Abstract
The cellular target of leptomycin B (LMB), a nuclear export inhibitor, has been identified as CRM1 (exportin 1), an evolutionarily conserved receptor for the nuclear export signal of proteins. However, the mechanism by which LMB inhibits CRM1 still remains unclear. CRM1 in a Schizosaccharomyces pombe mutant showing extremely high resistance to LMB had a single amino acid replacement at Cys-529 with Ser. The mutant gene, named crm1-K1, conferred LMB resistance on wild-type S. pombe, and Crm1-K1 no longer bound biotinylated LMB. 1H NMR analysis showed that LMB bound N-acetyl-l-cysteine methyl ester through a Michael-type addition, consistent with the idea that LMB binds covalently via its α,β-unsaturated δ-lactone to the sulfhydryl group of Cys-529. When HeLa cells were cultured with biotinylated LMB, the only cellular protein bound covalently was CRM1. Inhibition by N-ethylmaleimide (NEM), an alkylating agent, of CRM1-mediated nuclear export probably was caused by covalent binding of the electrophilic structure in NEM to the sulfhydryl group of Cys-529, because the crm1-K1 mutant showed the normal rate for the export of Rev nuclear export signal-bearing proteins in the presence of not only LMB but also NEM. These results show that the single cysteine residue determines LMB sensitivity and is selectively alkylated by LMB, leading to CRM1 inactivation.
Many cellular proteins either reside in the nucleus or shuttle between the nucleus and the cytoplasm by energy-dependent transport across the nuclear envelope. Specific sequences within a protein contain the information necessary for the nucleocytoplasmic transport: most nuclear proteins have nuclear localization sequences (NLS) rich in basic amino acids, whereas others carry short nuclear export sequences (NES) rich in leucine (1, 2). CRM1/exportin 1 was shown to be a receptor for the NES in both lower and higher eukaryotes (3–6). Genetic alterations in the CRM1 locus caused a defect in nuclear export of NES-bearing proteins in yeast (3, 5, 7, 8). Nuclear microinjection of a CRM1-specific antibody that prevents the in vitro NES binding inhibited in vivo protein nuclear export in mammalian cells (9). Thus, the NES-mediated nuclear export of proteins is a universal and conserved mechanism by which subcellular localization of proteins is controlled in cells.
CRM1 originally was identified as a protein essential for maintaining chromosome structure in the fission yeast Schizosaccharomyces pombe (10). The functional homologues that complement the fission yeast crm1 mutation were cloned from the budding yeast Saccharomyces cerevisiae (11) and from human cells (8, 12). We showed previously that a mutation (crm1-N1) of S. pombe crm1+ conferred resistance to leptomycin B (LMB) (13), which had been discovered as a potent antifungal antibiotic blocking the eukaryotic cell cycle (14, 15). In contrast, the cold-sensitive crm1-809 mutant strain was hypersensitive to LMB. Furthermore, morphological and biochemical phenotypes of crm1-809 mutant cells at nonpermissive temperature were identical to those of LMB-treated, wild-type cells. Taken together, we proposed that S. pombe Crm1 is the cellular target of LMB (13). Recently, Wolff et al. (16) rediscovered LMB as an agent that inhibits the nuclear export of HIV-1 Rev. We and others found that LMB abolished association of CRM1 with the NES by binding directly to CRM1, thereby inhibiting nuclear export of proteins (4–6, 9). Most recently, N-ethylmaleimide (NEM) was reported to block the CRM1 function (17), suggesting that CRM1 contains an essential cysteine residue.
During the course of screening for the intragenic or exogenic mutants that suppress LMB hypersensitivity of crm1-809, we happened to isolate a mutant of S. pombe that shows extremely high resistance to LMB. This mutant strain facilitated the identification of the amino acid residue in Crm1 (Cys-529) to which LMB as well as NEM bind via their electrophilic structures. We show evidence that LMB covalently and selectively binds the sulfhydryl group of Cys-529 via its α,β-unsaturated δ-lactone.
MATERIALS AND METHODS
Yeast Strains and Media.
The S. pombe strains used were JY266 (h+ leu1-32), JY182 (h90 mei1-B102 arg1 ura1), JY742 (h+ ade6-M216 leu1-32 ura4-D18), AC1 (h− leu1-32 crm1-809) (10), FN41 (h− ade6-M210 leu1-32 ura4-D18 crm1-F1), and KY21 (h+ ade6-M216 leu1-32 ura4-D18 crm1+∷crm1-K1≪ura4+). S. pombe routinely was grown in complete YPD medium (1% yeast extract/2% polypeptone/2% glucose). Minimal medium (18) was used for plasmid selection.
