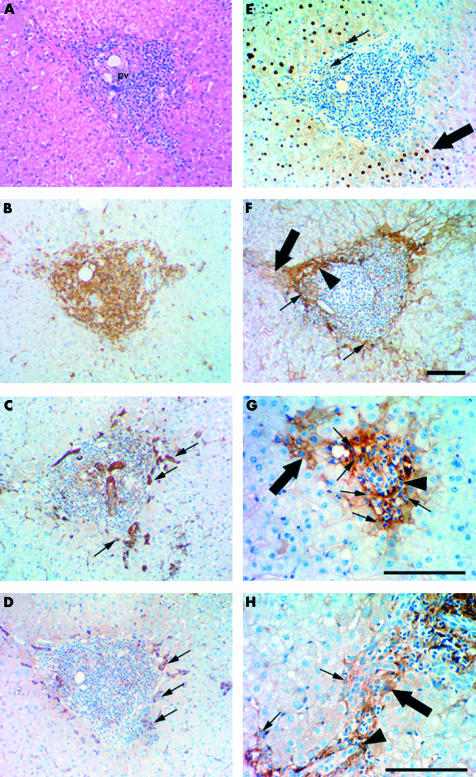
Immunohistochemistry on serial sections was performed to determine the tissue distribution of LT-β expression in HCV infected liver with moderate fibrosis (fig 2A–F ▶). A chronic immune response characterised by perivascular cuffing around the portal triads occurs during HCV infection (fig 2A ▶). The mass of small cells infiltrating the region surrounding the portal triads were shown to be leucocytes on the basis of LCA (CD45) expression (fig 2B ▶). The majority of these leucocytes were not proliferating, as they were PCNA negative (fig 2E ▶) and did not appear to be undergoing clonal expansion. Towards the periphery of the inflammatory cell mass, oval cells were identified by morphology and expression of the oval cell markers π-GST (fig 2C ▶, small arrows) and M2-PK (fig 2D ▶, small arrows).
Figure 2.
Immunohistochemical staining illustrating cellular localisation of lymphotoxin β (LT-β) in human hepatitis C virus (HCV) infected liver with bridging fibrosis. (A–F) Serial staining for haematoxylin and eosin (H&E), leucocyte common antigen (LCA) (CD45), π-glutathione S-transferase (GST), M2-pyruvate kinase (M2-PK), proliferating cell nuclear antigen (PCNA), and LT-β suggests oval cells in HCV infected liver express LT-β. (A) H&E staining illustrating periportal cuffing by lymphocytes in chronic hepatitis C. The portal vein is labelled (pv). (B) LCA (CD45) expression illustrates that the mass of small cells clustered around the portal vein are CD45 positive. Oval cells (small arrows) were identified towards the periphery of this region by expression of π-GST (C) and M2-PK (D). (E) The small cells clustered directly around the portal vein are PCNA negative while oval-like cells (small arrows) are weakly PCNA positive and large centrally located hepatocytes (large arrow) are strongly PCNA positive. (F) LT-β expression was restricted to the periphery of the inflammatory cell mass, suggesting that the majority of LCA positive cells are not expressing LT-β. Cell types expressing LT-β include small portal hepatocytes (large arrow), oval cells (small arrows), and inflammatory cells (arrowhead). Bile ducts and large hepatocytes are negative. (G) Periportal cuffing by lymphocytes in chronic hepatitis C, portal fibrosis. This region contains LT-β positive oval cells (small arrows) and small hepatocytes adjacent to the portal vein with membranous and cytoplasmic LT-β staining (large arrow). LT-β expression is observed in some inflammatory cells (arrowhead). (H) Chronic hepatitis C with cirrhosis shows similar LT-β staining patterns, with oval cells (small arrows) and some inflammatory cells (arrowhead) staining positive for LT-β. Hepatocytes adjacent to fibrous septae also express LT-β (large arrow). Magnification bars represent 100 μm.
