Abstract
Background: Bacterial community structures in human pancreatic and biliary tracts were evaluated.
Methods: Gall bladder stones from 153 patients, 20 gall bladder walls, six common duct stones, 52 biliary stents, 21 duodenal biopsies, nine pancreatic duct biopsies, and five bile ducts were investigated using fluorescence in situ hybridisation (FISH) with ribosomal RNA targeted Cy3/Cy5 (carbocyanine) labelled oligonucleotide probes.
Result: Duodenal, gall bladder, and bile duct walls were free of bacteria. A dense multispecies bacterial biofilm was present within the pancreatic duct of patients with calcific pancreatitis and within biliary stents, irrespective of diagnosis. The concentration, density, and amenability of the biofilm to FISH and DNA staining declined progressively with the grade of stent occlusion. The lowest detectable bacterial concentrations were found by FISH in completely occluded stents and brown/mixed gall stones. Bacteria were not detectable with FISH in cholesterol gall stones.
Conclusions: A wide range of different branches and groups of bacteria participate in the development of biofilms on the surfaces of foreign bodies, such as biliary stents, mixed gall stones, or calcific pancreatic ducts, but not on the surface of pure cholesterol gall stones. Occlusion of stents leads to progressive extinction of the biofilm and mummification of its components. Deposition of cholesterol or other substances within the biofilm matrix may be a novel mechanism of host defence against bacteria present in these biofilms.
Keywords: bacterial biofilms, cholesterol, pancreas, biliary tract, gall stone pathogenesis, stent occlusion
Bacterial biofilms are increasingly identified as a source of many recalcitrant bacterial infections such as periodontal disease, endocarditis, chronic obstructive lung disease, foreign body related infections, etc.1,2 Gall stones are naturally occurring foreign bodies. The incidence, distribution, and impact of bacteria in gall stone formation are poorly understood. Currently accepted concepts of bacterial involvement are derived mainly from culture studies from the 1960s, supported by electron microscopy studies3,4 and molecular-genetic methods using polymerase chain reaction (PCR), with subsequent cloning and sequencing in the 1990s.5
The main disadvantage of both culture and PCR techniques is that they do not provide any information about spatial organisation of the bacteria either to each other or to the surface to which the biofilm is attached. They are therefore of limited use in evaluation of complex biofilms.
Our aims were to study the composition and spatial organisation of bacterial biofilms on the surface of duodenal, pancreatic, and bile duct epithelium within biliary stents, common duct/gall bladder stones, and on the gall bladder wall. We used fluorescence in situ hybridisation (FISH) with rRNA targeted fluorescent probes at the domain and group levels6 to assess the potential impact of bacteria in stent obliteration and gall stone pathogenesis.
METHODS
All human samples were removed for clinical indications, and represented in most cases material that would otherwise have been discarded. The use of this histological material for research aims and obtaining additional duodenal biopsies was approved by the Institutional Human Research Committee of the Charite Hospital and all patients gave written informed consent.
Investigations were performed on gall bladder stones from 153 patients. Twenty corresponding gall bladder walls and six endoscopically removed common duct stones were also studied. In addition, we studied 52 endoscopically removed biliary stents, 12 corresponding duodenal biopsies, nine pancreatic duct and corresponding nine duodenal biopsies from patients with calcific pancreatitis, and five common bile ducts from patients operated on for cholangiocarcinoma. All patients with chronic pancreatitis had strictures of the pancreatic duct and had undergone previous manipulations, such as sphincterotomy in all and endoprosthesis placement in four. None had pancreatic surgery.
Tissue preparation
Surgically removed gall bladders and biopsies of the pancreas, duodenum, and bile duct tissues were fixed in Carnoy solution7 and initially sent to the Department of Pathology for histological investigation. Thereafter paraffin blocks of these tissues were subjected to FISH evaluation.
Gall stone processing
All gall stones were placed in 10% neutral buffered formalin immediately after surgery. A total of 132 patients had cholesterol gall stones (cholesterol content >70%), 19 had mixed pigment gall stones (cholesterol content 30–70%), and two had brown pigment gall stones (<30% cholesterol). Four of the six common duct gall stones were mixed cholesterol gall stones and two were brown pigment gall stones. A 100 mg section of gall stone was crushed in 1.5 ml of physiological saline and vigorously shaken for five minutes. The mixture was centrifuged at 700 g for 30 seconds. Then, 750 μl of supernatant were taken, added to 750 μl of fresh physiological saline, shaken, and centrifuged once more for 30 seconds. Then, 750 μl of supernatant were centrifuged at 6500 g for 10 minutes. The precipitate was resuspended in 75% ethanol and stored at −20°C. The remnants of the stones and intact stones were stored in absolute alcohol at −20°C and used to prepare smears and slices.
Biliary stents
Removed undamaged biliary stents (Wilson-Cook Medical, Limerick, Ireland) were placed in 10% neutral buffered formalin. Two 2 cm pieces were cut off from the liver and duodenal ends of the stent and inspected for signs of occlusion. Physiological saline (2 ml) was passed by syringe through the lumen of the 2 cm stent pieces. The volume of the collected sludge was measured. To estimate the inside volume of the stent piece, the stent piece was dipped in a dish of water. One of the ends of the stent piece was closed under the water surface. The stent piece was than withdrawn and placed in a 2 ml Eppendorf tube. After centrifugation, the volume of fluid in the Eppendorf tube equalled the inside volume of the stent piece. The volume of extracted sludge was divided by the inside volume of the stent to quantify the grade of stent occlusion. Stents with a sludge volume <30% of stent volume were classified as narrowed. Stents with a sludge volume of 30–70% were considered partially occluded. The stent portion was defined as occluded if the sludge volume was >70% of stent volume.