Plasmid Constructions.
pALPC1 was constructed by inserting a PstI–SmaI fragment encoding a 6.2-kb chromosomal crm1+ region of pKK1 (gift from M. Yanagida, Kyoto University) (19) between SphI and SmaI sites of pAL19, after both the PstI and SphI sites had been blunt-ended. The mutant crm1 genes were cloned by the gap-repair method according to “Fission Yeast Handbook” at the web site http://www.bio.uva.nl/pombe/handbook/. pALPC1 was digested by SphI and BglII to remove the entire crm1-coding region, and the purified linear ars-containing fragment was transformed according to the lithium acetate protocol (20). pALPCA1 and pALPCF1, containing crm1-809 and crm1-F1, were recovered from mutants AC1 and FN41, respectively. The nucleotide sequences of mutant crm1 genes were determined by using specific primers corresponding to crm1+ and Genetic Analyzer 310 (Applied Biosystems). pALPCK1, encoding crm1-K1, was constructed by replacing the 2.2-kb SphI–HpaI fragment from pALPC1 with the corresponding fragment of pALPCF1.
Genetic Methods and Mutagenesis.
Mutagenesis and screening for mutants that suppress LMB sensitivity were carried out as follows. S. pombe AC1 was grown in YPD medium aerobically at 32°C, and exponentially growing cells (106 to 107 cells per ml) were washed and suspended in minimal medium with 3% ethyl methanesulfonate (EMS) for 40–120 min. The cells were washed three times with 0.9% NaCl and plated on YPD agar containing 20 ng/ml LMB. After 7 days of incubation at 32°C, growing colonies were selected. To obtain the chromosomal crm1-K1 mutant, we constructed pBUPCK1.3 by inserting a 2.6-kb BglII fragment of pALPCK1 into the BamHI site of pBS-ura4+ (gift from M. Yamamoto, The University of Tokyo). After pBUPCK1.3 had been digested with Eco47III, S. pombe JY742 cells were transformed with the linear fragment, and Ura+ cells were selected. Successful gene replacement in the transformants was confirmed by Southern hybridization.
1H NMR Spectroscopy.
1H NMR and correlated spectroscopy spectra were recorded on a JEOL A600 spectrometer (JEOL) at 600 MHz. One milligram of LMB (100 μl of a 10-mg/ml ethanol solution) was incubated with 2.0 mg of l-cysteine, glutathione (GSH), and N-acetyl-l-cysteine methyl ester (200 μl of 10-mg/ml methanol solutions) in 1 ml of Tris buffer (220 mM Tris⋅HCl, pH 7.5/100 mM NaCl) at room temperature. After 18 h, the solution was subjected directly to reverse-phase HPLC (column: Senshu-Pak C18, 8 × 250 mm; mobile phase: gradient of acetonitrile and water, both of which contain 0.1% trifluoroacetic acid (TFA), from 50:50 to 100:0 in 30 min; flow rate: 1 ml/min). The peak having a retention time of 28.2 min was found to be the Michael adduct of LMB with N-acetyl-l-cysteine methyl ester (1.3 mg), which was confirmed by MS showing m/z 740 (M + Na)+ and 1H NMR.
MS.
One microgram of a peptide (5 μl of 0.2-mg/ml solution) corresponding to hCRM1 (residues 513–530) or its derivative containing Ser-528 instead of Cys was treated with 10 μg of LMB (1 μl of a 10-mg/ml ethanol solution) in 15 μl of buffer (20 mM Tris⋅HCl, pH 7.5/100 mM NaCl) at 37°C. After 24 h of incubation, an aliquot of 0.5 μl of this solution was mixed with 0.5 μl of the matrix solution (10 mg/ml α-cyano-4-hydroxycinnamic acid in 1:1 acetonitrile/water containing 0.1% TFA). The mixed solution was deposited on a stainless steel sample plate, and the solvent was allowed to evaporate at ambient temperature. To remove salts, which hamper matrix-assisted laser desorption ionization analysis, the dried sample was rinsed by placing 1 μl of water on the surface of the sample for 5 s and removing the liquid. This sample was analyzed with a Voyager DE STR MALDI-TOF mass spectrometer (PerSeptive Biosystems) in the delayed-extraction mode.
Assay for LMB Binding.
HeLa cells (2 × 107 cells) in culture were pretreated with 0.1% ethanol (control) or 100 nM of competitors for 1 h and then incubated with 10 nM biotinylated LMB for 2 h. After the cells had been lysed with TBS (50 mM Tris⋅HCl/150 mM NaCl, pH 7.5) containing Complete protease inhibitors (Boehringer Mannheim) and 0.1% NP-40, the supernatants were prepared by centrifugation. Immobilized streptavidin (25 μl) was added to the lysates prepared from the cells treated with biotinylated LMB, and the mixtures were incubated for 24 h at 4°C. The bound proteins were washed thoroughly and then eluted by 50 μl of SDS/PAGE sample buffer containing 1.7% SDS and 100 mM DTT by boiling. Each eluate (10 μl per lane for silver staining and anti-biotin immunostaining, or 0.5 μl per lane for anti-hCRM1 immunostaining) was applied to 5–20% Tris/glycine gradient gels (Bio-Rad). Proteins separated on one gel were silver-stained, and those on other two gels were transferred to poly(vinylidene difluoride) (PVDF) membranes (Millipore). The blots were incubated with a diluted (1:1,000) rabbit polyclonal anti-hCRM1 antibody and an anti-biotin antibody conjugated with horseradish peroxidase (New England Biolabs, 1:3,000), respectively, followed by detection using the Amersham enhanced chemiluminescence system.
In Vitro Transcription and Translation.
