Abstract
The major pathway of genetic recombination and DNA break repair in Escherichia coli requires RecBCD enzyme, a complex nuclease and DNA helicase regulated by Chi sites (5′-GCTGGTGG-3′). During its unwinding of DNA containing Chi, purified RecBCD enzyme has two alternative nucleolytic reactions, depending on the reaction conditions: simple nicking of the Chi-containing strand at Chi or switching of nucleolytic degradation from the Chi-containing strand to its complement at Chi. We describe a set of recC mutants with a novel intracellular phenotype: retention of Chi hotspot activity in genetic crosses but loss of detectable nucleolytic degradation as judged by the growth of mutant T4 and λ phages and by assay of cell-free extracts. We conclude that RecBCD enzyme's nucleolytic degradation of DNA is not necessary for intracellular Chi hotspot activity and that nicking of DNA by RecBCD enzyme at Chi is sufficient. We discuss the bearing of these results on current models of RecBCD pathway recombination.
HOMOLOGOUS genetic recombination is important for the faithful repair of double-strand breaks in DNA and for the generation of genetic diversity. In the bacterium Escherichia coli, the major (RecBCD) pathway of recombination and repair of linear double-strand (ds) DNA requires the unusually complex RecBCD enzyme. Composed of three subunits, the purified enzyme has at least six activities: DNA unwinding (helicase), ds exonuclease, single-stranded (ss) exonuclease, DNA-dependent ATPase, weak ss endonuclease, and loading of RecA protein onto ssDNA (reviewed by Kowalczykowski 2000; Smith 2001). The activity of RecBCD enzyme is regulated by Chi sites (5′-GCTGGTGG-3′), which were first recognized as hotspots of recombination in bacteriophage λ (reviewed by Smith 2001). The molecular basis of Chi's hotspot activity, the localized stimulation of intracellular recombination, is not clearly established because of the difficulty of inferring intracellular reactions from the properties of purified enzymes and DNA. Here, we describe multiple alleles of recC whose novel phenotype indicates that Chi hotspot activity can occur in the absence of extensive nucleolytic degradation. We discuss how these results bear on previously proposed models of Chi-stimulated recombination by the RecBCD pathway (Smith et al. 1981b; Thaler et al. 1988; Anderson and Kowalczykowski 1997a).
A combination of both genetic and physical studies of RecBCD enzyme has led to the following picture (Figure 1). RecBCD enzyme binds tightly to a dsDNA end, from which it commences unwinding (Taylor and Smith 1980, 1995a). The RecB and RecD subunits each contain an ATP-dependent helicase: RecB travels on one strand in the 3′ → 5′ direction, and RecD on the other strand in the 5′ → 3′ direction (Phillips et al. 1997; Dillingham et al. 2003; Taylor and Smith 2003). Since RecB travels more slowly than RecD, a single-stranded loop accumulates ahead of RecB and grows as the reaction proceeds (Taylor and Smith 2003). RecBCD is highly processive and can unwind >30 kb of DNA without dissociating from the DNA (Taylor and Smith 1980; Roman et al. 1992). A Chi site affects RecBCD only as the enzyme is unwinding DNA and only if it encounters 5′-GCTGGTGG-3′ from the right, as written here (Taylor et al. 1985). Current evidence suggests that the RecC subunit recognizes Chi, perhaps when the strand with Chi moves through the RecB helicase domain and into a tunnel in RecC (Figure 2A; Singleton et al. 2004).
Figure 1.—
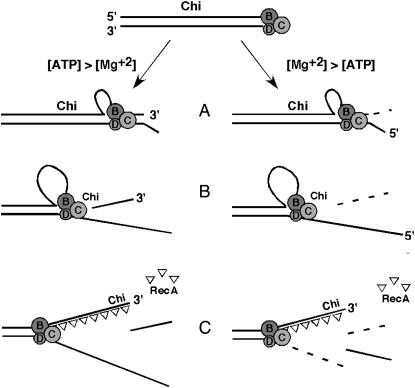
Two reactions of purified RecBCD enzyme at Chi, depending on the ATP:Mg2+ ratio. (Top) RecBCD binds to a dsDNA end, the RecB helicase being engaged with the 3′-end and the RecD helicase with the 5′-end. RecD is faster than RecB, and a ssDNA loop accumulates on the 3′ → 5′ (“top”) strand ahead of the RecB subunit (A). (Left) If the concentration of ATP is greater than that of Mg2+, RecBCD nuclease activity is low, and the enzyme nicks the top strand at Chi (B). (Right) If the concentration of Mg2+ is greater than that of ATP, RecBCD nuclease activity is high, and the enzyme degrades the top strand up to Chi (B), nicks the bottom strand, and degrades the bottom strand to the left of Chi (C). RecBCD loads RecA protein onto the top strand to the left of Chi (the “Chi tail”) (C). See Introduction and discussion for further description and references.
Figure 2.—
Structure of RecBCD bound to a dsDNA end (after Singleton et al. 2004). The RecB subunit is orange, RecC is blue, and RecD is green. (A) Surface representation. The 4 terminal base pairs of DNA are unwound, and the 3′-end lies within RecB. During unwinding, this strand is postulated to pass through a tunnel in RecB and RecC on its way to the nuclease domain of RecB. Chi is postulated to be recognized by parts of the tunnel in RecC (red arrow). The 5′-end of the DNA lies within RecC and extends toward an unordered part of RecD lying behind the surface shown. (B) Ribbon representation. The C terminus of RecC is red (residues 790–840) or magenta (residues 841–1122) to indicate the region deleted by recC2717 or truncated by the recC1041 nonsense mutation (see Figure 3). These mutants have the Chi+ Nuc− Rec+ (†) phenotype (see results). Trp841 lies behind a magenta helix but in front of the DNA (light blue line). The C-terminal domain of RecC may contact the unordered part of RecD (ends marked with black dots) and aid assembly of RecD into the holoenzyme complex (Amundsen et al. 2002). The orange dot near the bottom indicates the Ca2+ ion crystallized in the nuclease active site. Relative to the view in A, the top of the molecule is tilted slightly forward and rotated slightly clockwise, as viewed from the top. Images were constructed with PyMol and data for RecBCD from the Protein Databank (accession code 1W36).
When Chi is properly encountered, the activities of purified RecBCD are changed in a manner that depends on the reaction conditions (Figure 1). If the concentration of ATP is greater than that of Mg2+ ions (Figure 1, left), then RecBCD nicks the strand with Chi a few nucleotides to the 3′ side of this sequence; continued unwinding produces a ssDNA fragment with Chi at its 3′-end (Ponticelli et al. 1985; Taylor et al. 1985; Taylor and Smith 1995b). If the concentration of Mg2+ is greater than that of ATP (Figure 1, right), RecBCD degrades the 3′-ended strand up to the Chi sequence (Dixon and Kowalczykowski 1993), cuts the complementary strand (Taylor and Smith 1995b), and continues to degrade that strand (in the 5′ → 3′ direction) (Anderson and Kowalczykowski 1997a); this reaction also produces ssDNA with Chi at or near its 3′-end, called here the “Chi tail.” Thus, in the first reaction, Chi induces DNA cutting of the strand engaged by RecB to produce the Chi tail, whereas in the second reaction Chi blocks DNA cutting of that strand to produce the Chi tail. Upon exiting the tunnel in RecC noted above, the 3′-ended strand is postulated to enter one of two channels: one directed toward the nuclease domain and the other directed away from it (Singleton et al. 2004). The choice of channel may depend upon the reaction conditions and may change at Chi, to induce or block DNA cutting. Chi also triggers RecBCD to load RecA protein onto the ss Chi tail, at least under the second reaction condition (Anderson and Kowalczykowski 1997b); self-loading of RecA onto ssDNA under the first reaction condition precludes determining if purified RecBCD has that activity under the first condition as well.
In E. coli cells there is an excess of total Mg2+ over ATP, but most of the Mg2+ is bound, probably to nucleic acids (Moncany and Kellenberger 1981). The effective concentration of Mg2+ available to RecBCD enzyme is unknown, and therefore it is not clear how RecBCD acts at Chi in cells. To address this question, we have studied mutant forms of RecBCD enzyme that allow a comparison of the intracellular phenotypes and the properties of mutant RecBCD enzymes, either in unfractionated extracts or after purification. The novel phenotype of the class of recC mutants described here indicates that RecBCD exonuclease and therefore degradation up to Chi are not necessary for Chi's stimulation of recombination.
MATERIALS AND METHODS
Culture media:
Rich media (broth and agar plates), containing Difco tryptone–yeast extract (LB) or Difco tryptone (TB), minimal medium agar (OMBG), and phage suspension medium (SM) are described by Cheng and Smith (1989). Ampicillin (100 μg/ml), streptomycin (50 μg/ml), tetracycline (25 μg/ml), and kanamycin (25 μg/ml) were used where appropriate.
Bacterial and phage strains, plasmids, and genotype designations:
Supplemental Tables S1 and S2 at http://www.genetics.org/supplemental/ list the E. coli strains and plasmids with their genotypes and sources. Genealogies are available upon request. Table 1 lists the E. coli rec and related mutations used here. All strains carry the recF143 mutation so that recombination is limited to the RecBCD pathway (Smith 1988). The genotype recBCD+ indicates recB+ recC+ recD+. Genotypes such as recC1041 imply recB+ recC1041 recD+ unless designated otherwise. For visual clarity, proteins are indicated with superscript allele numbers, such as RecBC1041D. Phage T4 and its gene 2 (amN51) mutant are described by Epstein et al. (1964) and Silverstein and Goldberg (1976). λ phages for Chi hotspot crosses contained b1453 (an int red gam deletion) plus susJ6 cI857 χ+D123 (strain 1081), χ+D123 susR5 (1082), susJ6 χ+76 cI857 (1083), and susJ6 χ+76 (1084) and are described by Stahl and Stahl (1977).