Bacteria were extracted from the sludge and stored according to the protocol used for extraction of bacteria from gall stones.
A 3 mm section of the adjacent liver ends and duodenal ends of the biliary stents were cut and embedded in paraffin for further investigation of bacterial biofilm.
FISH
Fluorescence in situ hybridisation was performed on glass slides.8 Oligonucleotide probes were synthesised with a Cy3 or Cy5 (carbocyanine) reactive fluorescent dye at the 5′ end (MWG Biotech, Ebersberg, Germany). A set of 28 FISH probes at domain and group levels together with one species specific probe for Helicobacter pylori were used (table 1 ▶). Formamide concentration and hybridisation temperature were applied according to references, with the aim of achieving optimal stringency.
Table 1.
Fluorescence in situ hybridisation probes
| Name | Target | Reference No |
| Eub338 | Virtually all Bacteria, Kingdom (Eu)Bacteria | 9 |
| Alf1b | Alpha group of Proteobacteria: Rhodobacter, Acetobacter, Paracoccus, | 10 |
| Beta42a | Beta subclass of Proteobacteria: Rhodocyclus, Bordetella, Neisseria, Thiobacillus, Alcaligenes, and other | 10 |
| Gam42a | Gamma subclass of Proteobacteria: Enterobacteriaceae, Proteus, Legionella, Azotobacter | 10 |
| Ec1531 | Escherichia coli | 11 |
| Srb385 | Sulphate reducing bacteria | 12 |
| Hpy-1 | Helicobacter pylori epsilon subclass of Proteobacteria | 13 |
| Arc1430 | Arcobacter sp epsilon subclass of Proteobacteria | 8 |
| HGC | Gram positive bacteria with high G+C content: Actinobacteria | 14 |
| LGC | Gram positive bacteria with low G+C content: Firmicutes | 15 |
| Sfb | Segmented filamentous bacteria | 16 |
| Erec482 | Clostridium coccoides-Eubacterium rectale group | 17 |
| Chis150 | Clostridium histolyticum group | 17 |
| Clit135 | Clostridium. lituseburense group | 17 |
| Lab158 | Lactobacillus and Enterococcus group | 18 |
| Strc493 | Streptococcus | 17 |
| Ecyl | Eubacterium biforme, Clostridium innocuum, and other | 19 |
| Phasco | Acidaminococcus fermentans and other | 19 |
| Veil | Veillonella group | 19 |
| Rbvo,Rfla | Ruminococcus flavefaciens, Clostridium leptum | 19 |
| Bif164 | Bifidobacterium | 20 |
| Ato291 | Atopobium, Coriobacterium, Eggerthell, and Collinsella spp | 21 |
| CF319a | Cytophaga-Flavobacterium group | 22 |
| Bac303 | Bacteroides/Prevotella group | 22 |
| Bfra602 | Bacteroides fragilis group | 17 |
| Bdis656 | Bacteroides distasonis group | 17 |
| Fprau645 | Fusobacterium prausnitzii group | 23 |
| Non338 | Nonsense probe was used to test for non-specific binding | 10 |
Spatial arrangement and associative behaviour of bacteria were investigated on smears of sludge or stones or on 4 μm thick sections of corresponding material such as shock frozen gall stones, paraffin embedded gall bladder or bile duct walls, biliary stents, or pancreatic or duodenal biopsies.
To immobilise the amorphous cholesterol stone material and the heavy plastic parts of the stent sections, a temperature resistant Tesa sticky tape (NeoLab, Heidelberg, Germany) was spread over the shock frozen gall stone or stent and a 4 μm thick section was cut below the Tesa film. The sticky tape with the stone or stent section was then attached to a glass slide with the stone/stent surface lying above.
Quantification of bacteria extracted from stone/sludge
Bacteria extracted by dissolution and differential centrifugation of sludge were still organised in dense conglomerates bound by the organic matrix. For better quantification of cell numbers, 50 μl of the bacterial suspensions were dispersed in 980 μl of 0.1% sodium pyrophosphate by mild sonication (Branson Sonifer B-12; Danbury, USA). The sonicated suspension was centrifuged and dispersed in the initial volume and 5 μl of bacteria were placed in serial dilutions onto gelatine coated microscope slides (0.1% gelatine, 0.01% KCr(SO4)2).
When bacteria were visualised by FISH, 4,6-diamidino-2-phenylindole (DAPI), or Gram stain, three counts of 100 bacteria each from different sites were prepared and photo documented with a Nikon e600 fluorescence microscope (Nikon, Tokyo, Japan) for later comparison.
Biofilm structure
Group specific signals (orange fluorescence Cy3) were visualised simultaneously with an Eub338 probe targeting virtually all bacteria (far red fluorescence Cy5) or vice versa. The location of specific groups within the whole bacterial community was evaluated.
Ten digital pictures of bacteria at different sites on the microscopic slide were taken with a Nikon DXM1200 camera and software (Nikon, Tokyo, Japan).
Amenability to FISH and DNA stain
The number of ribosomes may reach 105 per metabolically active bacterial cell, permitting an intense fluorescence signal. The intensity of the hybridisation signal fades with falling activity or degradation of cell structures after cell death.24 Amenability of bacteria to FISH and other DNA stains is therefore a parameter characterising the state and integrity of bacteria.