[35S]Methionine-labeled CRM1 proteins were synthesized by using a TNT T7 Quick Coupled Transcription/Translation System (Promega) according to the manufacturer’s instructions. Templates for transcription and translation were prepared by PCR amplifications by using pALPC1 (crm1+) or pALPCK1 (crm1-K1) as templates and primers 5′-TAATACGACTCACTATAGGGAGACCACCATGGAGGGCATCCTGGCATTC-3′ (sense) and 5′-GGGCTCGAGCTGATCACCTCATCTAGATAGTTCTTCCTCCTCCATAGTAG-3′ (antisense). Sac. cerevisiae CRM1 was synthesized by using pCRM1 (11) as a template and 5′TAATACGACTCACTATAGGGAGACCACCATGGAAGGAATTTTGGATTTTTC-3′ (sense) and 5′-GGGCTCGAGCAGATCTCCTCAGCTAGCATCATCAAGTTCGGAAGGTTT-3′ (antisense) as primers. The sense primers contain a T7 promoter and Kozak sequence. An aliquot of each lysate containing [35S]methionine-labeled CRM1 was subjected to the LMB-binding assay by using biotinylated LMB. After the labeled CRM1 proteins had been incubated for 2 h with competitors at final concentrations of 20 μM for LMB and the inactive LMB analog, 10 mM for NEM, and 1 mM for GSH, then 200 nM biotinylated LMB was added to the mixture. The bound proteins were collected by immobilized streptavidin as described above and analyzed with a bioimaging analyzer BAS2000 (FUJIX, Tokyo). In the case of NEM treatment, the reaction was quenched by the addition of 20 mM DTT (17).
RESULTS
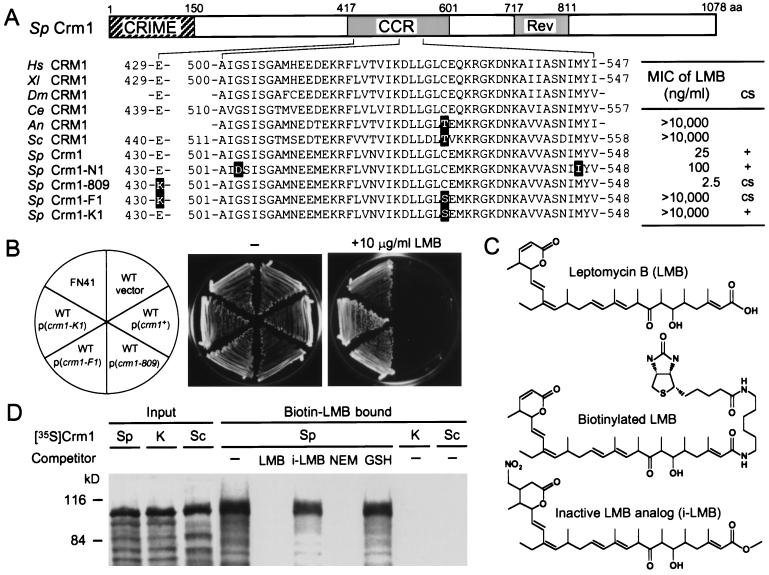
Strain AC1 with the cold-sensitive mutation crm1-809 is hypersensitive to LMB (13). By cloning and sequencing the crm1-809 allele, we found that Glu-430 in the central conserved region (CCR) was replaced by Lys (Fig. 1A). Taken together with our previous finding that an LMB-resistance mutation (crm1-N1) also occurred in CCR, it seemed probable that the region was involved in the interaction with LMB. To obtain a clue to understanding the functional role of CCR and the mechanism by which LMB exerts its inhibitory effect on Crm1, we mutagenized strain AC1 and searched for mutants that suppress the LMB sensitivity. This type of genetic approach may lead to identification of a protein(s) that interacts with CCR and/or is involved in Crm1 functions and LMB sensitivity. Among the suppressor mutants thus obtained, one mutant named FN41 was viable even in the presence of 10 μg/ml LMB, the highest concentration to be dissolved in the medium (Fig. 1B). This mutant was 4,000 times more resistant than the parental strain. The LMB resistance was dominant, as determined by making a stable diploid (data not shown). On the other hand, FN41 was still cold-sensitive and overproduced p25Apt1, whose expression is controlled by Pap1, a transcription factor negatively regulated by Crm1 (21, 22), as did parental crm1-809 mutant (data not shown). These observations suggest that FN41 acquired an additional dominant mutation that confers LMB resistance. Tetrad analysis after crossing FN41 with a strain containing the wild-type crm1+ background indicated that the LMB resistance mutation was present in the chromosomal crm1 locus (data not shown).
Figure 1.