TABLE 1.
rec and related mutations used in this study
| Allele | Description | Source or reference |
|---|---|---|
| recB21∷IS186 | IS186 insertion at codon 305 | Schultz et al. (1983); Amundsen et al. (2000) |
| recC73 | ΔT at codon 646 | Schultz et al. (1983); Arnold et al. (2000) |
| recC1041 | Trp841 → UGA841 | Amundsen et al. (2002) |
| recC1010 | Gly905 → Glu905 | Chaudhury and Smith (1984b); Amundsen et al. (2002) |
| recC2710–C2718 | ExoIII-generated deletions (see Figure 3) | Amundsen et al. (2002) |
| recC2725 | UGA841 UGA842 plus C-terminal deletiona | This study |
| recC2726 | UGA841 UAA842 plus C-terminal deletionb | This study |
| ΔrecC2730∷kan | Substitution of kan for recC ORF | This study |
| recD1013 | Gln4 → UAA4 | Chaudhury and Smith (1984b); Amundsen et al. (1986, 2002) |
| ΔrecBCD232 | thyA–recBCD–argA deletion | Chaudhury and Smith (1984a) |
| ΔrecBCD234 | recC–argA deletionc | Chaudhury and Smith (1984a) |
| ΔrecA∷kan | Substitution of kan for recA ORF | Reddy and Gowrishankar (2000) |
| recJ284∷Tn10 | Tn10 insertion | Lovett and Clark (1984) |
| ΔxonA∷FRTkanFRT | Substitution of kan for xonA ORF | Feschenko et al. (2003) |
| sbcB15 | Ala183 → Tyr183 | GenBank accession AM235176 |
The deletion leaves codons 1–840 intact.
The deletion leaves codons 1–840 intact. An additional 6 bp specifying the KpnI recognition site (5′-GGTACC-3′) are 3′ of UAA842.
The deletion ends between codons 605 and 630 of recC (Figure 3; Amundsen et al. 2002).
Strain constructions:
E. coli strains were constructed by phage P1 transduction, transformation with plasmids, or by “recombineering” using appropriate mutant oligonucleotides (Integrated DNA Technologies, Coralville, IA, or Invitrogen, Carlsbad, CA) and electroporation of strain DY378 or HME63 (Thomason et al. 2005). Plasmid mutations were made by PCR-based strategies, some with the QuikChange kit (Stratagene, La Jolla, CA), and appropriate mutant oligonucleotides; details are available upon request. Plasmid and chromosomal mutations were confirmed by nucleotide sequencing of appropriate PCR products. For experiments with plasmid-borne deletions recC2710–2718, C2725, or C2726 (see Figure 3 and Table 7), fresh transformants of strain V68 (recC73), V2830 (ΔrecC2730∷kan), or V2659 (ΔrecBCD234) were made with DNA propagated in strain DH5α, purified, and used immediately.
Figure 3.—
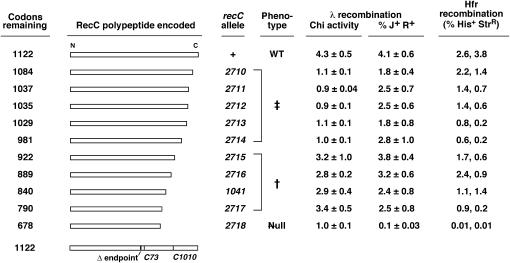
Intermediate length C-terminal deletions of recC retain Chi hotspot activity. Open bars indicate the extent of the RecC polypeptide remaining in each exonuclease III-generated deletion. The recC1041 nonsense mutation is included for reference. Chi activity and λ recombinant frequencies are the mean ± SEM from five independent experiments with plasmid-bearing derivatives of strain V68 (recC73) or strain V2830 (ΔrecC2730∷kan). The recombinant frequencies in Hfr crosses are from two independent experiments with plasmid-bearing derivatives of strain V68. The bar at the bottom indicates the positions of the recC73 frameshift mutation, the recC1010 missense mutation, and the left end of the Δ(recC–argA)234 deletion.
TABLE 7.
recC deletions mimicking recC1041 retain Chi activity
| rec allelea | Chi activity | λ recombinant frequency (% J+ R+) |
|---|---|---|
| + | 5.1 ± 0.2 | 6.6 ± 0.6 |
| C1041 | 2.5 ± 0.2 | 6.9 ± 0.4 |
| C2725 | 1.9 ± 0.3 | 1.8 ± 0.3 |
| C2726 | 2.2 ± 0.3 | 2.1 ± 0.3 |
Data are the mean ± SEM from 10 independent experiments.
Strains are transformants of strain V2660 [Δ(thyA–recBCD–argA)232 rpsL31] with the indicated rec alleles on derivatives of plasmid pDWS2 (thyA+–recBCD–argA+).
Phage λ crosses and Chi activity:
Crosses between λ phages 1081 and 1082 and between 1083 and 1084 were conducted, and Chi activity was determined as described by Stahl and Stahl (1977). The frequency of J+ R+ recombinant phages in the two crosses was determined by differential plating on strain C600 (supE44) for total phage and on strain 594 (sup+) for J+ R+ recombinants. The frequencies in the two crosses did not differ significantly: in 20 pairs of crosses the ratio of the frequencies was 1.1 ± 0.05 (mean ± SEM; our unpublished data).
E. coli Hfr crosses:
Crosses between the indicated recipient strain (F− hisG4 rpsL31) and donor strain V1306 (Hfr PO44 his+ rpsL+) were conducted as described by Schultz et al. (1983) with selection for His+ StrR (rpsL31) recombinants. The ratio of Hfr:F− cells in the matings was ∼1:10. Recombinant frequencies are expressed as the number of recombinants per Hfr cell in the mating, corrected for viability of the recipient cells.
Efficiency of plaque formation by phage:
Cells were grown in TB broth to ∼1 × l08/ml. Phage in SM (0.05 ml) were added to 0.1 ml of cell culture, and the mixture incubated at 37° for 15 min. Top agar (2.5 ml) was added, and the mixture poured onto a TB agar plate, which was incubated at 37° overnight. The efficiency of plaque formation is the titer on the strain tested divided by the titer on strain V67 (recB21) for testing haploid strains or strain V653 [ΔrecBCD232 (pSA21)] for testing plasmid-containing strains.
RecBCD enzyme and reactions with DNA:
Enzymes from wild-type and recC1041 cells were prepared as described by Amundsen et al. (2002), as were the DNA substrates and methods for assaying unwinding and nicking of linear dsDNA at Chi (Taylor et al. 1985).
RESULTS
Experimental background: recBCD mutant phenotypes and intracellular RecBCD enzyme activity:
Mutations in the three genes encoding RecBCD produce a spectrum of phenotypes. Null mutations, including complete deletions, insertions, or early nonsense mutations, in recB and recC eliminate all activities of RecBCD. These mutants are recombination deficient (Rec−), are sensitive to DNA-damaging agents, and form small colonies, presumably stemming from a deficiency in recombinational repair of DNA breaks such as may arise from replication (Kuzminov 1999). In contrast, null mutations in recD abolish RecBCD ds and ss exonuclease activity (Nuc−) but not the helicase activity or the ability to load RecA onto ssDNA (Amundsen et al. 1986; Korangy and Julin 1993; Chen et al. 1998; Churchill et al. 1999). Because the nuclease active site is in RecB (Yu et al. 1998b), the RecD subunit must regulate the exonuclease activity. recD null mutants are Rec+, are barely more sensitive to DNA-damaging agents than wild type, and form large colonies. In these mutants, Chi is inactive (Chi−), and the resultant enzyme, called RecBC, is insensitive to Chi (Chaudhury and Smith 1984b; Amundsen et al. 1986; Yu et al. 1998a). This phenotype, Chi− Nuc− Rec+, is designated “double dagger” (‡). Certain recC mutations also have the ‡ phenotype; these mutants are altered in or lack the C-terminal domain of RecC that is necessary for the assembly of RecD into the holoenzyme (Amundsen et al. 2002). The recC mutations described here have a related but novel phenotype, Chi+ Nuc− Rec+, which we designate “dagger” (†).
The intracellular phenotypes of recBCD mutants, and hence the intracellular activities of RecBCD, are readily assessed by growth (plaque formation) of certain phages and by genetic crosses. RecBCD's exonuclease activity blocks the growth of phage T4 gene 2 mutants: gene 2 protein is thought to bind to the ends of T4 DNA in the virion, and upon infection it protects the DNA from degradation by RecBCD exonuclease (Oliver and Goldberg 1977; Lipinska et al. 1989). Degradation of intracellular DNA is likely via a combination of the nuclease activity of the RecBCD molecule that unwinds the DNA (ds exonuclease) and that of other RecBCD molecules acting on the initially unwound DNA (ss exonuclease). Thus, T4 2− phage make plaques on cells lacking RecBCD nuclease activity, whether or not the cells are Rec+ (Amundsen et al. 1990).
Chi activity and recombination proficiency can be measured quantitatively in crosses between phage λ red gam mutants, which lack the RecBCD inhibitory protein Gam and lack the λ recombination enzymes Red and thus rely upon the E. coli RecBCD pathway for recombination (reviewed by Smith 1983). Wild-type λ lacks Chi sites (designated χ0) and recombines at a low rate by the RecBCD pathway, but Chi mutations (designated χ+) locally stimulate this pathway. Chi hotspot activity is defined as the ratio of the recombinant frequency in an interval with Chi to that in the same interval without Chi (supplemental Table S3 at http://www.genetics.org/supplemental/); an increased ratio reflects the localized stimulation of recombination at and near Chi, the definition of a recombination hotspot. In the Chi assays reported here, we used two crosses designed by Stahl and Stahl (1977) that allow for normalization of the increased burst size of phages with Chi. A qualitative assay of Chi activity is the larger plaque size of λ red gam χ+ compared to that of λ red gam χ0 on lawns of cells with RecBCD exonuclease activity, which blocks the late (rolling circle) form of λ-replication. In this situation, λ red gam plaque formation is dependent on the RecBCD pathway of recombination to produce DNA that can be packaged into viable (plaque-forming) particles. In the absence of recombination, such as in E. coli recA mutants, no visible plaques are formed; with recombination (e.g., in recA+ recBCD+ cells) there are small plaques without Chi and large plaques with Chi (Smith 1983). Recombination can be assayed quantitatively as the frequency of recombinants in λ red gam phage crosses or in Hfr cellular crosses. Here, we use these assays to assess the intracellular activities of RecBCD exonuclease and Chi.