Three different parameters were used to assess the amenability of cells to FISH: (1) ratio of Eub338 Cy3 positive and Gram positive cells; (2) hybridisation time necessary to achieve a fluorescence signal; and (3) time until complete exhaustion of the specific fluorescence.
In cases of low amenability of bacteria to FISH, horseradish peroxidase labelled oligonucleotide probes and tyramide were used to enhance signals.25
Statistics
Fisher’s exact test and the Mann-Whitney U test were used to determine the significance of differences.
RESULTS
Normal duodenum, gall bladder, and bile duct wall
No biofilm was detected on the surface of any of the 21 duodenal biopsies, 20 gall bladder walls, or five bile duct walls.
Calcific pancreatic duct
Bacteria were attached to the pancreatic duct epithelium in 7/9 pancreatic duct biopsies obtained from patients with calcific pancreatitis (figs 1 ▶–3 ▶). Dense biofilms covered significant portions of the pancreatic duct in three biopsies. Filaments of bacteria were arranged in palisade or curly hair-like layers lining the epithelial surface, intermixed or coated by singular cocci, or interposed by grape-like coccoid structures (figs 1 ▶–3 ▶).
Figure 1.
Biofilm from calcific pancreatic duct simultaneously hybridised with group specific (A) and universal (B) probes. Multispecies bacteria homogeneously woven into the biofilm.
Figure 3.
Biofilm from calcific pancreatic duct simultaneously hybridised with group specific (A) and universal (B) probes. Bacteria formed a patchy biofilm in which microcolonies composed of the same bacteria and possibly descendent from a single cell were interposed with microcolonies of another origin.
More than 19% of DAPI stained cells were also Eub338 positive. Approximately 50% of Eub338 positive bacteria could be affiliated to different groups (table 2 ▶).
Table 2.
Bacterial concentrations, amenability to fluorescence in situ hybridisation (FISH), biofilm structure, and composition
| Pancreatic duct (n = 9) | Narrowed stent ends (n = 44) | Partially occluded stent ends (n = 33) | Occluded stent ends (n = 19) | Brown/mixed gall stones (n = 27) | |
| Amenability to FISH | |||||
| % of bacteria amenable to FISH | 95 | 60–90 | 10–30 | <1 | <0.1 |
| Time necessary for FISH (h) | 1–2 | 3–5 | >12 | >12 | >12 |
| Time until exhaustion of Cy3 fluorescence (s) | 60 | 10–20 | <10 | <10 | <3 |
| Time necessary for DAPI stain (min) | 1 | 10–60 | 3–6 h | >12 h | >12 h |
| Biofilm structure | |||||
| Mean/max concentrations of all bacteria (/ml) | 2×108/1×1010 | 7×107/2×108 | 9×105/4×107 | 2×105/8×107 | |
| % of bacteria within a mixed multispecies biofilm | 30–40 | >90 | >90 | <30 | <10 |
| % of bacteria organised in islands of the same group | 30–70 | 5 | <1 | <1 | <1 |
| % singular bacteria scattered over microscopic field | <5 | <1 | <1 | 70–90 | >90 |
| Group specific probes | |||||
| Alf1b | |||||
| A | 4 (44%) | 7 (16%) | 2 (6%) | ||
| B | 5% | 0.01% | <0.01% | 0 | 0 |
| C | 40% | 1% | 0.1% | ||
| Beta42a | |||||
| A | 0 | 2 (5%) | 0 | 0 | 0 |
| B | <0.01% | ||||
| C | 0.1% | ||||
| Gam42a | |||||
| A | 1 (11%) | 26 (59%) | 17 (51%) | 10 (52%) | 9 (33%) |
| B | <0.1% | 19% | 11% | 14% | 4% |
| C | 5% | 80% | 80% | 80% | 30% |
| Ec1531 | |||||
| A | 0 | 19 (44%) | 15 (45%) | 9 (47%) | 9 (33%) |
| B | 10% | 6% | 8% | 3% | |
| C | 60% | 60% | 80% | 20% | |
| Srb385 | |||||
| A | 2 (22%) | 19 (44%) | 10 (30%) | 4 (21%) | 0 |
| B | 0.5% | 0.1% | <0.01% | <0.01% | |
| C | 5% | 3% | 0.1% | 0.1% | |
| HGC | |||||
