Abstract
As an ancient segmental tetraploid, the maize (Zea mays L.) genome contains large numbers of paralogs that are expected to have diverged by a minimum of 10% over time. Nearly identical paralogs (NIPs) are defined as paralogous genes that exhibit ≥98% identity. Sequence analyses of the “gene space” of the maize inbred line B73 genome, coupled with wet lab validation, have revealed that, conservatively, at least ∼1% of maize genes have a NIP, a rate substantially higher than that in Arabidopsis. In most instances, both members of maize NIP pairs are expressed and are therefore at least potentially functional. Of evolutionary significance, members of many NIP families also exhibit differential expression. The finding that some families of maize NIPs are closely linked genetically while others are genetically unlinked is consistent with multiple modes of origin. NIPs provide a mechanism for the maize genome to circumvent the inherent limitation that diploid genomes can carry at most two “alleles” per “locus.” As such, NIPs may have played important roles during the evolution and domestication of maize and may contribute to the success of long-term selection experiments in this important crop species.
THE grasses (Poaceae) are a highly adaptable family of monocotyledonous plants that have been independently domesticated by several human civilizations. Maize (Zea mays L.) is a hypothesized ancient segmental tetraploid, and it is estimated that nearly one-third of all modern maize genes have a paralogous sequence (Blanc and Wolfe 2004). More recently, the expected divergence of the segmental allotetraploid event has been revised from the original 15–30% (Gaut and Doebley 1997) to 10–20% (Blanc and Wolfe 2004) on the basis of maize ESTs.
Genomewide duplications are generally believed to provide raw material for evolutionary innovation (Ohno 1970) and as such they have played important roles in the evolution of both plants and vertebrates (reviewed by Durand 2003; Moore and Purugganan 2005). In contrast to the diverged paralogs produced via ancient duplications, detailed analyses of the human genome have identified nearly identical sequences that were inadvertently collapsed, or condensed into a single contiguous region, during genome assembly (Bailey et al. 2002; Cheung et al. 2003; She et al. 2004).
Tandem duplications are common among plant species (Zhang and Gaut 2003). Indeed, Messing et al. (2004) have estimated that approximately one-third of maize genes are tandemly duplicated. Few of these tandem duplications are similar enough that they would collapse during genome assembly. Several tandem duplications of maize have been well characterized, including, R-r (Robbins et al. 1991), Rp1 (Richter et al. 1995), P1 (Zhang and Peterson 2005), and A1-b (Yandeau-Nelson et al. 2006). Such duplications can be generated via unequal recombination (Richter et al. 1995; Yandeau-Nelson et al. 2006). In contrast, the transposition of Mu-like transposons in rice (Pack-MULEs; Jiang et al. 2004; Juretic et al. 2005) and Helitrons in maize (Lal et al. 2003; Brunner et al. 2005; Lai et al. 2005; Lal and Hannah 2005; Morgante et al. 2005), which have incorporated fragments of unrelated genes, can generate dispersed genic duplications. Although as many as 11% of all maize gene fragments are unique to a specific inbred line (Morgante et al. 2005), the extent to which these gene duplications are functional is not known.
Because the maize inbred line B73 is homozygous at essentially all loci and its “gene space” has been extensively sequenced, it is an ideal candidate for beginning to study the extent, causes, and evolutionary significance of recent duplications in this complex genome. Toward this end, assemblies of B73 ESTs and gene-enriched Genome Survey Sequences (GSSs) were examined for the appearance of “polymorphic” nucleotide positions, which we term candidate paramorphisms (CPs; Emrich et al. 2004; Fu et al. 2004). If a specific CP site is not due to a sequencing error or residual heterozygosity, we term this site a paramorphism (PM; Fu et al. 2004). A paramorphism provides evidence of the existence of highly similar genomic loci and is strong evidence of a recent duplication without respect to the underlying duplication mechanism. We have termed a subset of such regions nearly identical paralogs (NIPs) if they exhibit ≥98% identity, are genic, and are not transposons or other repetitive sequences.
On the basis of highly conservative criteria, we estimate that ∼1% of genes in the B73 maize genome have at least one NIP, and nearly all of these exhibit >99% identity. In addition, we determined that many of these highly similar loci in the maize genome are genetically linked. Because Mu elements do not preferentially move to linked sites (Lisch et al. 1995), this result implies either that Helitrons preferentially insert into neighboring locations or that other mechanisms were involved in the origins of these genetically linked NIPs. The observed frequency of NIPs is substantially higher in maize than in the model dicotyledon, Arabidopsis thaliana, suggesting that this phenomenon is not universal in plants. Most importantly, we also report that members of many NIP families are differentially expressed. We hypothesize that the high frequency of NIPs in combination with their diverse expression patterns may have provided a selective advantage during the domestication and the genetic improvement of maize by classical plant breeders and may play a fundamental role in the success of long-term selection experiments (e.g., Laurie et al. 2004).