Isolation of an LMB-insensitive mutant and identification of the mutation. (A) A schematic representation of CRM1 functional domains and summary of allelic crm1 mutants and their phenotypes. An LMB-insensitive mutant FN41 (crm1-F1) was isolated by mutagenesis of AC1 (crm1-809). crm1-F1 contains a C529S mutation (TGT → AGT) besides E430K derived from crm1-809 (GAA → AAA). CRIME, a domain homologous to importin-β family proteins (CRM1, importin-β, etc.); Rev, a region in C-terminal-labeled CRM1 protected by Rev binding from protease digestion (26); MIC, minimal inhibitory concentration; Hs, Homo sapiens; Xl, Xenopus laevis (S. Khochbin, personal communication); Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; An, Aspergillus nidulans; Sc, Sac. cerevisiae; and Sp, S. pombe. Accession numbers are: D89729 (Hs), AC004423 (Dm), U64855 (Ce), AA966051 (An), D13039 (Sc), X15482 (Sp crm1+), and D16355 (Sp crm1-N1). (B) The single C529S mutation is sufficient for LMB resistance. S. pombe wild-type cells (JY266) were transformed with plasmids encoding crm1+ or various allelic mutant crm1 genes. FN41 and transformants were spread on YPD (1% yeast extract/2% polypeptone/2% glucose)-agar medium (−) or medium containing 10 μg/ml LMB and incubated for 5 days at 30°C. Wild-type cells transformed with empty vector (pAL19) also were tested (vector). (C) Chemical structures of LMB, biotinylated LMB, and an inactive LMB analog (i-LMB). (D) Cys-529 is essential for binding to LMB. 35S-labeled wild-type Crm1 (Sp), Crm1-K1 (K), and Sac. cerevisiae CRM1 (Sc) were synthesized in rabbit reticulocyte lysates (input) and subjected to the biotinylated LMB-binding assay after preincubation with various competitors. The eluates from immobilized streptavidin were analyzed by SDS/6% PAGE (biotin–LMB-bound).
The crm1 gene isolated from FN41 contained a base exchange that causes an amino acid change from Cys-529 to Ser in addition to the mutation (E430K) of crm1-809. This mutant crm1 gene was named crm1-F1 (Fig. 1A). To test whether the C529S mutation found in crm1-F1 actually contributes to the high resistance of FN41, we constructed plasmids containing the crm1-F1 gene and its derivative that has only the single amino acid exchange from Cys-529 to Ser (crm1-K1) and introduced them into wild-type cells. Cells transformed with either crm1-F1 or crm1-K1 did grow in the presence of 10 μg/ml LMB, whereas the cells harboring crm1+ or crm1-809 did not (Fig. 1B). Thus, the single amino acid change from Cys-529 to Ser in Crm1 is sufficient to confer the high LMB resistance on wild-type S. pombe. Cys-529 in CRM1 is conserved in LMB-sensitive organisms such as humans, but not in LMB-insensitive organisms such as Sac. cerevisiae (Thr-539) (Fig. 1A).
The above results suggested that Cys-529 was involved in LMB binding. If Cys-529 is necessary for LMB binding, then the Ser mutant protein (Crm1-K1, the crm1-K1 product) or Sac. cerevisiae CRM1 should fail to bind LMB. To verify this possibility, 35S-labeled Crm1, Crm1-K1, and Sac. cerevisiae CRM1 were synthesized by in vitro transcription and translation and tested for the ability to bind biotinylated LMB. As shown in Fig. 1D, 35S-labeled wild-type Crm1 was precipitated with biotinylated LMB by the immobilized streptavidin matrix. This binding is specific, because it was titrated out by adding excess LMB but not an inactive LMB analog (i-LMB) as a competitor. On the other hand, Crm1-K1 and Sac. cerevisiae CRM1 prepared in the same way were not precipitated with biotinylated LMB at all. Therefore, we conclude that the Cys-529 is essential for LMB binding. It seems likely that Cys-529 is the acceptor site for LMB and the most important determinant for LMB sensitivity of the evolutionarily conserved CRM1 proteins.
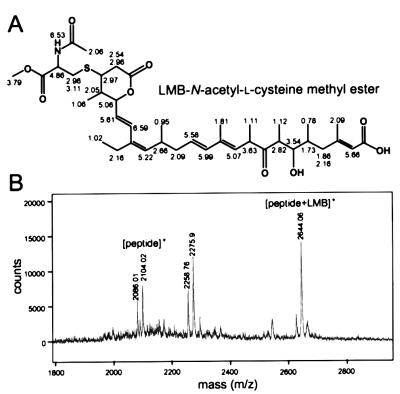
LMB has an electrophilic structure in its α,β-unsaturated δ-lactone, which is potentially reactive with biological nucleophiles, such as the sulfhydryls of cysteines. We showed that an LMB derivative that contains a saturated lactone ring lacking the electrophilicity (Fig. 1C) is biologically inactive (9) and failed to compete with biotinylated LMB for Crm1 binding (Fig. 1D). The unsaturated lactone in LMB therefore was expected to interact directly with the sulfhydryl group of Cys-529. This possibility was investigated by HPLC analysis of reaction products between LMB and several compounds containing free sulfhydryl groups. No reaction products were detected with l-cysteine or GSH. In addition, the presence of 1 mM GSH with biotinylated LMB did not inhibit Crm1 binding (Fig. 1D), suggesting that l-cysteine and GSH can barely interact with LMB. On the other hand, we found that N-acetyl-l-cysteine methyl ester formed an adduct with LMB. 1H NMR and correlated spectroscopy spectra of the purified adduct showed that LMB covalently binds the sulfhydryl group via its lactone by a Michael-type addition (Fig. 2A). Furthermore, we examined whether LMB can react with a synthetic peptide corresponding to the 18 amino acids of hCRM1 containing Cys-528 (residues 513–530) by mass spectrometric analysis. A peak of the 2644.06 Da molecular mass was detected in addition to a peak of 2104.02 Da for the free peptide (Fig. 2B). On the other hand, no peak of the adduct was detected with a peptide containing Ser-528 instead of Cys (data not shown). The difference in the masses between the modified peptide and the parental one was 540, which corresponds to the molecular weight of LMB, supporting the idea that a Michael addition occurs with polypeptides containing cysteines. The formation of the adducts with N-acetyl-l-cysteine methyl ester or the peptide required millimolar concentrations of LMB—much higher than that for the in vivo binding to CRM1 (10 nM; see below).