The recC1041 nonsense allele lacks exonuclease activity but retains Chi hotspot activity:
A strain with the recC1041 allele was isolated in a search for recBCD alleles with novel phenotypes (Amundsen et al. 2002). This isolate was Rec+ but sensitive to T4 2− phage and supported large plaque formation by λ red gam phage, both χ0 and χ+. The mutation was isolated on the F′15 factor, bearing the recBCD genes; after transfer of the mutation to the chromosome or to a high-copy-number plasmid, the phenotypes remained the same (see below). These phenotypes indicated that recC1041 cells lack RecBCD exonuclease activity, and, as expected, cell-free extracts lack detectable ATP-dependent ds exonuclease activity, the hallmark of RecBCD enzyme (Amundsen et al. 2002). Complementation and nucleotide sequence analysis showed that the mutation is a nonsense mutation in recC (Trp841 → UGA841), and the mutation was assigned to the category ‡ (Chi− Nuc− Rec+).
Further quantitative crosses of phage λ showed, however, that the recC1041 mutant retains Chi activity (Table 2). In recBCD+ cells, Chi activity was ∼5, indicating that the recombinant frequency was approximately five times higher in an interval with Chi than in the same interval without Chi (Stahl and Stahl 1977). In recB21∷IS186 (null) cells, the hotspot value was essentially unity, indicating that Chi has no effect on the recombinant frequency in the intervals measured, as reported previously (Stahl and Stahl 1977). In recC1041 cells, the hotspot value was ∼3. The hotspot values were similar whether recC1041 was on the chromosome, on an F′ factor, or on a derivative of plasmid pBR322 bearing the recBCD genes. In 43 experiments, the hotspot values in cells with plasmid-borne recC1041 ranged from 1.4 to 4.8; the mean ± SEM was 2.9 ± 0.2 (Tables 2 and 5–7; supplemental Table S3 at http://www.genetics.org/supplemental/, and additional data). This value was significantly >1 (P ≪ 0.001; one-sided t-test), indicating that Chi was active in these cells. As reported previously (Amundsen et al. 2002), recC1041 cells were recombination proficient in Hfr crosses and allowed plaque formation by phage T4 and its gene 2 mutant with high efficiency (Tables 2, 3, and 6).
TABLE 2.
recC1041 retains Chi hotspot activity and recombination proficiency but not exonuclease activity in vivo
| E. coli strain | λ recombinant frequency (% J+ R+)a | Hfr recombinant frequency (% His+ [StrR]) | Efficiency of plaque formation
|
|||
|---|---|---|---|---|---|---|
| rec alleles | Chi activitya | T4 | T4 2− | |||
| Plasmid-borne recBCD allelesb | ||||||
| —b | + | 5.0 ± 0.06 | 6.3 ± 0.6 | 3.8 | 1.1 | 3 × 10−7 |
| — | B21∷IS186 | 0.96 ± 0.03 | 0.13 ± 0.03 | 0.002 | 1 | 1 |
| — | C1041 | 3.4 ± 0.44 | 4.4 ± 0.5 | 1.9 | 1.1 | 1.1 |
| — | C1010 | 1.1 ± 0.04 | 3.6 ± 0.4 | 1.9 | 0.9 | 0.5 |
| — | D1013 | 1.0 ± 0.06 | 7.9 ± 0.9 | 1.6 | NDc | 1.4 |
| — | C1041 D1013 | 1.0 ± 0.06 | 3.8 ± 0.3 | 1.6 | 1.2 | ND |
| — | C1041 ΔDd | 1.1 ± 0.04 | 3.2 ± 0.1 | ND | ND | 0.9 |
| F′-borne recBCD allelese | ||||||
| —e | C1041 | 2.8 ± 0.17 | 3.4 ± 0.7 | ND | ND | ND |
| Chromosomal recBCD allelesf | ||||||
| V66 | + | 5.9 ± 0.8 | 7.8 ± 0.7 | 3.9 ± 0.5 | 1.1 | 4 × 10−7 |
| V67 | B21∷IS186 | 1.0 ± 0.1 | 0.13 ± 0.02 | 0.006 ± 0.002 | 1 | 1 |
| V2658 | C1041 | 2.6 ± 0.3 | 3.4 ± 0.4 | 0.5 ± 0.05 | 1.0 | 1.1 |
| V219 | C1010 | 1.0 ± 0.1 | 3.7 ± 1.4 | 3.7 ± 1.4 | 0.8 | 0.9 |
| V222 | D1013 | 0.95 ± 0.05 | 6.2 ± 1.0 | 2.0 ± 0.2 | 0.9 | 1.1 |
Data are the mean ± SEM from three to five independent experiments.
Strains are transformants of strain V2660 [Δ(thyA–recBCD–argA)232 rpsL31] with the indicated rec alleles on derivatives of plasmid pDWS2 (thyA+–recBCD–argA+).
Not determined.
This strain is a derivative of strain V330 [Δ(recBCD–argA)234] with recC1041 on plasmid pSA164 bearing the 11.7-kb BseRI fragment with recC and recB.
The F′15 factor (thyA+–recBC1041D–argA+) is in strain V2660.
Strains are isogenic derivatives of strain V66 (rpsL31). Hfr recombination data are the mean ± SEM from three independent experiments.
TABLE 5.
Chi is active in recC1041 cells carrying rpsL (StrR) alleles
| E. coli strain | rec allele | rpsL allelea | Chi activity | λ recombinant frequency (% J+ R+) |
|---|---|---|---|---|
| Plasmid-borne recBCD allelesb | ||||
| —b | + | + | 4.9, 4.1 | 9.8, 8.3 |
| — | + | 31 | 4.8, 5.1 | 8.7, 5.7 |
| — | + | 222 | 5.2, 4.1 | 10.2, 6.2 |
| — | C1041 | + | 3.1, 4.1, 3.2 | 4.1, 5.7, 4.1 |
| — | C1041 | 31 | 3.9, 2.1, 3.1 | 5.2, 4.9, 3.1 |
| — | C1041 | 222 | 2.5, 1.9, 2.0 | 4.1, 5.1, 4.2 |
| Chromosomal recBCD allelesc | ||||
| V66 | + | 31 | 5.1, 4.7 | 3.5, 2.9 |
| V2935 | + | 222 | 4.1, 4.6 | 4.2, 3.8 |
| V2658 | C1041 | 31 | 2.9, 2.1 | 3.9, 2.2 |
| V2936 | C1041 | 222 | 1.9, 1.8 | 0.3, 0.9 |
rpsL31 is A → C (Lys43 → Thr43); rpsL222 is A → C (Lys43 → Asn43) and C → T (Asp109 → Asp109) (our unpublished data).
Strains are transformants of strain V186 [Δ(thyA–recBCD–argA)232 rpsL+], V2660 [Δ(thyA–recBCD–argA)232 rpsL31], or V2950 [Δ(thyA–recBCD–argA)232 rpsL222] with the indicated rec alleles on derivatives of plasmid pDWS2 (thyA+–recBCD–argA+). Data are from two or three independent experiments.
Strains are isogenic derivatives of strain V66 (rec+).
TABLE 6.
recC1041 (Chi+) is epistatic to distal recC1010 (Chi−)
| Chi activity | λ recombinant frequency (% J+ R+) | Efficiency of plaque formation
|
||
|---|---|---|---|---|
| rec allelesa | T4 | T4 2− | ||
| Nonsuppressing strains | ||||
| + | 5.9, 4.1 | 6.7, 6.8 | 1.0 | 4 × 10−7, 9 × 10−7 |
| C1041 | 4.4, 3.1 | 3.5, 5.6 | 1.0 | 0.7, 0.9 |
| C1010 | 1.2, 1.3 | 5.9, 2.3 | 0.9 | 0.7, 0.8 |
| C1041 C1010 | 3.5, 3.2 | 2.1, 2.9 | NDb | 0.6, 0.7 |
| UGA-suppressing strainsc | ||||
| + | 4.2, 5.0 | 7.3, 6.2 | 0.9 | 6 × 10−7, 8 × 10−7 |
| C1041 | 4.1, 2.3 | 4.9, 3.4 | 0.8 | 6 × 10−5, 9 × 10−5 |
| C1010 | 1.2, 1.0 | 4.9, 3.1 | 1.0 | 0.6, 0.7 |
| C1041 C1010 | 1.2, 1.3 | 3.1, 4.2 | ND | 0.7, 0.8 |
Strains are transformants of strain V2660 [Δ(thyA–recBCD–argA)232 rpsL31] with the indicated rec alleles on derivatives of plasmid pDWS2 (thyA+–recBCD–argA+). Data are from two independent experiments.
Not determined.
The Trp-inserting UGA-suppressor Su9 was on plasmid pACYC360 (Martin et al. 1988).