MATERIALS AND METHODS
Locating and validating NIPs in collections of maize ESTs and GSSs:
EST sequences were generated from three B73 cDNA libraries constructed by Fang Qiu (Iowa State University) with the advice of the Bento Soares laboratory (University of Iowa). A total of 32,229 EST sequences and their corresponding trace files were deposited in GenBank after removing short inserts and other irregularities. These B73 EST sequences were first assembled with CAP3 (Huang and Madan 1999) using >98% similarity in detected overlaps, a minimum overlap size of 50 bp, and 60 bp as the clipping parameter. Potential NIPs were then identified by detecting contigs with CPs composed of at least two different nucleotides, each of which is supported by two independent EST reads, within CAP3 multiple sequence alignments.
We later endeavored to locate NIPs within “gene-enriched” maize genomic data (Palmer et al. 2003; Whitelaw et al. 2003) using an updated version of our maize assembled genomic islands (MAGIs; Emrich et al. 2004; Fu et al. 2005). We use the same CP-detection heuristic described above for EST NIPs, but we restricted these analyses to only methyl-filtered (MF) clones because ∼40% of current high-C0t clones contain cloning artifacts (Fu et al. 2004). In addition, we required that each CP variant be supported by at least two independent MF clones. On the basis of the criteria used to assemble the MAGIs (Fu et al. 2005), only CP-competent intervals that exhibit ≥98% identity are recovered.
Even with the conservative criteria described above, it was possible that some CPs resulted from sequencing errors. Primer3 (Rozen and Skaletsky 2000) was used to design primers ∼250 bp from each side of targeted CP sites. Genomic DNA was isolated from B73 seedling leaves using the protocol of Dietrich et al. (2002) and was PCR amplified using these CP-flanking primers. The resulting PCR products were analyzed via agarose gel electrophoresis. Single-band PCR products were then subjected to direct sequencing using the same CP-flanking PCR primers or were subcloned using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA) followed by sequencing with the T7 and T3 primers.
Annotation of NIPs:
GBrowse (V1.61) was downloaded from the Generic Model Organism Database website and installed using a MySQL database at its core. The CAP3 assembly output files, CP-competent intervals, CP sites, primers used to validate CPs, GeneSeqer alignments (at least one exon of similarity of ≥95% identity, ≥50 bp length), FGENESH predictions, and BLASTX hits (PIR-PSD v.79.00; E-value ≤1e-10) were converted into GFF files using PERL and AWK scripts for display on the MAGI website (http://magi.plantgenomics.iastate.edu/). CP-competent intervals were deemed genic if the MAGI contained a nonrepetitive gene model within 500 bp of the CP prediction. Repetitive models were excluded on the basis of protein matches to well-characterized transposons in GenBank.
NIP expression assays:
Forty-six validated MAGI–NIPs with at least one predicted exon were analyzed; 42 yielded a single genomic PCR band with the expected size. These were then subjected to touchdown RT–PCR using the pooled inbred line B73 cDNA, very similar to that described previously (Fu et al. 2005). In addition, RNA samples were also isolated from various tissues, organs, and developmental stages of the B73 inbred line similar to those described by Qiu et al. (2003). Reactions that yielded single bands that were not larger than the genomic PCR product were sequenced. If the sequence of a RT–PCR product had a double peak at the paramorphic site, we concluded that both members of the NIP family are expressed. If in a given source of RNA only a single peak was observed at a paramorphic site, we concluded that only that member was expressed in that sample. Only if identical results were obtained from two independent biological replications did we conclude that the two members of a NIP family were differentially expressed. In almost all instances, the results from the two replications were consistent.
Genetic mapping of NIPs:
NIPs were genetically mapped using 91 recombinant inbreds (RIs) of the intermated B73 × Mo17 (IBM) mapping population (Lee et al. 2002). CP validation primers that amplified B73 but not Mo17 DNA templates (i.e., plus/minus markers) were identified via gel electrophoresis. If a pair of NIPs is tightly linked genetically, the RIs will segregate 1:1 for the presence and absence of the B73-derived PCR product; conversely, if a pair of NIPs is unlinked genetically, the RIs will segregate 3:1 for the presence and absence of the B73-derived PCR product. NIPs with segregation ratios that fall between 1:1 and 3:1 were deemed to be loosely linked genetically. To position the tightly linked NIPs on the genetic map, the RI genotype scores for each NIP-derived marker were directly compared to the RI scores of all of the ∼3500 genetic markers on a genetic map developed by us (IBM_IDP+MMPmap4; Fu et al. 2006).