Figure 2.
Covalent binding of leptomycin B to N-acetyl-l-cysteine methyl ester and a peptide containing a cysteine. (A) Chemical structure of an LMB complex with N-acetyl-l-cysteine methyl ester and the chemical shifts of 1H NMR. An adduct generated in the mixture containing LMB and N-acetyl-l-cysteine methyl ester was purified by HPLC, and the structure was determined by 1H NMR and correlated spectroscopy. The chemical shifts are presented with the structure. (B) An addition reaction with a peptide containing a cysteine residue. An 18-aa peptide corresponding to hCRM1 residues 513–530 that contains Cys-528 was incubated with LMB for 24 h at 37°C and analyzed by matrix-assisted laser desorption ionization–TOF MS. A peak of 2,275.9 Da may be a product of the reaction between the peptide and α-cyano-4-hydroxycinnamic acid (αCHCA) used as the matrix. Calculated molecular mass (Da) is 540 for LMB, 2,104.54 for the peptide, and 189.04 for αCHCA.
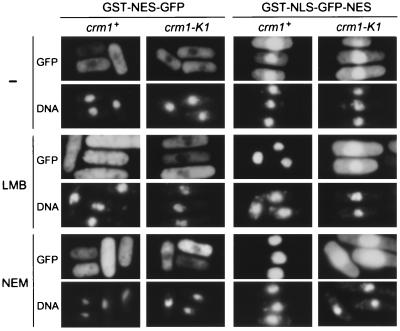
We next constructed the chromosomal crm1-K1 mutant by one-step gene replacement and tested for its ability to export the Rev NES-fusion proteins expressed under the inducible nmt1 promoter. Glutathione S-transferase (GST)-NES-green fluorescent protein (GFP) was excluded from the nucleus in both the wild-type and crm1-K1 backgrounds as shown in Fig. 3. Treatment of the wild-type cells with 50 ng/ml LMB for 6 h caused redistribution of the protein throughout the cell, because of inhibition of the Crm1-mediated nuclear export by LMB. In contrast, the crm1-K1 mutant could fully export GST-NES-GFP from the nucleus even in the presence of LMB, indicating that the nuclear export of NES-bearing proteins by Crm1-K1 is resistant to LMB. To test whether LMB partly affects the export rate in the crm1-K1 mutant cells, we expressed a GST-GFP fusion protein containing both Rev NES and simian virus 40 large T NLS (GST-NLS-GFP-NES), which is able to shuttle between the nucleus and the cytoplasm (9). Equilibrium between the import and export rates determines the subcellular distribution of this protein. In S. pombe, the import rate of simian virus 40 large T NLS is slightly higher than the export rate of Rev NES, leading to slightly stronger nuclear staining (Fig. 3). When wild-type cells were treated with LMB, GST-NLS-GFP-NES became highly accumulated in the nucleus and essentially was undetectable in the cytoplasm. In the crm1-K1 mutant cells, however, distribution of the protein was totally unaffected by LMB treatment. These results indicate that Crm1-K1 can export Rev NES at almost the same rate as wild-type Crm1, regardless of the presence of LMB.
Figure 3.
Nuclear export by Crm1-K1 is resistant to LMB and NEM. S. pombe strains JY266 (crm1+) and KY21 (crm1-K1) were transformed with pR1GrevNESF1 for GST-Rev NES-GFP (GST-NES-GFP) and pR1GsvNLSFrevNES1 for GST-SV40 T NLS-GFP-Rev NES (GST-NLS-GFP-NES), as described (9). These genes are under the control of the wild-type nmt1 promoter, which can strongly direct transcription when thiamin is absent in the medium. Cells were cultured in the absence of thiamin for 12 h at 30°C for induction of the proteins and then cultivated further for 6 h in the same medium with or without LMB (50 ng/ml) or NEM (100 μM) at 30°C. Cells were stained with 4′,6-diamidino-2-phenylindole for DNA and observed under a Zeiss Axiophot-2 microscope.