LT-β protein was not detected in histological sections of normal liver (data not shown). However, in subjects with chronic hepatitis C, LT-β protein was observed (fig 2F–H ▶), concentrated around the periphery of the inflammatory cell mass (fig 2F ▶). Specifically, LT-β was observed in oval cells (fig 2F–H ▶, small arrow), small portal hepatocytes (fig 2F–H ▶, large arrow), and some inflammatory cells (fig 2F–H ▶, arrowhead). Oval cells were identified by their characteristic oval shape and basophilic nucleus and were located around the periphery of the inflammatory cell mass; the same region that showed expression of oval cell markers π-GST (fig 2C ▶, small arrow) and M2-PK (fig 2D ▶, small arrow). Inflammatory cells were identified by densely staining irregularly shaped nuclei. LT-β staining was more extensive in the livers of patients with fibrosis, with hepatocytes adjacent to fibrous septae (fig 2H ▶, large arrow) expressing LT-β.
Noticeably, the small LT-β positive hepatocytes showed membranous and cytoplasmic LT-β staining (fig 2F, G ▶, large arrow), consistent with previous reports that LT-β exists as a cell surface bound protein associated with LT-α. Oval-like cells surrounding the inflammatory lesion were weakly PCNA positive (fig 2E ▶, arrow), and large centrally located hepatocytes were strongly PCNA positive, indicating that both cell types were proliferating. However, small hepatocytes were PCNA negative. Due to the portal location of the non-proliferating small LT-β positive hepatocytes, it is possible that these hepatocytes have arisen by differentiation of LT-β positive oval cells.
DISCUSSION
The biological significance of LT-β signalling remains focused on its role in the lymphoid system even though LT-βR expression by non-lymphoid cell types suggests a role outside of immune function and development.19 In this study we show for the first time that LT-β is detectable in HCV infected liver. LT-β protein was detected in many liver cell types including oval cells, small hepatocytes, some inflammatory cells, and possibly sinusoidal and vascular endothelial cells. The majority of LT-β positive cells were located around the periphery of portal tract infiltrates in early fibrotic states and extended into central regions, along fibrotic septae, as the grade of fibrosis increased.
We have observed elevated levels of LT-β localised to oval cells induced by a CDE diet (unpublished data) in animal models of chronic liver damage. Experiments using both a T cell line10 or an oval cell derived line indicate that basal activity of the human LT-β promoter can be induced by TNF (manuscript in preparation). A compromised oval cell response in TNFRI knockout mice shows that TNF interaction with TNFRI is an integral mediator of oval cell proliferation.4 As TNF/TNFRI signalling is also responsible for liver regeneration following partial hepatectomy,28,29 divergence of downstream signalling pathways must occur as the oval cell compartment is not recruited during the regenerative response following partial hepatectomy. It is possible that TNF signalling may also activate the LT-β signalling pathway in oval cells, thus providing the divergent signal necessary to distinguish oval cell mediated liver regeneration from hepatocyte mediated liver regeneration. This is supported by the observation that proliferating oval cells, identified by expression of PCNA, were located near small non-proliferating hepatocytes that expressed LT-β. The possibility that these hepatocytes have arisen by differentiation of LT-β positive oval cells is the focus of current investigations.
Expression of LT-β on inflammatory cells is consistent with previous findings.30 One characteristic of chronic HCV infection is the organisation of inflammatory infiltrates into lymphoid follicles containing B cells, T cells, and dendritic cells, providing a microenvironment for recruitment and retention of lymphocytes to the site of chronic HCV infection.31 These lymphoid follicles arise by a mechanism similar to secondary lymphoid organogenesis, in which LT-α1β2/ LT-βR signalling is essential.32 One requirement for lymphocyte recruitment and retention is expression of adhesion molecules and chemokines. In the liver, inflammation is associated with expression of adhesion molecules and chemokines known to be induced by LT-β, including interferon gamma inducible protein 10, RANTES (regulated on activation, normal T cell expressed and secreted), interleukin 8, monocyte chemoattractant protein 1, and macrophage inflammatory protein 1α.33–35 Thus our finding of LT-β expression in cells surrounding portal tract infiltrates supports a role for LT-α1β2/ LT-βR interaction in cellular trafficking during chronic HCV infection.