| A | 13 (30%) | 10 (30%) | 7 (36%) | 8 (29%) | |
| B | 0 | 1.3% | 2.4% | 2.0% | 1% |
| C | 30% | 40% | 20% | 10% | |
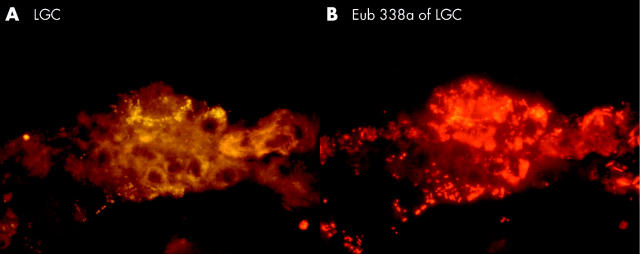
| LGC* | |||||
| A | 6 (66%) | 24 (54%) | 21 (63%) | 12 (63%) | 22 (81%) |
| B | 8% | 6.8% | 15% | 22% | 30% |
| C | 30% | 80% | 60% | 60% | 80% |
| Lab158 | |||||
| A | 4 (44%) | 20 (45%) | 21 (63%) | 9 (47%) | 10 (37%) |
| B | 1% | 2.5% | 2% | 10% | 5% |
| C | 10% | 20% | 40% | 25% | 40% |
| Strc493* | |||||
| A | 6 (66%) | 30 (68%) | 20 (60%) | 14 (73%) | 20 (74%) |
| B | 3% | 1% | 6% | 24% | 30% |
| C | 20% | 10% | 50% | 50% | 60% |
| Bif164 | |||||
| A | 2 (22%) | 9 (20%) | 12 (36%) | 5 (18%) | 0 |
| B | <0.01% | 1% | <0.1% | <0.01% | |
| C | 0.01% | 5% | 3% | 1% | |
| Erec482 | |||||
| A | 0 | 3 (7%) | 2 (6%) | 0 | 0 |
| B | 0.3% | 0.4% | |||
| C | 10% | 10% | |||
| Chis150 | |||||
| A | 1 (11%) | 12 (27%) | 10 (30%) | 11 (58%) | 0 |
| B | <0.1%/ | 0.2% | 0.1% | 0.03 | |
| C | 0.1% | 5% | 3% | 0.5% | |
| Clit135 | |||||
| A | 0 | 22 (50%) | 10 (30%) | 11 (58%) | 0 |
| B | 0.2% | 0.1% | 1% | ||
| C | 3% | 5% | 8% | ||
| CF319a | |||||
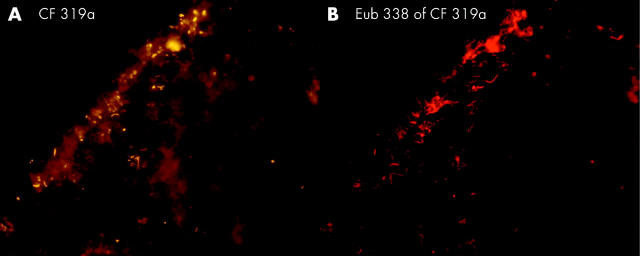
| A | 4 (44%) | 24 (55%) | 20(60%) | 5 (26%) | 0 |
| B | 8% | 1.5% | 1% | 0.3% | |
| C | 40% | 20 | 10% | 1% | |
| Bac303 | |||||
| A | 3 (33%) | 34 (77%) | 25 (76%) | 11 (58%) | 22 (81%) |
| B | 5% | 25% | 12% | 12% | 8% |
| C | 15% | 80% | 50% | 30 | 80 |
A = number (%) of patients colonised with a specific group.
B = mean per cent of all bacteria amenable to FISH positively hybridised with a group specific probe.
C = maximal per cent of all bacteria amenable to FISH (Eub 338 + 4,6-diamidino-2-phenylindole (DAPI) stain) positively hybridised with a group specific probe.
*Numbers are given after lysozyme treatment.
The associative behaviour of bacteria was manifold (figs 1 ▶–3 ▶). Thirty to 40% of bacteria were homogeneously woven into the biofilm. Most often, however, the biofilm was composed of patchy islands in which microcolonies, composed of the same bacteria and possibly descendent from a single cell, were interposed with microcolonies of another origin (table 2 ▶, fig 3 ▶).
Bacteria in pancreatic duct biopsies were highly fluorescent after one hour of hybridisation and the hybridisation signal was still measurable even after 60 seconds of exposure. Bacteria were seen after only one minute with the DAPI stain.
Biliary stents
No sludge was detected in four of the 52 stents. Both ends were occluded in five stents. Nine stents were occluded at the liver side and partially occluded on the duodenal side. Six stents were partially occluded on both sides. Twelve stents were partially occluded at the liver side and narrowed at the duodenal side. Sixteen stents were narrowed in their lumen without relevant occlusion.
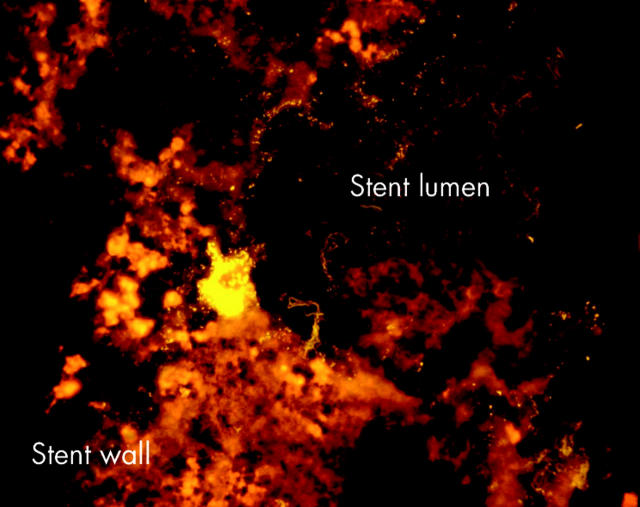
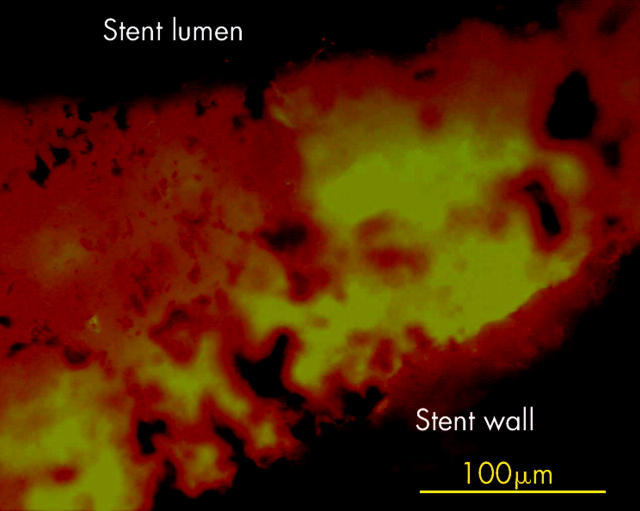
Four biliary stents without sludge had no bacteria in the fluid with which they were rinsed. Nevertheless, a thin “sessile” bacterial biofilm was found attached to the stent wall when microtome sections of stents were investigated under the microscope. In situ hybridisation of narrowed and occluded stent sections showed interwoven multispecies biofilms consisting of fuzzy long filaments, more or less evenly interposed with rods and cocci. The biofilm was either directly attached to the inner surface of the stent or spread over the sludge matrix, which continuously covered the inner side of the stent (figs 4 ▶–7 ▶). Bacteria were always above and never below the sludge matrix.