Recently, it was shown that the CRM1-mediated nuclear export is sensitive to NEM, an agent alkylating the sulfhydryl groups of cysteines in proteins (17). In fact, 100 μM NEM inhibited the nuclear export of GST-NES-GFP or GST-NLS-GFP-NES in S. pombe (Fig. 3), suggesting that some of the cysteine residues in Crm1 that can be modified by NEM participate in the nuclear export function of Crm1 in S. pombe. Surprisingly, the nuclear export activity of Crm1-K1 was fully resistant not only to LMB but also to NEM, because the nuclear localization of the NES-bearing proteins by NEM was not observed in the crm1-K1 mutant cells. Furthermore, the in vitro binding assay with biotinylated LMB showed that NEM competed with biotinylated LMB for Crm1 binding (Fig. 1D). These results confirm that CRM1 is also responsible for the inhibition of nuclear export of proteins by NEM. Because the electrophilic structure in NEM readily binds covalently with the sulfhydryl groups of cysteines, we assume that Cys-529 of CRM1 is the target of modification by NEM.
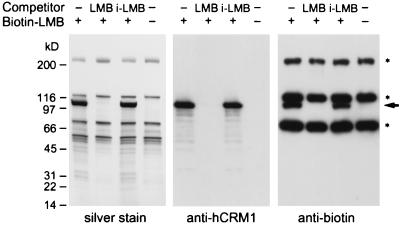
A question arose about whether LMB can specifically modify the sulfhydryl group of the cysteine residue at 529 in CRM1, because the Michael-type addition should occur in the cell with other proteins containing cysteine residues on their surface. To address this question, we analyzed in vivo LMB-binding proteins by using a biotinylated LMB derivative. HeLa cells were cultured with 10 nM biotinylated LMB for 2 h, the biotin-containing complexes were isolated by streptavidin-conjugated agarose beads, and the bound proteins were analyzed by SDS/PAGE followed by silver staining and Western blotting. The silver-stained gel (5–20% gradient of acrylamide) of the eluate from the streptavidin beads without biotinylated LMB showed a number of proteins that probably bind to biotin or streptavidin (Fig. 4, silver stain). However, the samples containing biotinylated LMB revealed one additional polypeptide of 102 kDa (Fig. 4, arrow), whereas no such protein was detected when a 10-fold molar excess of LMB was added to the cell culture 1 h before the biotin–LMB addition (LMB as a competitor). The protein bound to biotinylated LMB also was detected when the inactive LMB analog was added to the cell culture instead of active LMB (i-LMB as a competitor), indicating that LMB binding to the protein is specific. Western blot analysis using an anti-hCRM1 antiserum revealed the identity of p102 as hCRM1 (Fig. 4, anti-hCRM1). The same band was also reactive with an anti-biotin antibody conjugated with horseradish peroxidase (Fig. 4, anti-biotin), indicating that the interaction between biotinylated LMB and hCRM1 is stable in the presence of SDS and DTT even after heat denaturation. Besides three intrinsic biotin-binding proteins, no proteins other than hCRM1 were detected as proteins that bound covalently to biotin upon treatment of cells with biotinylated LMB. These results demonstrate that LMB can selectively alkylate CRM1 in the cell.
Figure 4.
Covalent binding of biotinylated LMB to hCRM1 in vivo. HeLa cells were preincubated with 0.1% ethanol (− competitor), 100 nM LMB (LMB), or 100 nM inactive LMB analog (i-LMB) for 1 h and then treated with or without 10 nM biotinylated LMB for 2 h. After cells were lysed, the proteins bound to biotinylated LMB were isolated by using immobilized streptavidin and analyzed by electrophoresis in an SDS/5–20% polyacrylamide gradient gel followed by silver staining (Left). A single protein emerged by incubation with biotinylated LMB in the absence of LMB competitor or in the presence of inactive LMB (arrow). The proteins resolved in the polyacrylamide gel were transferred to two poly(vinylidene difluoride) membranes, one that was immunostained with a rabbit polyclonal antiserum raised against hCRM1 (Center) and another that was stained with a horseradish peroxidase-conjugated anti-biotin antibody (Right) by using a chemiluminescence system. Asterisks indicate intrinsic biotin-binding proteins such as carboxylases.
DISCUSSION
Despite the widespread use of LMB as a specific nuclear export inhibitor, the molecular details behind the inhibition of CRM1 have not been elucidated. In this study, we show evidence that LMB covalently binds to a single cysteine residue to inactivate CRM1. LMB can bind in vitro the sulfhydryl group of N-acetyl-l-cysteine methyl ester or a cysteine in the synthetic peptide corresponding to hCRM1 (residues 513–530) through its α,β-unsaturated δ-lactone by a Michael-type addition. Although we have not obtained final proof of the reaction with the full-length protein, in vivo covalent binding of biotinylated LMB to hCRM1 (Fig. 4) strongly suggests that the Michael-type addition reaction also occurs in the cell. The S. pombe mutant Crm1 protein containing an amino acid change from Cys-529 to Ser (Crm1-K1) lost the ability to interact with LMB, and cells expressing Crm1-K1 became insensitive to LMB. The cysteine is located in CCR and is conserved in those LMB-sensitive organisms such as humans and S. pombe, but not in those LMB-insensitive organisms such as Sac. cerevisiae and Aspergillus nidulans (Fig. 1A). Thus, Cys-529 is the most important determinant of LMB sensitivity of the CRM1 proteins.