TABLE 3.
recC1041 lacks detectable RecBCD exonuclease activity on λ red gam phage
| Efficiency of plaque formationb
|
||||||
|---|---|---|---|---|---|---|
| E. coli straina | rec alleles | λ+ | λ red gam χ0 | λ red gam χ+ | T4+ | T4 2− |
| V66 | + | 1.5, 1.1 | 1.0, 0.9 | 1.3, 1.1 | 1.2, 1.1 | 1.2 × 10−7, 1.0 × 10−7 |
| V2881 | ΔrecA∷kan | 0.9, 1.2 | 3 × l0−6, 5 × 10−6 | 4 × 10−6, 4 × 10−6 | 0.9, 0.8 | 1.3 × l0−7, 1.9 × 10−7 |
| V2658 | recC1041 | 1.5, 1.0 | 1.3, 0.9 | 1.1, 1.1 | 0.8, 0.9 | 0.8, 0.7 |
| V2884 | recC1041 ΔrecA∷kan | 1.2, 0.9 | 0.7, 1.2 | 1.1, 0.9 | 0.8, 0.7 | 0.8, 0.9 |
| V67 | recB21 | 1 | 1 | 1 | 1 | 1 |
Strains are isogenic derivatives of strain V66 (rpsL31) with the indicated recB and recC alleles both on the chromosome and on derivatives of plasmid pDWS2 (thyA+–recBCD–argA+).
Titer of phage relative to that on strain V67 (pSA21) (rpsL31). Data are from two independent experiments. All λ plaques were large except those of λ red gam χ0 on strain V66 (rec+), which were small.
The lower but readily observable Chi hotspot value in recC1041 cells compared to that in recBCD+ cells may be a consequence of a mixture of Chi-independent and Chi-dependent loading of RecA protein by enzyme from recC1041 cells (Amundsen et al. 2002). Like RecBC enzyme, enzyme from recC1041 cells loads RecA protein, even in the absence of Chi, onto the DNA end at which unwinding commences (Churchill et al. 1999; Amundsen et al. 2002). Furthermore, in the absence of RecBCD exonuclease, λ red gam phages can replicate to high levels in the rolling-circle mode; these forms may recombine by a Chi-independent pathway (Stahl and Stahl 1977; Smith 1983). Thus, RecBC1041D may form Chi-independent recombinants, as does RecBC (Chaudhury and Smith 1984b), If so, these recombinants would dilute the Chi-dependent recombinants, and the measured hotspot value would be decreased relative to that in recBCD+ cells.
As noted above, the hotspot values and the recombinant frequencies were not significantly different if the recBCD alleles were on the chromosome or on a derivative of pBR322 (Table 2). Extracts of cells with the pBR322-borne recBCD+ genes contain ∼25 times as much RecBCD enzyme activity as those of cells with the recBCD+ genes on the chromosome (Ponticelli et al. 1985). Thus, changing the level of RecBC1041D enzyme by a factor of ∼25 (from plasmid-borne level to chromosomal level) did not detectably change Chi activity.
As a direct assay of RecBCD exonuclease on λ red gam phages in recC1041 cells, we tested the ability of these phages to grow on recA mutant derivatives. λ red gam cannot recombine in recA mutants, and phage growth is blocked by RecBCD exonuclease (Smith 1983). We found that λ red gam χ0 and χ+ made large plaques with near unit efficiency on recC1041 ΔrecA∷kan cells (Table 3), indicating the lack of detectable RecBCD exonuclease even in cells with recC1041 on a derivative of the high-copy-number plasmid pBR322. As expected, λ red gam phage growth was blocked in ΔrecA∷kan recBCD+ cells, and T4 2− growth in recC+ or recC1041 cells was not affected by the ΔrecA∷kan mutation.
Chi hotspot activity in recC1041 requires RecD polypeptide but not RecJ or ExoI ss exonuclease:
Removal of RecD from wild-type RecBCD enzyme abolishes Chi activity (Chaudhury and Smith 1984b; Amundsen et al. 1986). To determine if RecD is also required for Chi activity in recC1041 cells, we coupled the recD1013 mutation with recC1041. recD1013 is a nonsense mutation (Gln4 → UAA4), which effectively eliminates RecD (Table 1). Chi was inactive in recC1041 recD1013, recC1041 ΔrecD, and recC+ recD1013 cells (Table 2). Thus, RecD is required for Chi activity in all situations reported to date (Chaudhury and Smith 1984b; Amundsen et al. 1986; see discussion).
Although RecBCD is the most potent exonuclease in extracts of E. coli (our unpublished data), there are other nucleases that might influence Chi activity. Of particular importance is RecJ, a 5′→ 3′ ss exonuclease that is required for recombination in recD mutants (Lovett et al. 1988) and for recombination in the absence of RecBCD (and the presence of appropriate sbc suppressor mutations that activate the RecE or RecF pathway) (Lovett and Clark 1984). The recJ284∷Tn10 (null) mutation did not significantly alter Chi hotspot activity in recC+ or recC1041 mutant cells (Table 4). Exonuclease I (ExoI), a 3′→5′ ss exonuclease, blocks recombination in recB or recC null mutants; the sbsB15 mutation, in conjunction with elimination of the SbcCD nuclease, suppresses this recombination deficiency to allow full activity of the RecF pathway (Smith 1988). The sbcB15 missense mutation greatly reduces ExoI nuclease activity (Phillips et al. 1988), but the mutant protein may retain the DNA-binding activity and thereby block access of other nucleases, whereas the ΔxonA allele deletes the gene (Feschenko et al. 2003). The sbsB15 or ΔxonA mutation had no effect on, or only slightly reduced, Chi hotspot activity in recBCD+, recC1041, recJ284, or recC1041 recJ284 strains (Table 4). Razavy et al. (1996) also reported high Chi activity in recBCD+ strains bearing recJ284, ΔxonA, and sbcB15 mutations, singly or in combinations. Thus, RecBCD ds exonuclease and the RecJ and ExoI ss exonucleases are not required for Chi activity in recC1041 cells.
TABLE 4.
RecJ and ExoI are not required for the Chi+ Nuc− Rec+ (†) phenotype
| Allelesa
|
||||
|---|---|---|---|---|
| E. coli strain | recC | Other | λ recombinant frequency (% J+ R+) | Chi activity |
| V66 | + | — | 6.6 ± 0.7 | 5.0 ± 0.2 |
| V2757 | + | recJ284∷Tn10 | 5.4 ± 0.8 | 5.1 ± 0.1 |
| V2991 | + | sbcB15 | 4.2 ± 0.3 | 4.7 ± 0.2 |
| V2997 | + | ΔxonA | 4.0 ± 0.1 | 4.0 ± 0.2 |
| V2996 | + | recJ284∷Tn10 sbcB15 | 4.1 ± 0.2 | 4.1 ± 0.2 |
| V2999 | + | recJ284∷Tn10 ΔxonA | 4.4 ± 0.2 | 4.1 ± 0.3 |
| V2658 | recC1041 | — | 2.7 ± 0.1 | 2.6 ± 0.2 |
| V2758 | recC1041 | recJ284∷Tn10 | 3.7 ± 0.5 | 2.4 ± 0.2 |
| V2992 | recC1041 | sbcB15 | 3.0 ± 0.2 | 2.3 ± 0.1 |
| V2998 | recC1041 | ΔxonA | 3.0 ± 0.1 | 2.2 ± 0.1 |
| V2993 | recC1041 | recJ284∷Tn10 sbcB15 | 2.6 ± 0.4 | 2.2 ± 0.1 |
| V3000 | recC1041 | recJ284∷Tn10 ΔxonA | 2.6 ± 0.2 | 2.1 ± 0.1 |
Strains are isogenic derivatives of strain V66 (rpsL31) with the indicated alleles on the chromosome. Data are the mean ± SEM from three to five independent experiments.
An rpsL mutation does not alter the recC1041 phenotype:
Although changing the level of RecBC1041D had no significant effect on Chi activity (see above), we entertained the possibility of low-level translational “readthrough” of the recC1041 UGA codon. Such readthrough would produce full-length RecC polypeptide and potentially wild-type RecBCD enzyme, which would be expected to manifest Chi activity and exonuclease activity. The strains used in Tables 2, 3, and 4 and in supplemental Table S3 at http://www.genetics.org/supplemental/ contain the rpsL31 mutation; rpsL mutations confer resistance to streptomycin and often reduce readthrough (Dong and Kurland 1995). We found that these strains and the rpsL+ and rpsL222 derivatives of them all had Chi hotspot activity (Table 5) and were sensitive to T4 2−. The rpsL222 mutation reduces readthrough of lacZ UGA nonsense mutations by a factor of 10–40 (Dong and Kurland 1995). The lack of a sufficiently sensitive assay for RecC polypeptide precludes a direct measure of low-level readthrough of recC1041 (also see below). These results argue against the observed Chi activity being due to readthrough of recC1041 that produces a low level of wild-type RecBCD enzyme.
recC1041 is epistatic to recC1010, a distal Chi− missense mutation:
The recC1010 mutation (Gly905 → Glu905) alters a domain in RecC required for the assembly of RecD into the RecBCD holoenzyme complex and consequently confers the Chi− Nuc− Rec+ (‡) phenotype (Amundsen et al. 2002). As an additional test of the possibility of readthrough from recC1041, we coupled recC1041 (Trp841 → UGA841) and recC1010, which are in the order 5′–C1041–C1010–3′. In this double mutant, any readthrough RecC polypeptide would have Glu905 and would be expected to have the Chi− Nuc− Rec+ phenotype. We found, however, that Chi was active in this double mutant (Table 6). As expected, introduction of a UGA suppressor (Su9) to permit readthrough led to the loss of Chi activity but retention of high efficiency of plaque formation by T4 2− phage, i.e., the recC1010 Chi− Nuc− Rec+ (‡) phenotype. These results further indicate that there is little, if any, readthrough of recC1041.
A set of recC C-terminal deletions with Chi hotspot activity:
Since recC1041 is a nonsense mutation, deletions of the 3′-end of recC are expected to produce the same phenotype. We previously described a set of ExoIII-generated deletions of recC (Amundsen et al. 2002), some of which are illustrated in Figure 3. Cells with each of these deletions allow plaque formation by T4 2− mutant phage and large plaque formation by λ red gam χ0 or χ+ phage, indicating that all of these deletions abolish RecBCD exonuclease activity (Amundsen et al. 2002). Cells with the five shortest deletions, recC2710–2714, lacked Chi activity and mimicked recC1010 (i.e., they had the Chi− Nuc− Rec+ ‡ phenotype). Cells with three longer deletions, recC2715–recC2717, however, had a phenotype like that of recC1041 (i.e., they had the Chi+ Nuc− Rec+ † phenotype). Cells with an even longer deletion, recC2718, had the null (Rec−) phenotype, as reported previously (Amundsen et al. 2002). These results show that, although all the deletions abolish exonuclease activity, the short recC deletions abolish Chi activity but intermediate-length deletions retain Chi activity, in parallel with the phenotypes of recC1010 and recC1041, respectively. Since these plasmids lack the 3′-end of the recC gene, there is no possibility of full-length RecC polypeptide being synthesized.