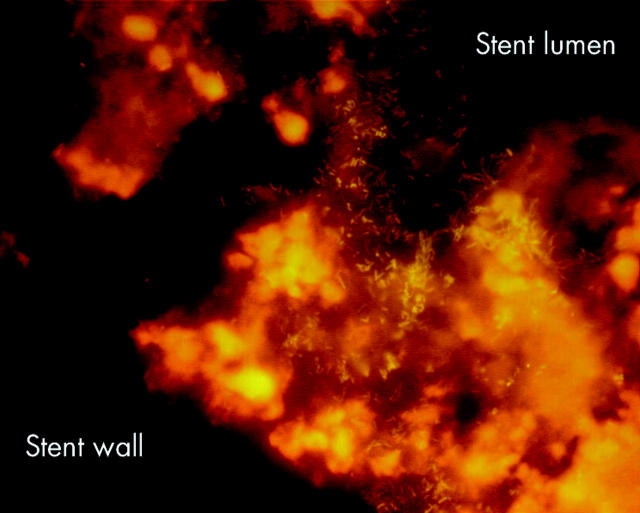
Figure 4.
Whole stents were hybridised with the universal Eub338 probe. Bacteria (green-orange signal) were spread over the sludge located always above and never below the sludge matrix. Bacteria vanished from the lumen with growing occlusion of the bile stent. Duodenal part of a partially occluded stent.
Figure 7.
Whole stents were hybridised with the universal Eub338 probe. Bacteria (green-orange signal) were spread over the sludge located always above and never below the sludge matrix. Bacteria vanished from the lumen with growing occlusion of the bile stent. Liver side of a completely occluded stent.
As immobilisation of the heavy plastic stent section on glass proved to be difficult, the composition of the bacterial community in stent biofilms was studied on smears of sludge. A rich complex bacterial biofilm was found in the sludge from all narrowed or partially occluded stents.
More than 95% of bacteria in the sludge were organised in clot-like structures and only a small part was singularly spread over the glass surface (table 2 ▶, figs 8 ▶–10 ▶).
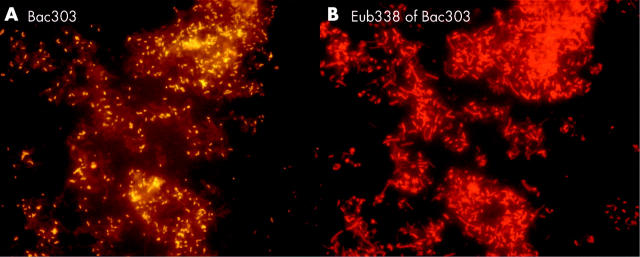
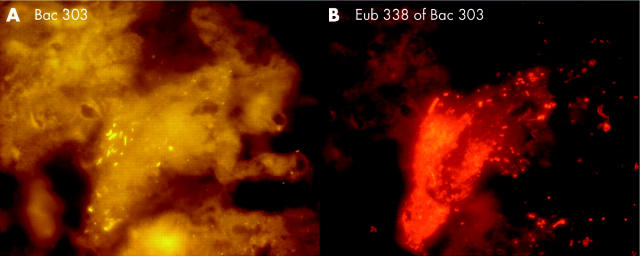
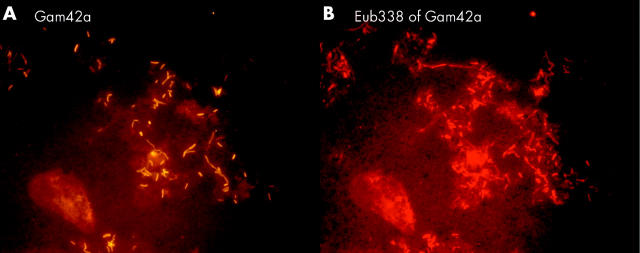
Figure 8.
Stent sludge hybridised with group specific and universal fluorescence in situ hybridisation probes. The same field is shown hybridised with the indicated group specific probe (A) and the Eub338 probe that detects all bacteria (B). Most of the bacteria in the biliary stents were primarily organised in heterogeneously mixed multispecies biofilms.
Figure 10.
Stent sludge hybridised with group specific and universal fluorescence in situ hybridisation probes. The same field is shown hybridised with the indicated group specific probe (A) and the Eub338 probe that detects all bacteria (B). Most of the bacteria in the biliary stents were primarily organised in heterogeneously mixed multispecies biofilms.