We assume that the inhibitory effect of NEM on the nuclear export of proteins carrying the Rev NES in S. pombe (Fig. 3) results from the covalent binding of NEM with the sulfhydryl group of Cys-529, because the nuclear export by Crm1-K1 was resistant to NEM. Competition by NEM with biotinylated LMB for Crm1 binding supports this idea. However, Cys-529 itself appears to be nonessential for the CRM1 function, and it can be replaced by at least serine (Crm1-K1) or probably threonine (Sac. cerevisiae CRM1), because the crm1-K1 mutant strain is viable without showing any temperature or cold sensitivity and could export the proteins carrying NES from the nucleus. These amino acid residues are similar not only in size, but also in that they have hydroxy groups that can form hydrogen bonds, as does the sulfhydryl groups of cysteines. Therefore, we speculate that alkylation of the cysteine residue disrupts the hydrogen bond that is important for the active CRM1 conformation or interaction with other proteins. This hypothesis will be verified partly by introducing other amino acids at position 529.
Because LMB binding causes a significant mobility shift of the CRM1 protein in native gels (4), it seems probable that alkylation at the cysteine residue in CCR induces a dramatic alteration of the three-dimensional structure of the protein. The S. pombe mutants showing altered sensitivity to LMB, crm1-809, crm1-N1, and crm1-K1, all were mapped onto CCR (Fig. 1), indicating that subtle changes in the conformation of this region greatly affects the affinity to LMB. However, the precise function of CCR is still obscure. RanGTP binding is proposed to occur in the N-terminal CRIME domain (23), which shares low but distinct homology with importin β that imports the complex of importin α and the NLS-containing proteins into the nucleus in a Ran-dependent manner (24, 25). Therefore, it seems unlikely that CCR plays a direct role in interacting with Ran. One possibility is that CCR is responsible for the interaction with NES. LMB may bind in the NES-binding pocket in CCR, thereby sterically blocking NES binding. Recently, Askjaer et al. (26) reported that a C-terminal region encompassing Asp-716 and Lys-810 of hCRM1 was involved in Rev binding, which was determined by a protein-footprinting assay. They also showed that CRM1 could associate directly with Rev in the absence of RanGTP and that LMB did not interfere with this association. However, these observations do not seem to rule out the possibility of the involvement of CCR in NES binding, because the weak CRM1/Rev association in the absence of RanGTP also occurred with not only wild-type Rev but also two NES-mutated Rev proteins, RevM10 and RevM32 (26), the mutations in the conserved leucines of which render Rev functionally inactive in transport (27, 28). On the other hand, RanGTP binding enhances the CRM1/Rev association in an LMB-sensitive, functional NES-dependent manner (26). It therefore is conceivable that LMB competes with NES for RanGTP-dependent formation of a stable CRM1/NES complex. It is also possible that CCR is important for mediating the conformational change to form a stable CRM1/NES complex upon RanGTP binding to the N-terminal CRIME domain. LMB may prevent the physical movement required for the conformational change probably because of the disruption of a hydrogen bond caused by selective alkylation at the cysteine residue, which is important for the CCR function. These two possibilities for CCR function, NES binding and mediating the conformational change, may not be mutually exclusive.
We showed previously that CRM1 was the major protein that bound LMB in vitro (9). In the present study, we demonstrated that CRM1 was the only protein detected as the in vivo LMB-binding protein (Fig. 4). Chemicals containing electrophilic structures such as α,β-unsaturated carbonyl groups can react by a Michael-type addition to biological nucleophiles, especially the sulfhydryl groups of cysteines (29). However, it is still unclear how LMB can work in the presence of high concentrations of l-cysteine and GSH in the cells and how LMB can selectively modify a specific, single cysteine residue on CRM1 among the many other cysteines in CRM1 and other proteins. Because l-cysteine and GSH contain free sulfhydryl groups, LMB conceivably might react with these compounds, thereby becoming unavailable to react with CRM1. However, an addition of a high concentration of GSH did not prevent in vitro Crm1 binding to biotinylated LMB (Fig. 1D). We could not detect LMB adducts with l-cysteine or GSH in vitro but we could detect it with N-acetyl-l-cysteine methyl ester. It therefore is possible that l-cysteine and GSH are too hydrophilic to gain access to the highly hydrophobic LMB, but that acetylation and methylation of l-cysteine sufficiently increase its hydrophobicity. The hydrophobic acyl side chain of LMB is also partly responsible for the specific alkylation of CRM1, because its shortening greatly reduced the biological activity despite its intact electrophilic structure (unpublished results). Because there is a distinct hydrophobic amino acid stretch (residues 517–528) in the N-terminal flanking region of Cys-529, it seems conceivable that the hydrophobic interaction between CRM1 and LMB contributes to the high affinity. However, the hydrophobic interaction alone cannot fully account for the in vivo specific binding at low nanomolar concentrations of LMB, because the reaction between LMB and N-acetyl-l-cysteine methyl ester or the synthetic peptide containing the hydrophobic stretch was not efficient and required a millimolar concentration of LMB. It therefore is more likely that a region containing Cys-529 forms a pocket into which LMB is readily incorporated, just like an enzyme–substrate interaction. An attractive scenario would be that this hydrophobic pocket is also responsible for binding to hydrophobic NES. The role of CCR in NES binding currently is under investigation.