We next constructed a deletion whose endpoint is immediately after the recC1041 UGA841nonsense codon and an additional nonsense codon UGA842. As expected, cells with this deletion, recC2725, had Chi hotspot activity (Table 7) and were sensitive to T4 2− phage. Another deletion, recC2726, identical to recC2725 except for the stronger stop codon UAA842 and 6 bp with a KpnI site used for cloning, also conferred Chi hotspot activity. These results confirm that elimination of the RecC C terminus can abolish RecBCD exonuclease activity without abolishing Chi hotspot activity.
In some cases, cultures of cells with these plasmid-borne recC deletions were unstable, as is often the case for plasmids in recBCD Nuc− mutants (Biek and Cohen 1986; Cohen and Clark 1986). These cultures sometimes appeared Rec− and lacked Chi activity (i.e., had the recC null phenotype) or failed to grow in ampicillin-containing media selective for the plasmids, but in no case were the cells Rec+ Chi−. For example, among 10 parallel cultures started from isolated colonies of a strain with the recC2725 mutation, 4 had Chi activity, ranging from 2.1 to 3.4 (mean ± SEM = 2.5 ± 0.3); these cultures also produced a higher frequency of λ J+ R+ recombinants than the other cultures (2.6 ± 0.6% vs. 1.2 ± 0.4%). Among subcultures of these 10 cultures, tested the next day, one remained Chi+ and two previously without Chi activity gained it, ranging from 2.3–3.3 (2.9 ± 0.3); again, the Chi+ cultures were more recombination proficient (3.1 ± 0.1% vs. 1.1 ± 0.2%). We suspect that, in the absence of RecBCD exonuclease activity, these plasmids enter rolling-circle replication, with consequent loss of recBCD or bla (ampicillin-resistance determinant) genes or their expression or consequent sequestration of RecBCD enzyme by the linear DNA. Cultures with plasmid-borne recC1041 were more stable. For example, 10 parallel cultures all had Chi activity, ranging from 1.6 to 3.2 (2.5 ± 0.2) on the first day and from 1.4 to 4.2 (3.3 ± 0.3) on the subsequent day. The basis of instability of the recC deletion plasmids is unclear, but all cultures tested allowed plaque formation by T4 2−, indicating that they lacked RecBCD exonuclease activity even when Chi activity was observed.
Enzyme purified from recC1041 cells has Chi-nicking and RecA-loading activities but no detectable exonuclease activity:
Enzyme purified from recC1041 cells has stoichiometric amounts of the RecB and RecC polypeptides but <0.03 mol-equivalents of RecD polypeptide (our unpublished data), as expected from the lack of the RecC domain required for RecD assembly into the holoenzyme (Amundsen et al. 2002). The requirement for RecD for hotspot activity in recC1041 cells (Table 2) presents a paradox that we address in the discussion. The mutant RecC polypeptide has an apparent mass of ∼95 kDa, as determined by SDS–polyacrylamide gel electrophoresis and as expected from the truncation at codon 840 of 1122 in recC+ (Amundsen et al. 2002). This purified enzyme has no detectable exonuclease activity (Amundsen et al. 2002).
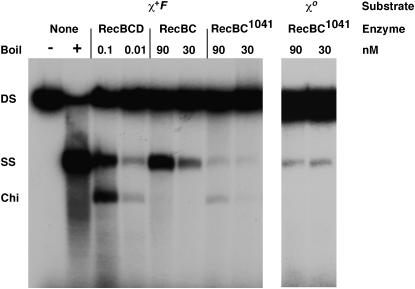
The retention of Chi hotspot activity but not RecBCD exonuclease in recC1041 cells suggested that the enzyme in recC1041 cells has Chi-nicking and RecA-loading activities. With enzyme purified from these cells, Chi nicking activity was observed on two different linear DNA substrates derived from pBR322, one with χ+F (Figure 4) and the other with χ+E (our unpublished data). With each substrate, the DNA-unwinding activity was weaker than that of wild-type RecBCD enzyme. This is due in part to the lower affinity for DNA of the mutant enzyme than that of wild-type RecBCD enzyme. The apparent KD of the mutant enzyme was comparable to that of the RecBC enzyme (∼50 nm) but much higher than that of RecBCD (∼0.2 nm) (Taylor and Smith 1995a; our unpublished data). At equal protein concentrations, the mutant enzyme unwound less DNA than did RecBC, suggesting that the KM or the fraction of protein unable to unwind DNA was higher for the mutant enzyme than for RecBC. But of the DNA unwound, a comparable amount was cut at or near the Chi site by both wild-type and recC1041 mutant enzymes and on both substrates; as expected, cutting was not detected on DNA without Chi (Figure 4). We conclude that, although the recC1041 helicase activity in vitro is weak, DNA unwinding by the recC1041 enzyme is still coupled to high-frequency Chi nicking. Because the recC1041 enzyme preparation was deficient in RecD, in a limited number of experiments we added extracts of RecD-overproducing cells to the preparation and saw slight stimulation of Chi nicking (our unpublished data); reconstitution of RecBCD holoenzyme from individual components is problematic (Chen et al. 1997; Singleton et al. 2004). As expected from the recombination proficiency of recC1041 cells, recC1041 enzyme loads RecA protein onto the initial DNA end in the absence of Chi (Amundsen et al. 2002) and onto the Chi tail in the presence of Chi (our unpublished data). Below, we discuss the implications of these results for the mechanism of Chi-stimulated recombination.
Figure 4.—
Enzyme from recC1041 cells has DNA-unwinding and Chi-cutting activities. RecBCD, RecBC, and RecBC1041 enzyme were assayed using HindIII-linearized, 5′ 32P-labeled pBR322 χ+F225 or χ0 DNA (Amundsen et al. 2000). The DNA substrate (0.8 nm) and indicated concentration of purified enzyme were incubated at 37° for 3 min. The reaction products were analyzed by electrophoresis in a 1% agarose gel. The positions of dsDNA substrate (DS), unwound ssDNA (SS; boiled), and the major product of Chi-dependent nicking (Chi) are shown.
DISCUSSION
Current models for the action of RecBCD enzyme at Chi and the consequent local stimulation of recombination (Chi hotspot activity) differ in the role of the RecBCD nuclease. In one view, RecBCD enzyme nicks DNA at Chi (Smith et al. 1981b; Smith 1991, 2001), whereas, in another view, the enzyme uses its ds exonuclease activity to degrade one strand up to Chi and then switches the nuclease to the other strand (Thaler et al. 1988; Stahl et al. 1990; Kuzminov et al. 1994; Myers and Stahl 1994; Anderson and Kowalczykowski 1997b). Here, we show that a novel class of recBCD mutants lacking detectable ds exonuclease activity retains Chi activity. These mutants and their phenotypes have multiple implications for the function of RecBCD enzyme and its role in recombination.
Nucleolytic degradation is not necessary for Chi hotspot activity:
The properties of the recC Chi+ Nuc− Rec+ (†) mutants described here are not expected from the view that Chi stops DNA degradation by RecBCD, since Chi activity was retained in mutants that lack detectable ds exonuclease activity (Tables 2–7 and supplemental Table S3 at http://www.genetics.org/supplemental/; Amundsen et al. 2002). Although a very low level of RecBCD exonuclease in these mutants cannot be excluded, there is not sufficient exonuclease activity to block the growth of T4 gene 2− phage or λ red gam phage in recA derivatives (Tables 2, 3, and 6). Since there is ∼5 kb of redundant DNA at the ends of T4 virion DNA, DNA degradation must be <5 kb to allow plaque formation by a phage DNA molecule injected into the cell. The amount of DNA degradation required to block the switch from θ-form replication to rolling-circle (σ) replication of λ is unknown but may be <1 kb. In the λ phages used to measure Chi activity, the Chi sites (χ+D and χ+76) were located 3.5 and 15.5 kb from the entry site for RecBCD (Stahl and Stahl 1977; Smith et al. 1981a; Kobayashi et al. 1982), and both were active in the recC Chi+ Nuc− Rec+ (†) mutants (Tables 2 and 4–7; supplemental Table S3 at http://www.genetics.org/supplemental/). Thus, RecBC1041D enzyme can travel >15 kb from a DNA end and productively interact with Chi. These data argue against there being sufficient RecBCD exonuclease activity to degrade DNA up to Chi in crosses in the recC† mutants studied here.
Previous evidence also argues that the reaction condition under which purified RecBCD nicks at Chi (excess ATP) more nearly reflects the behavior of RecBCD and Chi in cells. Purified RecBCD enzyme, after acting at a Chi site, loses nuclease and Chi-nicking activity on a second DNA substrate, provided the reaction contains excess ATP over Mg2+ (Taylor and Smith 1992, 1999), the condition that favors nicking at Chi. The three subunits disassemble, perhaps at the end of the first DNA substrate, and the enzyme remains inactive for >2 hr but can be reactivated by reassembly upon addition of excess Mg2+. If Mg2+ is in excess from the start of the reaction, no inactivation is observed, and the enzyme acts at Chi on a second DNA substrate. In E. coli cells, if RecBCD is exposed to linearized plasmid DNA with Chi, it manifests reduced Chi activity in λ crosses (Myers et al. 1995). Similarly, treatment of recBCD+ cells with bleomycin, which can produce dsDNA breaks and allow RecBCD to interact with the chromosomal Chi sites, subsequently leads to productive T4 2− infections and loss of Chi activity in λ crosses (Köppen et al. 1995). Thus, the reaction condition favoring nicking at Chi readily accounts for the reduction or loss of RecBCD activities in cells.