Bacteroides, Gamma-Proteobacteria, Firmicutes, and Clostridium lituseburense groups were most predominant, comprising up to 60% of all Eub338 positive bacteria. Together with HGC, Ato291, Clit135, Lab158, Bif164, and Erec482 group probes, more than 90% of all FISH accessible bacteria could be affiliated to groups (table 2 ▶). Apart from these main groups, all probes (table 1 ▶) hybridised positively in at least 10 specimens. Although the percentage was often less than 1% of all FISH accessible bacteria in a sample, the overall concentrations of the “underrepresented” bacteria within the biofilm were >106/ml and therefore significant. Helicobacter was not found. None of the patients had less than 10 different bacterial groups in their biofilm.
Degree of occlusion of the stent
The composition, density, and amenability of the biofilm depended strongly on the degree of stent occlusion.
Narrowed stents
The highest bacterial concentration was found in stents that were narrowed but not occluded at either end (table 2 ▶). Bacterial concentration was higher in the liver end than in the duodenal end of the stent. The biofilm was mostly of heterogeneously mixed multispecies origin. However, as was noted in studies of the calcific pancreatic duct, islands of single monobacterial species and long chains were occasionally found embedded within the biofilm layer. These “islands” hybridised most often with Strc493, Gam42a, Bac303, and LGC probes. The amenability of bacteria to FISH and DAPI was high. The numbers of bacteria detected on Gram stain were similar to those enumerated by hybridisation with the Eub338 probe (table 2 ▶).
Partially occluded stents
Mean bacterial concentration in partially occluded stents was significantly lower than in narrowed stents (p<0.05). Visually, bacterial biofilms were much less dense. All of the bacteria demonstrated a characteristic associative behaviour forming nearly equidistributed multispecies conglomerates. The islands of bacterial “monocultures” were no longer seen. The amenability of bacteria to FISH and DAPI stain was notably diminished. Overnight hybridisation was necessary for stable signals. After overnight hybridisation, fluorescence expired in <10 seconds. The numbers of bacteria seen on Gram stain were twice as high as the numbers enumerated by FISH. More than three hours were necessary for a positive DAPI stain (table 2 ▶).
Occluded stents
Mean concentration of bacteria detectable with FISH declined significantly in occluded stents compared with narrowed stents (p<0.001). Positive fluorescence was achieved only after 12 hours of hybridisation. The fluorescence signal extinguished after 3–6 seconds of exposure. Less than 10% of the bacteria seen by autofluorescence or on Gram stain were amenable to FISH.
The groups of bacteria found in narrowed and occluded biliary stents were similar (table 2 ▶) but the proportion of major bacterial groups within the biofilm diminished relative to the proportion of less numerous groups.
No biofilm, but scattered bacteria in negligible concentrations, was found in four liver ends of five completely occluded stents.
Gall stones
Bacterial concentrations of 105/ml were observed in eight of 23 mixed cholesterol gall stones and in all four brown pigment gall stones (p<0.03). Mean concentrations of bacteria detected with FISH did not differ statistically from those observed in occluded parts of the stents. Gram stain and autofluorescence revealed a large number of structures resembling bacteria. Most of them however could not be definitively identified as such, and only a small part of them gave a positive hybridisation signal. Fluorescence was extremely weak, often vanishing while the picture was being focused. The specificity of hybridisation with the group specific probes was difficult to prove because of the weakness of the fluorescence signal. We found no notable difference in organisation of microbiota between occluded stents and gall stones or between gall bladder and choledochal stones.
No bacteria were detected by FISH in elutes of all 132 cholesterol stones while a background of “shadows resembling bacteria” were seen in elutes of 15 of the 23 mixed stones (p<0.001). Single bacteria in low concentrations within these shadows hybridised positively with the Eub338 probe. Gram stain was difficult to interpret, and the DAPI stain was not helpful. FISH using horseradish peroxidase labelled oligonucleotide probes and tyramide signal amplification did not result in increased signal intensities.25
Because of the low concentrations of positively hybridised bacteria and their diverse morphological appearance, no further differentiation with group specific probes was made.
DISCUSSION
Our data are the first to demonstrate in situ the existence of rich microbial biofilms within diseased pancreatic and biliary tracts. As no biofilm was found in corresponding duodenal biopsies, or on the epithelium of gall bladder or bile duct walls, we must assume that bacteria detected within biliary stents, calcific pancreatic ducts, or gall stones are opportunistic newcomers taking advantage of foreign bodies or destructive anatomical changes and not sessile residents specialised for life within the pancreatic-biliary tree. Similar to the situation in purulent processes, we would have expected that occlusion of the stent would hinder the host to suppress the bacterial biofilm, and expected higher bacterial concentrations in occluded compared with open parts of the stent. The opposite was found. Bacterial concentrations, density, and amenability to FISH and DNA stain decreased gradually with increasing obliteration of the stent. Mean bacterial concentrations were four logs lower in completely occluded than in narrowed parts of stents. Low bacterial concentrations similar to those in occluded parts of the stents were found in mixed cholesterol and brown pigment gall stones. The lowest bacterial concentrations, almost zero, were found in the liver end of totally occluded stents and in cholesterol gall stones. Although a large number of structures resembling bacteria on Gram stain and autofluorescence were still seen in this cholesterol-rich material, most did not or only poorly hybridised with bacterial probes.