Acknowledgments
We thank Dr. M. Yanagida, Graduate School of Biostudies, Kyoto University, and Drs. M. Yamamoto and S. Y. Shinozaki, Graduate School of Science, The University of Tokyo, for plasmids and S. pombe strains and Dr. T. Akisawa, Setsunan University, for synthesizing the peptides. We are also grateful to Dr. S. Khochbin, Institut National de la Santé et de la Recherche Médicale U309, Institut Albert Bonniot, and Drs. S. Aoki and M. Kobayashi, Osaka University, for sharing unpublished information and to Dr. M. Nishiyama, Biotechnology Research Center, The University of Tokyo, and Dr. Y. Yoshida, Tokyo Metropolitan Institute of Medical Science, for discussion. This work was supported in part by a special grant for Advanced Research on Cancer from the Ministry of Education, Culture, and Science of Japan, Takeda Science Foundation to M.Y., and Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists to N.K.
ABBREVIATIONS
- CCR
central conserved region
- CRM1
chromosomal region maintenance 1
- CRIME
CRM1, importin-β, etc
- GFP
green fluorescent protein
- GSH
glutathione
- LMB
leptomycin B
- NEM
N-ethylmaleimide
- NES
nuclear export signal
- NLS
nuclear localization signal
- GST
glutathione S-transferase
Footnotes
References
- 1.Mattaj I W, Englmeier L. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- 2.Pemberton L F, Blobel G, Rosenblum J S. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- 3.Stade K, Ford C S, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 4.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 5.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 6.Ossareh-Nazari B, Bachelerie F, Dargemont C. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 7.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 8.Kudo N, Khochbin S, Nishi K, Kitano K, Yanagida M, Yoshida M, Horinouchi S. J Biol Chem. 1997;272:29742–29751. doi: 10.1074/jbc.272.47.29742. [DOI] [PubMed] [Google Scholar]
- 9.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 10.Adachi Y, Yanagida M. J Cell Biol. 1989;108:1195–1207. doi: 10.1083/jcb.108.4.1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toda T, Shimanuki M, Saka Y, Yamano H, Adachi Y, Shirakawa M, Kyogoku Y, Yanagida M. Mol Cell Biol. 1992;12:5474–5484. doi: 10.1128/mcb.12.12.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishi K, Yoshida M, Fujiwara D, Nishikawa M, Horinouchi S, Beppu T. J Biol Chem. 1994;269:6320–6324. [PubMed] [Google Scholar]
- 14.Hamamoto T, Gunji S, Tsuji H, Beppu T. J Antibiot. 1983;36:639–645. doi: 10.7164/antibiotics.36.639. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida M, Nishikawa M, Nishi K, Abe K, Horinouchi S, Beppu T. Exp Cell Res. 1990;187:150–156. doi: 10.1016/0014-4827(90)90129-x. [DOI] [PubMed] [Google Scholar]
- 16.Wolff B, Sanglier J-J, Wang Y. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 17.Holaska J M, Paschal B M. Proc Natl Acad Sci USA. 1998;95:14739–14744. doi: 10.1073/pnas.95.25.14739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alfa C, Fantes P, Hyams J, McLeod M, Warbrick E. Experiments with Fission Yeast: A Laboratory Course Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 133–136. [Google Scholar]
- 19.Kumada K, Yanagida M, Toda T. Mol Gen Genet. 1996;250:59–68. doi: 10.1007/BF02191825. [DOI] [PubMed] [Google Scholar]
- 20.Ito H, Fukuda Y, Murata K, Kimura A. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toone W M, Kuge S, Samuels M, Morgan B A, Toda T, Jones N. Genes Dev. 1998;12:1453–1463. doi: 10.1101/gad.12.10.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kudo N, Taoka H, Toda T, Yoshida M, Horinouchi S. J Biol Chem. 1999;274:15151–15158. doi: 10.1074/jbc.274.21.15151. [DOI] [PubMed] [Google Scholar]
- 23.Görlich D, Dabrowski M, Bischoff F R, Kutay U, Bork P, Hartmann E, Prehn S, Izaurralde E. J Cell Biol. 1997;138:65–80. doi: 10.1083/jcb.138.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Görlich D, Vogel F, Mills A D, Hartmann E, Laskey R A. Nature (London) 1995;377:246–248. doi: 10.1038/377246a0. [DOI] [PubMed] [Google Scholar]
- 25.Moroianu J, Hijikata M, Blobel G, Radu A. Proc Natl Acad Sci USA. 1995;92:6532–6536. doi: 10.1073/pnas.92.14.6532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Askjaer P, Jensen T H, Nilsson J, Englmeier L, Kjems J. J Biol Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- 27.Malim M H, Bohnlein S, Hauber J, Cullen B R. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 28.Malim M H, McCarn D F, Tiley L S, Cullen B R. J Virol. 1991;65:4248–4254. doi: 10.1128/jvi.65.8.4248-4254.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lyss G, Knorre A, Schmidt T J, Pahl H L, Merfort I. J Biol Chem. 1998;273:33508–33516. doi: 10.1074/jbc.273.50.33508. [DOI] [PubMed] [Google Scholar]