A consideration of the reciprocality of recombination also argues, albeit not conclusively, against degradation up to Chi. For a single recombination event between two parental DNA molecules to produce the two reciprocal recombinant types, neither DNA substrate can be extensively degraded, since that would result in marker loss. The reaction condition favoring degradation up to Chi destroys one or the other strand to each side of Chi (Figure 1, right); reconstruction of an intact DNA molecule would require capture of the ssDNA fragment to the right of Chi and additional DNA synthesis. Unlike the Chi tail to the left of Chi (Figure 1), the DNA to the right of Chi is not loaded with RecA protein (Anderson and Kowalczykowski 1997b). RecA–RecBCD-dependent recombination of λ has been reported in some studies to be reciprocal (Sarthy and Meselson 1976; Stahl et al. 1982, 1984; Kobayashi et al. 1984) but in others not (Lam et al. 1974; Stahl et al. 1980) or in others both (Ennis et al. 1987; Stahl et al. 1990, 1995). Apparent nonreciprocality seems to be observed only when phage replication is blocked. Because all of the substrate and product molecules cannot be accounted for with certainty in phage crosses (unlike fungal meiotic crosses), a firm conclusion cannot be drawn. Nevertheless, the simplest way to account for the apparent reciprocality, where reported, seems to be via nondestructive modes of DNA metabolism. Apparent nonreciprocality, where reported, is consistent either with degradation up to Chi (Figure 1, right) or with break-copy resolution after nicking at Chi (Figure 5, bottom right).
Figure 5.—
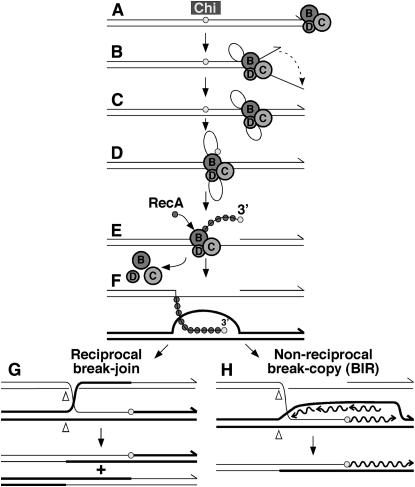
Nick-at-Chi model of RecBCD-promoted recombination (after Smith et al. 1981b; Smith 2001). RecBCD binds a dsDNA end (A) and unwinds DNA to produce a loop-tail structure (B) (Figures 1 and 2). The ssDNA ends can anneal to form a “rabbit ear” structure (C). At Chi (small shaded circles), RecBCD nicks the top strand (D) and loads RecA onto the 3′ Chi tail (E) (Figure 1), which undergoes strand exchange with a homologous duplex to form a D-loop (F). At some point after Chi, RecBCD leaves the DNA and the three subunits disassemble. Two fates of the D-loop joint molecule are shown at the bottom. (Bottom left) Cutting of the D-loop and reciprocal strand exchange form a Holliday junction (G), whose migration and resolution (involving cutting at Δ, strand swapping, and ligation) by some combination of RuvABC and RecG proteins produce reciprocal recombinants. (Bottom right) The 3′ Chi tail primes leading-strand DNA synthesis, which is converted into a replication fork (H); resolution at Δ produces one recombinant, plus one parental and one fragmented molecule not shown. This break-copy scheme is identical to break-induced replication (BIR). Alternative resolutions are not shown.
A model for recombination incorporating nicking at Chi and loading of RecA protein:
The properties of the recC Chi+ Nuc− Rec+ (†) mutants are consistent with models in which RecBCD enzyme nicks DNA at Chi (Smith et al. 1981b; Smith 1991, 2001) and thereafter loads RecA protein onto the 3′ Chi tail (Anderson and Kowalczykowski 1997b), since these activities were retained in the mutants (Figure 4; Amundsen et al. 2002). In this model, RecBCD engages a dsDNA end, the right end in Figures 1 and 5, and rapidly unwinds the DNA with the production of a ssDNA loop, which grows with time and moves along the DNA. Upon encountering Chi, the enzyme nicks the “top” strand (that with 5′-GCTGGTGG-3′) and loads RecA protein onto that strand to the left of Chi. RecBCD continues to unwind the DNA and may load additional RecA molecules that nucleate further RecA polymerization in the 5′ → 3′ direction to the end of the Chi tail. This ssDNA–RecA filament then engages a homologous duplex and forms a D-loop. Two pathways are possible after D-loop formation (Figure 5, bottom). In one, the D-loop is cut and anneals with the gap remaining after unwinding of the Chi tail. The resultant Holliday junction is resolved into recombinant molecules by some combination of RuvABC and RecG proteins. This pathway may involve local (repair) DNA synthesis. In the second pathway, the 3′-end of the Chi tail primes DNA synthesis, which in conjunction with PriA and other replication proteins may lead to a complete replication fork; this reaction may be the mechanism by which RecBCD and RecA promote replication restart after fork disruption (Smith 1991; Kowalczykowski 2000). Both pathways may involve mismatch repair.
Support for this model comes from a variety of genetic and enzymological studies. In genetic assays, mutants lacking the enzymes noted above are deficient for recombination involving linear dsDNA, as occurs in λ red gam crosses, Hfr crosses, and P1 transductions (reviewed by Smith 1983, 1988). Chi is active in such crosses, and mutations in recC or in the Chi sequence specifically block Chi hotspot activity (Stahl et al. 1975; Dower and Stahl 1981; Schultz et al. 1981, 1983). Biochemical studies with purified proteins have demonstrated the activities of the individual enzymes noted above (reviewed by Smith 1988, 2001; Kowalczykowski et al. 1994; Kowalczykowski 2000). Mutations that alter RecBCD or Chi coordinately alter Chi nicking (Ponticelli et al. 1985; Taylor et al. 1985; Cheng and Smith 1987) or switching of DNA degradation from one strand to the other at Chi (Dixon and Kowalczykowski 1993; Taylor and Smith 1995b; Anderson and Kowalczykowski 1997a). These results are consistent with both models in Figure 1. RecBCD and RecA have been coupled to produce joint DNA molecules, thought to be D-loops (Figure 5F), and this reaction is stimulated by Chi (Dixon and Kowalczykowski 1991; Anderson and Kowalczykowski 1997b). Further coordination of enzymes to produce recombinants in vitro remains to be achieved and may be difficult if, as postulated, recombination is closely linked to DNA replication. Until that goal is achieved, models of recombination must be considered tentative but useful guides toward that goal.
The intracellular role of RecBCD exonuclease:
If RecBCD's exonuclease is not necessary for Chi activity, what is its physiological role? Degradation of intracellular DNA by RecBCD has been observed in several situations. These include degradation of λ DNA subjected to restriction endonucleases (Simmon and Lederberg 1972), T4 gene 2 mutant DNA (Oliver and Goldberg 1977), and UV-irradiated DNA in recA mutants (“reckless degradation”; Clark et al. 1966). In each of these situations, the DNA cannot recombine with intact DNA. We have proposed that RecBCD's exonuclease degrades DNA that cannot recombine (Smith 2001). DNA that does recombine would become circular, losing the DNA end required for RecBCD enzyme activity (Goldmark and Linn 1972).
It has been proposed that Chi protects intracellular DNA from degradation by RecBCD. For example, switching a plasmid into rolling-circle replication leads to high-molecular-weight forms of the DNA only if the plasmid contains Chi; this result was interpreted to mean that Chi protects the linear branch of the rolling circle from degradation by RecBCD (Dabert et al. 1992). However, formation of high-molecular-weight DNA also requires RecA protein. We propose that in these cells a nascent rolling circle recombines with another circular plasmid, in a Chi–RecBCD–RecA-dependent manner, to produce a “dumbbell-shaped” molecule, which would lack a DNA end and be resistant to RecBCD exonuclease. In another example, Chi partially protects linearized plasmid DNA from RecBCD, both in cis (i.e., the DNA with Chi) and in trans (i.e., the DNA of a separate linearized plasmid; Kuzminov et al. 1994). As in the previous example, Chi's protection was seen only in recA+ cells and may reflect recombination between plasmids.
The role of RecD polypeptide in RecBCD enzyme:
Since its discovery (Amundsen et al. 1986; Biek and Cohen 1986), the RecD polypeptide has presented mysteries. The sequence of the recD gene suggested that RecD is a helicase (Gorbalenya et al. 1988; Hodgman 1988), but that activity was demonstrated only much later (Dillingham et al. 2003; Taylor and Smith 2003). The initial reports of recD mutants showed that they lack RecBCD nuclease activity (Chaudhury and Smith 1984b; Amundsen et al. 1986). This result was interpreted by some investigators to mean that the nuclease active site resides in RecD, but that site clearly resides in RecB (Yu et al. 1998a,b). Nevertheless, RecBC enzyme has <1/1000 of the ds and ss exonuclease activity of the wild-type enzyme (Yu et al. 1998a). Thus, RecD must regulate the nuclease activity. RecA loading by RecBCD is inhibited by RecD until the enzyme responds to a Chi- and nuclease-dependent signal (Churchill et al. 1999; Amundsen et al. 2000). The physical basis by which RecD regulates RecBCD remains unknown.
To our knowledge, RecD is required for Chi hotspot activity in all situations reported to date. RecD is required for Chi activity in otherwise wild-type cells (Chaudhury and Smith 1984b; Amundsen et al. 1986) and in the recC Chi+ Nuc− Rec+ (†) mutants (Table 2). In extracts of wild-type cells, RecD is associated with the holoenzyme complex, but in extracts of the recC† mutants, it is not detectably associated with the RecBC† complex (Amundsen et al. 1986, 2002). These results are consistent with the recC† mutants lacking a domain of RecC required for the assembly of RecD into the holoenzyme (Amundsen et al. 2002; Figure 2). But the role of RecD, which is required for Chi activity in these cells, is a puzzle. RecD may be weakly associated with the RecBC† complex and dissociate upon opening of the cells. This possibility is consistent with the tenuous connection between RecD and RecC, part of which is removed by the RecC C-terminal truncations (†) studied here (Figures 2B and 3). Alternatively, in these cells RecD may associate with the RecBC† complex when this complex binds to a DNA end, or RecD may play a role without binding to the RecBC† complex. Further investigations will be needed to answer this question.