The number of ribosomes that can be targeted by hybridisation probes declines with degradation of cell structures. Changes in permeability of the cell walls can also hinder probe penetration of the cell and reaching the target molecule. The observed changes in the intensity of the fluorescence signal and DAPI stain in cholesterol-rich gall stones may indicate a shrinking vitality, growing degradation of bacteria within biofilms, and mummification of bacterial walls and structures. The lack of hybridisation signal in cholesterol gall stones and some completely occluded pieces of stents does not mean that bacteria were physically absent. In our previous studies using quantitative PCR with subsequent cloning and sequencing, we showed that nearly all cholesterol gall stones contained DNA sequences of diverse bacterial groups,5 and that concentrations of bacterial DNA declined within weeks on storage of frozen stones.26 Thus bacteria were obviously there but were not viable and were undergoing degradation at the time of investigation.
Gall stones are traditionally divided into three main groups: cholesterol (>70% of cholesterol), brown pigment gall stones (<30% of cholesterol), and black pigment gall stones (nearly no cholesterol).26 The high frequency of mixed cholesterol gall stones in our series of common duct gall stones was due to the difficulties in recovering amorphous brown pigment gall stones with the endoscope. The number of “metabolic” black pigment stones is usually very small, and we had none in our series. Bacteria were readily found in brown pigment stones but not in cholesterol gall stones, which had led to the assumption that the brown pigment stones are of infectious origin whereas cholesterol stones are due to metabolic disturbances.27
It was demonstrated in the 1960s that beta glucuronidase from E coli deconjugates bilirubin diglucuronide to free bilirubin in bile which then precipitates as insoluble calcium bilirubinate, the main component of brown gall stones. Bacteria induced deconjugation of bilirubin is still thought to be the main mechanism of brown stone formation. There is however broad structural overlap between brown and cholesterol gall stones. A large number of gall bladder stones are mixed or at least a partial composite of cholesterol and brown pigment gall stones. This has led to speculation that the pathogenesis of brown and cholesterol gall stones may be similar but due to different pathways. Cholesterol gall stones were therefore screened for bacteria with every new emerging technique. Electron microscopy and molecular genetic methods were applied. Bacterial remnants were found in cholesterol stones but a viable bacterial population was missing.27 The hypothesis of bacterial involvement in cholesterol gall stone formation was therefore never popular. Mechanisms of bile cholesterol saturation were the mainstream of research. A very important question was never answered. How do cholesterol stones stay sterile? The uneven cleft surface of cholesterol gall stone offers ideal shelter for microbial biofilm. Organic components of the gall stones could be an energy source. The biliary tract is bordered on one side by the intestine, a large culture fermenter of complex microbiota, and on the other side by the liver, which filters and secretes bacteria from blood. Gall stones should provide an ideal environment for development of biofilms because they persist over dozens of years in the human body. But cholesterol stones are mostly sterile at the time of elective cholecystectomy in spite of documented episodes of purulent cholangitis in between. Why is this?
The overall conditions at the surface of non-occluded biliary stents and on the surface of brown and cholesterol gall stones are the same. Either lack of nutrients or presence of soluble inhibitors within bile could explain the observed depletion of bacterial biofilm from bile stents to cholesterol gall stones.
We hypothesise that both cholesterol stone formation and stent occlusion may represent physiological responses that provide resistance to biofilm propagation on foreign bodies within the biliary tract. However, the specific role of cholesterol and other substances has yet to be defined. These studies may help in understanding new mechanisms with which the body can protect itself against otherwise extremely recalcitrant bacterial biofilms.
Figure 2.
Biofilm from calcific pancreatic duct simultaneously hybridised with group specific (A) and universal (B) probes. Multispecies bacteria homogeneously woven into the biofilm.
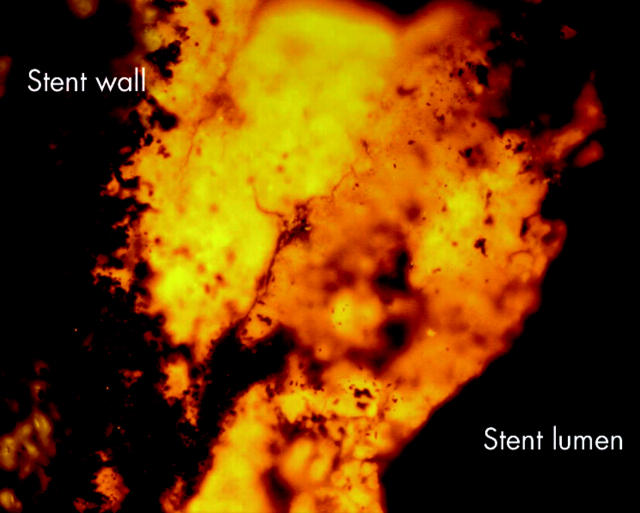
Figure 5.
Whole stents were hybridised with the universal Eub338 probe. Bacteria (green-orange signal) were spread over the sludge located always above and never below the sludge matrix. Bacteria vanished from the lumen with growing occlusion of the bile stent. Liver side of a partially occluded stent.
Figure 6.
Whole stents were hybridised with the universal Eub338 probe. Bacteria (green-orange signal) were spread over the sludge located always above and never below the sludge matrix. Bacteria vanished from the lumen with growing occlusion of the bile stent. Duodenal side of a completely occluded stent.
Figure 9.
Stent sludge hybridised with group specific and universal fluorescence in situ hybridisation probes. The same field is shown hybridised with the indicated group specific probe (A) and the Eub338 probe that detects all bacteria (B). Most of the bacteria in the biliary stents were primarily organised in heterogeneously mixed multispecies biofilms.