The role of chemokines in attracting cells appears to be a universal mechanism in events that require cell movement and various chemokines involved in homing of inflammatory cells to sites of damage and inflammation are expressed in pathologies associated with oval cell infiltration.34,35 These chemokines may be responsible for recruitment and migration of oval cells during oval cell mediated liver regeneration. In light of recent findings identifying bone marrow as a source of oval cell precursors that migrate to the liver to facilitate liver regeneration,36–39 it is likely that LT-β has a central role in mediating chemokine expression leading to liver repopulation.
Expression of LT-β by inflammatory cells may contribute to, but does not entirely account for, the increased hepatic expression of LT-β mRNA. Increased hepatic LT-β mRNA levels are not specific to hepatitis C as elevated levels have also been observed in subjects with hereditary haemochromatosis in which inflammation is often not observed (unpublished data). Oval cells however have been previously reported in the livers of subjects with hereditary haemochromatosis.1 Oval cell numbers increase significantly with fibrosis and cirrhosis in both hepatitis C and hereditary haemochromatosis.1 This study shows that the increase in oval cell numbers parallels the increase in LT-β mRNA levels observed when fibrosis and cirrhosis are present, further supporting a role for LT-β in oval cell mediated liver regeneration.
We conclude that LT-β expression is observed on multiple hepatic cell types, predominantly oval cells, in chronic HCV infection. LT-β gene expression and oval cell numbers are increased when fibrosis or cirrhosis is present, suggesting a role for LT-β in the pathogenesis of chronic hepatitis C and possibly oval cell mediated liver regeneration. It is likely that LT-α plays a similar role as it is always associated with LT-β as a trimer.
Acknowledgments
This study was supported by the National Health and Medical Research Council of Australia, Cancer Foundation of Western Australia, and Raine Medical Research Foundation. Dr E Croager is the recipient of a Healy Postdoctoral Research Fellowship. We thank Dr Jeffrey Browning for his kind gift of the mouse anti-human LT-β antibody.
Abbreviations
HCV, hepatitis C virus
HCC, hepatocellular carcinoma
TNF, tumour necrosis factor
TNFRI, tumour necrosis factor receptor 1
LT, lymphotoxin
LT-βR, LT-β receptor
NFκB, nuclear factor κB
CDE, choline deficient ethionine supplemented
PCR, polymerase chain reaction
GAPDH, glyceraldehyde-3-phosphate dehydrogenase
M2-PK, M2-pyruvate kinase
GST, glutathione S-transferase
LCA, leucocyte common antigen
PCNA, proliferating cell nuclear antigen
REFERENCES
- 1.Lowes KN, Brennan BA, Yeoh GC, et al. Oval cell numbers in human chronic liver diseases are directly related to disease severity. Am J Pathol 1999;154:537–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vessey CJ, Hall PM. Hepatic stem cells: a review. Pathology 2001;33:130–41. [PubMed] [Google Scholar]
- 3.Sell S. Heterogeneity and plasticity of hepatocyte lineage cells. Hepatology 2001;33:738–50. [DOI] [PubMed] [Google Scholar]
- 4.Knight B, Yeoh GCT, Husk KL, et al. Impaired preneoplatic changes and liver tumour formation in tumour necrosis factor receptor type 1 knockout mice. J Exp Med 2000;192:1809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Evarts RP, Hu Z, Fujio K, et al. Activation of the hepatic stem cell compartment in the rat: role of transforming growth factor, hepatocyte growth factor, and acidic fibroblast growth factor in early proliferation. Cell Growth Diff 1993;4:555–61. [PubMed] [Google Scholar]