A segment of RecC that inhibits Chi activity:
The region of RecC between amino acids ∼790 and ∼922 appears to inhibit Chi activity, at least in the absence of the RecC C-terminal ∼150 amino acids. Chi was inactive in recC mutants lacking the extreme C terminus, defined by the endpoints of recC2714 and recC2710, but not in more extensive deletions (Figure 3). We infer that these more extensive deletions, defined by the endpoints of deletions recC2717 and recC2715, remove an inhibitor of Chi activity. Since part of RecC appears to recognize Chi (see Introduction and Figure 2), we suppose that some Chi-dependent signal is sent from RecC to the RecB and RecD subunits to change the nuclease and RecA-loading activities of the enzyme at Chi. If this is the case, the region from amino acids ∼790 to ∼922 may block transduction of this signal unless the RecC subunit is intact. Although this proposal is speculative, it suggests that the interplay between this region and the C-terminal region of RecC is critical in Chi activity. Additional mutational and physical analyses of RecBCD enzyme may support this proposal and reveal the nature of the change of RecBCD enzyme at Chi.
Acknowledgments
We are grateful to Sue Lovett for the ΔxonA∷kan allele; to Gareth Cromie, Jon Gallant, Manjula Reddy, Dennis Schultz, and Andrew Taylor for discussions and valuable suggestions; to Andrew Taylor for Figure 2; and to Gareth Cromie, Luther Davis, Joe Farah, and Andrew Taylor for helpful comments on the manuscript. This research was supported by grant R01 GM031693 from the National Institute of General Medical Sciences to G.R.S.
References
- Amundsen, S. K., A. F. Taylor, A. M. Chaudhury and G. R. Smith, 1986. recD: the gene for an essential third subunit of exonuclease V. Proc. Natl. Acad. Sci. USA 83: 5558–5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundsen, S. K., A. M. Neiman, S. M. Thibodeaux and G. R. Smith, 1990. Genetic dissection of the biochemical activities of RecBCD enzyme. Genetics 126: 25–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundsen, S. K., A. F. Taylor and G. R. Smith, 2000. The RecD subunit of the Escherichia coli RecBCD enzyme inhibits RecA loading, homologous recombination and DNA repair. Proc. Natl. Acad. Sci. USA 97: 7399–7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundsen, S. K., A. F. Taylor and G. R. Smith, 2002. A domain of RecC required for assembly of the regulatory RecD subunit into the Escherichia coli RecBCD holoenzyme. Genetics 161: 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, D. G., and S. C. Kowalczykowski, 1997. a The recombination hot spot χ is a regulatory element that switches the polarity of DNA degradation by the RecBCD enzyme. Genes Dev. 11: 571–581. [DOI] [PubMed] [Google Scholar]
- Anderson, D. G., and S. C. Kowalczykowski, 1997. b The translocating RecBCD enzyme stimulates recombination by directing RecA protein onto ssDNA in a χ regulated manner. Cell 90: 77–86. [DOI] [PubMed] [Google Scholar]
- Arnold, D. A., N. Handa, I. Kobayashi and S. C. Kowalczykowski, 2000. A novel, 11 nucleotide variant of χ, χ*: one of a class of sequences defining the Escherichia coli recombination hotspot χ. J. Mol. Biol. 300: 469–479. [DOI] [PubMed] [Google Scholar]
- Biek, D. P., and S. N. Cohen, 1986. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J. Bacteriol. 167: 594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A. M., and G. R. Smith, 1984. a Escherichia coli recBC deletion mutants. J. Bacteriol. 160: 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, A. M., and G. R. Smith, 1984. b A new class of Escherichia coli recBC mutants: implications for the role of RecBC enzyme in homologous recombination. Proc. Natl. Acad. Sci. USA 81: 7850–7854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H-W., B. Ruan, M. Yu, J. Wang and D. A. Julin, 1997. The RecD subunit of the RecBCD enzyme from Escherichia coli is a single-stranded DNA-dependent ATPase. J. Biol. Chem. 272: 10072–10079. [DOI] [PubMed] [Google Scholar]
- Chen, H.-W., D. E. Randle, M. Gabbidon and D. A. Julin, 1998. Functions of the ATP hydrolysis subunits (RecB and RecD) in the nuclease reactions catalyzed by the RecBCD enzyme from Escherichia coli. J. Mol. Biol. 278: 89–104. [DOI] [PubMed] [Google Scholar]
- Cheng, K. C., and G. R. Smith, 1987. Cutting of Chi-like sequences by the RecBCD enzyme of Escherichia coli. J. Mol. Biol. 194: 747–750. [DOI] [PubMed] [Google Scholar]
- Cheng, K. C., and G. R. Smith, 1989. Distribution of Chi-stimulated recombinational exchanges and heteroduplex endpoints in phage lambda. Genetics 123: 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill, J. J., D. G. Anderson and S. C. Kowalczykowski, 1999. The RecBC enzyme loads RecA protein onto ssDNA asymmetrically and independently of χ resulting in constitutive recombination activation. Genes Dev. 13: 901–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, A. J., M. Chamberlin, R. P. Boyce and P. Howard-Flanders, 1966. Abnormal metabolic response to ultraviolet light of a recombination deficient mutant of Escherichia coli K12. J. Mol. Biol. 19: 442–454. [DOI] [PubMed] [Google Scholar]
- Cohen, A., and A. J. Clark, 1986. Synthesis of linear plasmid multimers in Escherichia coli K12. J. Bacteriol. 167: 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabert, P., S. D. Ehrlich and A. Gruss, 1992. χ sequence protects against RecBCD degradation of DNA in vivo. Proc. Natl. Acad. Sci. USA 89: 12073–12077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham, M. S., M. Spies and S. C. Kowalczykowski, 2003. RecBCD enzyme is a bipolar DNA helicase. Nature 432: 893–897. [DOI] [PubMed] [Google Scholar]
- Dixon, D. A., and S. C. Kowalczykowski, 1991. Homologous pairing in vitro stimulated by the recombination hotspot, Chi. Cell 66: 361–371. [DOI] [PubMed] [Google Scholar]
- Dixon, D. A., and S. C. Kowalczykowski, 1993. The recombination hotspot χ is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell 73: 87–96. [DOI] [PubMed] [Google Scholar]
- Dong, H., and C. G. Kurland, 1995. Ribosome mutants with altered accuracy translate with reduced processivity. J. Mol. Biol. 248: 551–561. [DOI] [PubMed] [Google Scholar]
- Dower, N. A., and F. W. Stahl, 1981. χ activity during transduction-associated recombination. Proc. Natl. Acad. Sci. USA 78: 7033–7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis, D. G., S. K. Amundsen and G. R. Smith, 1987. Genetic functions promoting homologous recombination in Escherichia coli: a study of inversions in phage λ. Genetics 115: 11–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein, R. H., A. Bolle, C. Steinberg, E. Kellenberger, E. Boy de la Tour et al., 1964. Physiological studies of conditional lethal mutants of bacteriophage T4D. Cold Spring Harbor Symp. Quant. Biol. 28: 375–392. [Google Scholar]
- Feschenko, V. V., L. A. Rajman and S. T. Lovett, 2003. Stabilization of perfect and imperfect tandem repeats by single-stranded DNA exonucleases. Proc. Natl. Acad. Sci. USA 100: 1134–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark, P. J., and S. Linn, 1972. Purification and properties of the RecBC DNase of Escherichia coli K-12. J. Biol. Chem. 247: 1849–1860. [PubMed] [Google Scholar]
- Gorbalenya, A. E., E. V. Koonin, A. P. Donchenko and V. M. Blinov, 1988. A novel superfamily of nucleoside triphosphate-binding motif containing proteins which are probably involved in duplex unwinding in DNA and RNA replication and recombination. FEBS Lett. 235: 16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgman, T., 1988. A new superfamily of replicative proteins. Nature 333: 22–23. [DOI] [PubMed] [Google Scholar]
- Kobayashi, I., H. Murialdo, J. M. Crasemann, M. M. Stahl and F. W. Stahl, 1982. Orientation of cohesive end site cos determines the active orientation of χ sequence in stimulating recA•recBC-mediated recombination in phage λ lytic infections. Proc. Natl. Acad. Sci. USA 79: 5981–5985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi, I., M. M. Stahl, F. R. Fairfield and F. W. Stahl, 1984. Coupling with packaging explains apparent nonreciprocality of Chi-stimulated recombination of bacteriophage λ by RecA and RecBC functions. Genetics 108: 773–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köppen, A., S. Krobitsch, B. Thoms and W. Wackernagel, 1995. Interaction with the recombination hot spot χ in vivo converts the RecBCD enzyme of Escherichia coli into a χ-independent recombinase by inactivation of the RecD subunit. Proc. Natl. Acad. Sci. USA 92: 6249–6253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korangy, F., and D. A. Julin, 1993. Kinetics and processivity of ATP hydrolysis and DNA unwinding by the RecBC enzyme from Escherichia coli. Biochemistry 32: 4873–4880. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski, S. C., 2000. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 25: 156–165. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski, S. C., D. A. Dixon, A. K. Eggleston, S. D. Lauder and W. M. Rehrauer, 1994. Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58: 401–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov, A., 1999. Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev. 63: 751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov, A., E. Schabtach and F. W. Stahl, 1994. χ sites in combination with RecA protein increase the survival of linear DNA in Escherichia coli by inactivating exoV activity of RecBCD nuclease. EMBO J. 13: 2764–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam, S. T., M. M. Stahl, K. D. McMilin and F. W. Stahl, 1974. Rec-mediated recombinational hot spot activity in bacteriophage λ. II. A mutation which causes hot spot activity. Genetics 77: 425–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipinska, B., A. S. Rao, B. M. Bolten, R. Balakrishnan and E. B. Goldberg, 1989. Cloning and identification of bacteriophage T4 gene 2 product gp2 and action of gp2 on infecting DNA in vivo. J. Bacteriol. 171: 488–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett, S. J., and A. J. Clark, 1984. Genetic analysis of the recJ gene of Escherichia coli K12. J. Bacteriol. 157: 190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett, S. T., C. Luisi-DeLuca and R. D. Kolodner, 1988. The genetic dependence of recombination in recD mutants of Escherichia coli. Genetics 120: 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, R., M. Hearn, P. Jenny and J. Gallant, 1988. Release factor competition is equivalent at strong and weakly suppressed nonsense codons. Mol. Gen. Genet. 213: 144–149. [DOI] [PubMed] [Google Scholar]