Acknowledgments
We thank Broad Medical Research Program of the Eli and Edythe L Broad Foundation for supporting this project.
Abbreviations
Cy3/Cy5, different carbocyanine dyes used in fluorescence microscopy to label the oligonucleotide probes
DAPI, 4,6-diamidino-2-phenylindole
FISH, fluorescence in situ hybridisation
PCR, polymerase chain reaction
Conflict of interest: None declared.
REFERENCES
- 1.Tool GO, Kaplan HB, Kolter R. Biofilm formation as microbial development. Ann Rev Microbiol 2000;54:49–79. [DOI] [PubMed] [Google Scholar]
- 2.Costerton W, Veeh R, Shirtliff M, et al. The application of biofilm science to the study and control of chronic bacterial infections. J Clin Invest 2003;112:1466–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stewart L, Smith AL, Pellegrini AC, et al. Pigment gallstones form as a composite of bacterial microcolonies and pigment solid. Ann Surg 1987;206:242–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung JWC, Sung JY, Costerton JW. Bacteriological and electron microscopy examination of brown pigment stones. J Clin Microb 1989;27:915–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Swidsinski A, Wolfgang L, Pahlig H, et al. Molecular genetic evidence of bacterial colonization of cholesterol gallstones. Gastroenterology 1995;108:860–4. [DOI] [PubMed] [Google Scholar]
- 6.Amann RI, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbial Rev 1995;59:143–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuo K, Ota H, Akamatsu T, et al. Histochemistry of the surface mucous gel layer of the human colon. Gut 1997;40:782–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Snaidr J, Amann R, Huber I, et al. Phylogenetic analysis and in-situ identification of bacteria in activated sludge. Appl Environ Microbiol 1997;63:2884–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amann R, Krumholz L, Stahl DA. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J Bacteriol 1990;172:762–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manz W, Amann R, Ludwig W, et al. Phylogenetic oligodeoxynucleotide probes for the major subclasses of Proteobacteria: problems and solutions. System Appl Microbiol 1992;15:593–600. [Google Scholar]
- 11.Poulsen LK, Licht TR, Rang C, et al. Physiological state of Escherichia coli BJ4 growing in the large intestines of streptomycin-treated mice. J Bacteriol 1995;177:5840–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amann R, Stromley J, Devereux R, et al. Molecular and microscopic identification of sulfate-reducing bacteria in multispecies biofilms. Appl Environ Microbiol 1992;58:614–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Feydt-Schmidt A, Rüssmann H, Lehn N, et al. Fluorescence in situ hybridization vs. epsilometer test for detection of clarithromycin-susceptible and clarithromycin-resistant Helicobacter pylori strains in gastric biopsies from children. Aliment Pharmacol Ther 2002;16:2073–9. [DOI] [PubMed] [Google Scholar]
- 14.Roller C, Wagner M, Amann R, et al. In situ probing of gram-positive bacteria with high DNA G + C content using 23S rRNA-targeted oligonucleotides. Microbiology 1994;140:2849–58. [DOI] [PubMed] [Google Scholar]
- 15.Meier H, Amann R, Ludwig W, et al. Specific oligonucleotide probes for in situ detection of a major group of gram-positive bacteria with low DNA G + C content. Syst Appl Microbiol 1999;22:186–96. [DOI] [PubMed] [Google Scholar]
- 16.Urdaci MC, Regnault B, Grimont PAD. Identification by in situ hybridization of segmented filamentous bacteria in the intestine of diarrheic trout (Oncorhynchus mykiss). Res Microbiol 2001;152:67–73. [DOI] [PubMed] [Google Scholar]
- 17.Franks AH, Harmsen HJ, Raangs GC, et al. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol 1998;64:3336–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harmsen HJ, Elfferich P, Schut F, et al. A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in fecal samples by fluorescent in situ hybridization. Microbiol Ecol Health Dis 1999;11:3–12. [Google Scholar]
- 19.Harmsen HJ, Raangs GC, He T, et al. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl Environ Microbiol 2002;68:2982–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langendijk PS, Schut F, Jansen GJ, et al. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16SrRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol 1995;61:3069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harmsen HJ, Wildeboer-Veloo AC, Grijpstra J, et al. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobiumcluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl Environ Microbiol 2000;66:4523–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manz W, Amann R, Ludwig W, et al. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 1996;142:1097–106. [DOI] [PubMed] [Google Scholar]
- 23.Suau A, Rochet V, Sghir A, et al. Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Syst Appl Microbiol 2001;24:139–45. [DOI] [PubMed] [Google Scholar]
- 24.Møller S, Sternberg C, Andersen JB, et al. In situ gene expression in mixed-culture biofilms: evidence of metabolic interactions between community members. Appl Environ Microbiol 1998;64:721–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pernthaler A, Pernthaler J, Amann R. Fluorescence in situ hybridization and catalyzed reporter deposition for the identification of marine bacteria. Appl Environ Microbiol 2002;68:3094–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Swidsinski A, Khilkin M, Pahlig H, et al. Time dependent changes in the concentration and type of bacterial sequences found in cholesterol gallstones. Hepatology 1998;27:662–5. [DOI] [PubMed] [Google Scholar]
- 27.Swidsinski A, Lee SP. The role of bacteria in gallstone pathogenesis. Front Biosci 2001;6:93–103. [DOI] [PubMed] [Google Scholar]