- 6.Matsusaka S, Tsujimura T, Toyosaka A, et al. Role of c-kit receptor tyrosine kinase in development of oval cells in rat 2-acetylaminofluorene/partial hepatectomy model. Hepatology 1999;29:670–6. [DOI] [PubMed] [Google Scholar]
- 7.Hu Z, Evarts RP, Fujio K, et al. Expression of transforming growth factor alpha/epidermal growth factor receptor, hepatocyte growth factor/c-met and acidic fibroblast growth factor/fibroblast growth factors during hepatocarcinogenesis. Carcinogenesis 1996;17:931–8. [DOI] [PubMed] [Google Scholar]
- 8.Bisgaard HC, Muller S, Nagy P, et al. Modulation of the gene network connected to IFN-γ in liver regeneration from oval cells. Am J Pathol 1999;155:1075–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagy P, Kiss A, Schnur J, et al. Dexamethasone inhibits the proliferation of hepatocytes and oval cells but not bile duct cells in rat liver. Hepatology 1998;28:423–9. [DOI] [PubMed] [Google Scholar]
- 10.Voon DC, Subrata LS, Abraham LJ. Regulation of lymphotoxin β by tumour necrosis factor, phorbol myristate acetate, and ionomycin in Jurkat T cells. J Interferon Cytokine Res 2001;21:921–30. [DOI] [PubMed] [Google Scholar]
- 11.Browning JL, Ngam-ek A, Lawton P, et al. Lymphotoxin β, a novel member of the TNF family that forms a heteromeric complex with lymphotoxin α on the cell surface. Cell 1993;72:847–56. [DOI] [PubMed] [Google Scholar]
- 12.Rennert PD, Browning JL, Mebius R, et al. Surface lymphotoxin alpha/beta complex is required for the development of peripheral lymphoid organs. J Exp Med 1996;184:1999–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koni PA, Sacca R, Lawton P, et al. Distinct roles in lymphoid organogenesis for lymphotoxin α and β revealed in lymphotoxin β-deficient mice. Immunity 1997;6:491–500. [DOI] [PubMed] [Google Scholar]
- 14.Iizuka K, Chaplin DD, Wang Y, et al. Requirement for membrane lymphotoxin in natural killer cell development. Proc Natl Acad Sci U S A 1999;96:6336–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu Q, Wang Y, Wang J, et al. The requirement of membrane lymphotoxin for the presence of dendritic cells in lymphoid tissues. J Exp Med 1999;190:629–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elewaut D, Brossay L, Santee SM, et al. Membrane lymphotoxin is required for the development of different subpopulations of NK T cells. J Immunol 2000;165:671–9. [DOI] [PubMed] [Google Scholar]
- 17.VanArsdale TL, VanArsdale SL, Force WR, et al. Lymphotoxin β receptor signalling complex: Role of tumour necrosis factor receptor-associated factor 3 recruitment in cell death and activation of NF-κB. Proc Natl Acad Sci U S A 1997;94:2460–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakano H, Oshima H, Chung W, et al. TRAF5, an activator of NF-κB and putative signal transducer for the lymphotoxin receptor. J Biol Chem 1996;271:14661–4. [DOI] [PubMed] [Google Scholar]
- 19.Degli-Esposti MA, Davis-Smith T, Din WS, et al. Activation of the lymphotoxin β receptor by cross-linking induces chemokine production and growth arrest in A375 melanoma cells. J Immunol 1997;158:1756–62. [PubMed] [Google Scholar]
- 20.Ngo VN, Korner H, Gunn MD, et al. Lymphotoxin alpha/beta and tumour necrosis factor are required for stromal cell expression of homing chemokines in B and T cell areas of the spleen. J Exp Med 1999;189:403–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.You LR, Chen CM, Lee YH. Hepatitis C virus core protein enhances NF-κB signal pathway triggering by lymphotoxin β receptor ligand and TNFα. J Virol 1999;73:1672–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harada K, Tsuneyama K, Hiramatsu K, et al. Significance of CD30-positive lymphocytes in livers in primary biliary cirrhosis. J Gastroenterol Hepatol 1999;14:1197–202. [DOI] [PubMed] [Google Scholar]