- Moncany, M. L., and E. Kellenberger, 1981. High magnesium content of Escherichia coli B. Experientia 37: 846–847. [DOI] [PubMed] [Google Scholar]
- Myers, R. S., and F. W. Stahl, 1994. χ and the RecBC D enzyme of Escherichia coli. Annu. Rev. Genet. 28: 49–70. [DOI] [PubMed] [Google Scholar]
- Myers, R. S., A. Kuzminov and F. W. Stahl, 1995. The recombination hot spot χ activates RecBCD recombination by converting Escherichia coli to a recD mutant phenocopy. Proc. Natl. Acad. Sci. USA 92: 6244–6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver, D. B., and E. B. Goldberg, 1977. Protection of parental T4 DNA from a restriction exonuclease by the product of gene 2. J. Mol. Biol. 116: 877–881. [DOI] [PubMed] [Google Scholar]
- Phillips, G. J., D. C. Prasher and S. R. Kushner, 1988. Physical and biochemical characterization of cloned sbcB and xonA mutations from Escherichia coli K-12. J. Bacteriol. 170: 2089–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips, R. J., D. C. Hickelton, P. E. Boehmer and P. T. Emmerson, 1997. The RecB protein of Escherichia coli translocates along single-stranded DNA in the 3′ to 5′ direction: a proposed ratchet mechanism. Mol. Gen. Genet. 254: 319–329. [DOI] [PubMed] [Google Scholar]
- Ponticelli, A. S., D. W. Schultz, A. F. Taylor and G. R. Smith, 1985. Chi-dependent DNA strand cleavage by RecBC enzyme. Cell 41: 145–151. [DOI] [PubMed] [Google Scholar]
- Razavy, H., S. K. Szigety and S. M. Rosenberg, 1996. Evidence for both 3′ and 5′ single-strand DNA ends in intermediates in Chi-stimulated recombination in vivo. Genetics 142: 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy, M., and J. Gowrishankar, 2000. Characterization of the uup locus and its role in transposon excisions and tandem repeat deletions in Escherichia coli. J. Bacteriol. 182: 1978–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roman, L. J., A. K. Eggleston and S. C. Kowalczykowski, 1992. Processivity of the DNA helicase activity of Escherichia coli recBCD enzyme. J. Biol. Chem. 267: 4207–4214. [PubMed] [Google Scholar]
- Sarthy, P. V., and M. Meselson, 1976. Single burst study of rec- and red-mediated recombination in bacteriophage lambda. Proc. Natl. Acad. Sci. USA 73: 4613–4617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, D. W., J. Swindle and G. R. Smith, 1981. Clustering of mutations inactivating a Chi recombinational hotspot. J. Mol. Biol. 146: 275–286. [DOI] [PubMed] [Google Scholar]
- Schultz, D. W., A. F. Taylor and G. R. Smith, 1983. Escherichia coli RecBC pseudorevertants lacking Chi recombinational hotspot activity. J. Bacteriol. 155: 664–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstein, J. L., and E. B. Goldberg, 1976. T4 DNA injection. I. Growth cycle of a gene 2 mutant. Virology 72: 195–211. [DOI] [PubMed] [Google Scholar]
- Simmon, V. F., and S. Lederberg, 1972. Degradation of bacteriophage lambda deoxyribonucleic acid after restriction by Escherichia coli K-12. J. Bacteriol. 112: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton, M. R., M. S. Dillingham, M. Gaudier, S. C. Kowalczykowski and D. B. Wigley, 2004. Crystal structure of RecBCD enzyme reveals a machine for processing DNA breaks. Nature 432: 187–193. [DOI] [PubMed] [Google Scholar]
- Smith, G. R., 1983. General recombination, pp. 175–209 in Lambda II, edited by R. W. Hendrix, J. W. Roberts, F. W. Stahl and R. A. Weisberg. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Smith, G. R., 1988. Homologous recombination in procaryotes. Microbiol. Rev. 52: 1–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G. R., 1991. Conjugational recombination in E. coli: myths and mechanisms. Cell 64: 19–27. [DOI] [PubMed] [Google Scholar]
- Smith, G. R., 2001. Homologous recombination near and far from DNA breaks: alternative roles and contrasting views. Annu. Rev. Genet. 35: 243–274. [DOI] [PubMed] [Google Scholar]
- Smith, G. R., M. Comb, D. W. Schultz, D. L. Daniels and F. W. Blattner, 1981. a Nucleotide sequence of the Chi recombinational hot spot χ+D in bacteriophage lambda. J. Virol. 37: 336–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, G. R., D. W. Schultz, A. F. Taylor and K. Triman, 1981. b Chi sites, RecBC enzyme, and generalized recombination. Proceedings of the 13th Stadler Genetics Symposium, Columbia, MO, pp. 25–37.
- Stahl, F. W., and M. M. Stahl, 1977. Recombination pathway specificity of Chi. Genetics 86: 715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, F. W., J. M. Crasemann and M. M. Stahl, 1975. Rec-mediated recombinational hot spot activity in bacteriophage lambda. III. Chi mutations are site-mutations stimulating Rec-mediated recombination. J. Mol. Biol. 94: 203–212. [DOI] [PubMed] [Google Scholar]
- Stahl, F. W., M. M. Stahl, R. E. Malone and J. M. Crasemann, 1980. Directionality and nonreciprocality of Chi-stimulated recombination in phage λ. Genetics 94: 235–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, F. W., M. M. Stahl, L. Young and I. Kobayashi, 1982. Chi-stimulated recombination between phage λ and the plasmid λdv. Genetics 102: 599–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, F. W., M. Lieb and M. M. Stahl, 1984. Does Chi give or take? Genetics 108: 795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, F. W., L. C. Thomason, I. Siddiqi and M. M. Stahl, 1990. Further tests of a recombination model in which χ removes the RecD subunit from the RecBCD enzyme of Escherichia coli. Genetics 126: 519–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, F. W., C. E. Shurvinton, L. C. Thomason, S. Hill and M. M. Stahl, 1995. On the clustered exchanges of the RecBCD pathway operating on phage λ. Genetics 139: 1107–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, A., and G. R. Smith, 1980. Unwinding and rewinding of DNA by the RecBC enzyme. Cell 22: 447–457. [DOI] [PubMed] [Google Scholar]
- Taylor, A. F., and G. R. Smith, 1992. RecBCD enzyme is altered upon cutting DNA at a Chi recombination hotspot. Proc. Natl. Acad. Sci. USA 89: 5226–5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, A. F., and G. R. Smith, 1995. a Monomeric RecBCD enzyme binds and unwinds DNA. J. Biol. Chem. 270: 24451–24458. [DOI] [PubMed] [Google Scholar]
- Taylor, A. F., and G. R. Smith, 1995. b Strand specificity of nicking of DNA at Chi sites by RecBCD enzyme: modulation by ATP and magnesium levels. J. Biol. Chem. 270: 24459–24467. [DOI] [PubMed] [Google Scholar]
- Taylor, A. F., and G. R. Smith, 1999. Regulation of homologous recombination: Chi inactivates RecBCD enzyme by disassembly of the three subunits. Genes Dev. 13: 890–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, A. F., and G. R. Smith, 2003. RecBCD enzyme is a DNA helicase with fast and slow motors of opposite polarity. Nature 423: 889–893. [DOI] [PubMed] [Google Scholar]
- Taylor, A. F., D. W. Schultz, A. S. Ponticelli and G. R. Smith, 1985. RecBC enzyme nicking at Chi sites during DNA unwinding: location and orientation dependence of the cutting. Cell 41: 153–163. [DOI] [PubMed] [Google Scholar]
- Thaler, D. S., E. Sampson, I. Siddiqi, S. M. Rosenberg, F. W. Stahl et al., 1988. A hypothesis: Chi-activation of RecBCD enzyme involves removal of the RecD subunit, pp. 413–422 in Mechanisms and Consequences of DNA Damage Processing, edited by E. Friedberg and P. Hanawalt. Alan R. Liss, New York.
- Thomason, L., D. L. Court, M. Bubunenko, N. Costantino, H. Wilson et al., 2005. Recombineering: genetic engineering in bacteria using homologous recombination, pp. 1–21 in Current Protocols in Molecular Biology, Unit 1.16, edited by F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman et al. John Wiley & Sons, New York. [DOI] [PubMed]
- Yu, M., J. Souaya and D. A. Julin, 1998. a The 30-kDa C-terminal domain of the RecB protein is critical for the nuclease activity, but not the helicase activity, of the RecBCD enzyme from Escherichia coli. Proc. Natl. Acad. Sci. USA 95: 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, M., J. Souaya and D. A. Julin, 1998. b Identification of the nuclease active site in the multifunctional RecBCD enzyme by creation of a chimeric enzyme. J. Mol. Biol. 283: 797–808. [DOI] [PubMed] [Google Scholar]