- 23.Tilg H, Diehl AM. Cytokines in alcoholic and non-alcoholic steatohepatitis. N Engl J Med 2000;343:1467–76. [DOI] [PubMed] [Google Scholar]
- 24.Hayashi N, Mita E. Involvement of Fas system-mediated apoptosis in pathogenesis of viral hepatitis. J Viral Hepatol 1999;6:357–65. [DOI] [PubMed] [Google Scholar]
- 25.Neuman M, Angulo P, Malkiewicz I, et al. Tumour necrosis factor-alpha and transforming growth factor-beta reflect severity of liver damage in primary biliary cirrhosis. J Gastroenterol Hepatol 2002;17:196–202. [DOI] [PubMed] [Google Scholar]
- 26.Chen C, You L, Hwang L, et al. Direct interaction of hepatitis C virus core protein with the cellular lymphotoxin β receptor modulates the signal pathway of the lymphotoxin β receptor. J Virol 1997;71:9417–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brunt EM. Grading and staging the histopathological lesions of chronic hepatitis: the Knodell histology activity index and beyond. Hepatology 2000;31:241–6. [DOI] [PubMed] [Google Scholar]
- 28.Akerman PA, Cote P, Yang SQ. Antibodies to tumour necrosis factor α inhibit liver regeneration after partial hepatectomy. Am J Physiol 1992;263:G579–85. [DOI] [PubMed] [Google Scholar]
- 29.Yamada Y, Kirillova I, Peschon JJ, et al. Initiation of liver growth by tumour necrosis factor: deficient liver regeneration in mice lacing type I tumour necrosis factor receptor. Proc Natl Acad Sci U S A 1997;94:1441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ware CF, Crowe PD, Grayson MH, et al. Expression of surface lymphotoxin and tumor necrosis factor on activated T, B, and natural killer cells. J Immunol 1992;149:3881–8. [PubMed] [Google Scholar]
- 31.Lalor PF, Shields P, Grant A, et al. Recruitment of lymphocytes to the human liver. Immunol Cell Biol 2002;80:52–64. [DOI] [PubMed] [Google Scholar]
- 32.Kuprash DV, Alimzhanov MB, Tumanov AV, et al. TNF and lymphotoxin β co-operate in the maintenance of secondary lymphoid tissue microarchitecture but not in the development of lymph nodes. J Immunol 1999;163:6575–80. [PubMed] [Google Scholar]
- 33.Adams DH, Hubscher S, Fear J, et al. Hepatic expression of macrophage inflammatory protein-1 alpha and macrophage inflammatory protein-1 beta after liver transplantation. Transplantation 1996;61:817–25. [DOI] [PubMed] [Google Scholar]
- 34.Afford SC, Fisher NC, Neil DA, et al. Distinct patterns of chemokine expression are associated with leukocyte recruitment in alcoholic hepatitis and alcoholic cirrhosis. J Pathol 1998;186:82–9. [DOI] [PubMed] [Google Scholar]
- 35.Shields PL, Morland CM, Salmon M, et al. Chemokine and chemokine receptor interactions provide a mechanism for selective T cell recruitment to specific liver compartments within hepatitis C-infected liver. J Immunol 1999;163:6236–43. [PubMed] [Google Scholar]
- 36.Petersen BE, Bowen WC, Patrene KD, et al. Bone marrow as a potential source of hepatic oval cells. Science 1999;14:1168–70. [DOI] [PubMed] [Google Scholar]
- 37.Alison MR, Poulsom R, Jeffery R, et al. Hepatocytes from non-hepatic stem cells. Nature 2000;406:257. [DOI] [PubMed] [Google Scholar]
- 38.Thiese ND, Nimmakayalu M, Gardner R, et al. Liver from bone marrow in humans. Hepatology 2000;32:11–16. [DOI] [PubMed] [Google Scholar]
- 39.Lagasse E, Connors H, Al-Dhalimy M, et al. Purified hematopoietic stem cells can differentiate into hepatocytes in vitro. Nat Med 2000;6:1229–34. [DOI] [PubMed] [Google Scholar]