Locating NIPs within Arabidopsis:
A total of 190,978 A. thaliana ESTs were downloaded from dbEST (GenBank) in June 2004, and 50 bp were trimmed from each end to reduce false positives associated with low-quality sequences. These ESTs were then clustered using PaCE (Kalyanaraman et al. 2003) under default parameters, and contigs were generated using CAP3 from each resulting cluster as previously described. Polymorphic sites with representation in ≥25% of participating ESTs, which also violated random expectation for sequencing errors (P < 0.01), were selected; 28 primer pairs were designed to flank the 24 previously unreported duplications using Primer3. Successful reactions, which yielded a single band (N = 25), were sequenced and the corresponding trace files were analyzed.
In addition, all 68 low-copy Arabidopsis gene pairs that have rates of synonymous substitution (Ks) <2% (Lynch and Conery 2000; Moore and Purugganan 2003) were analyzed. Using the 02/28/2004 Arabidopsis gene annotation from The Arabidopsis Information Resource (http://www.arabidopsis.org), each potential NIP pair was checked to ensure that both members were genic and were annotated as distinct loci. Pairs that met these initial criteria were then compared using BLAST; candidates without a highly similar (>98% identity) continuous alignment were manually aligned and validated where possible. The genetic distances between members of a NIP family were determined by multiplying the physical distance that separates them by the centimorgan/megabase values reported by Zhang and Gaut (2003).
RESULTS
In silico detection of maize NIPs:
Nearly identical sequences are subject to being erroneously “collapsed” into single sequences during genome assembly. Collapsed segmental duplications within the human genome assembly were identified by virtue of their overrepresentation among randomly generated sequences (Bailey et al. 2002), and it has been estimated that >8% of public human single nucleotide polymorphisms (SNPs) are potentially paramorphisms rather than actual SNPs (Cheung et al. 2003).
Evidence for the existence of NIPs in the inbred maize B73 genome was first sought in EST data. A total of 32,229 3′ EST sequences generated by us from the B73 inbred line were assembled into 3975 contigs and 6804 singleton ESTs. To be considered a CP, each of the two nucleotides must be supported by at least two independent sequence reads. Because this conservative heuristic qualifies only a subset of an assembly for locating putative NIPs, we term such regions “CP competent.” Of the 3975 EST contigs generated by CAP3 (Huang and Madan 1999), 1659 were CP competent. To further analyze the correctness of these CP predictions, all 1659 candidates were manually inspected and the respective trace files were analyzed; following these analyses, 78 contigs were deemed promising.
Experimental validation of EST-based CP sites:
In silico predicted CP sites could arise erroneously due to sequencing errors. We therefore endeavored to experimentally validate many of the putative NIPs. A total of 75 primer pairs flanking predicted CP sites were designed from the 78 EST contigs; 54 of these primer pairs amplified a single band from B73 genomic DNA. These PCR products were sequenced. Only those CP sites that exhibited overlapping sequence trace peaks were considered to be “validated.” Overlapping trace peaks were mostly of equal intensity, although in a few instances the relative intensities were consistent with differential NIP copy number in the maize genome. Of the 54 sequenced EST contigs that contained putative CPs, 9 could be validated in this manner.
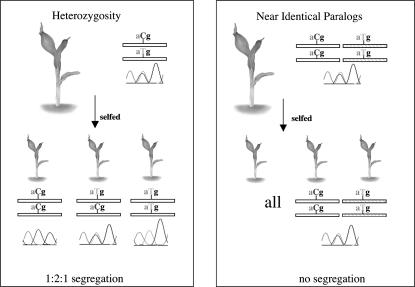
Those CP sites that were validated via sequencing provide evidence in B73 of either residual heterozygosity or NIPs. The strategy outlined in Figure 1 was employed to distinguish between these possibilities. All nine validated EST contigs were analyzed in 20 individual selfed progeny from their B73 parent plant and in a pool of 20 individual progeny from 4 additional B73 parent plants (a total of 80 plants). If the validated CPs arose via the presence of residual heterozygosity, overlapping and nonoverlapping sequence trace peaks should segregate among the selfed progeny. No evidence of residual heterozygosity was detected. We therefore conclude that B73 exhibits a very low level of residual heterozygosity. We further conclude that 0.5% (9/1659) of the analyzed EST contigs is derived from NIPs.
Figure 1.—
Strategy used for determining whether a CP is indicative of residual heterozygosity or the existence of a NIP. Because alleles segregate during meiosis, CPs associated with residual heterozygosity are expected to segregate in a 1:2:1 ratio among selfed progeny. In contrast, NIPs would not be expected to segregate among the selfed progeny of an inbred line.
NIPs discovered within a partial maize genome assembly:
For purposes of NIP detection, ESTs are valuable because they are expressed and therefore inherently meet one of the criteria for classifying a duplicated sequence as a NIP (i.e., expression). On the other hand, because introns may be more diverged than ESTs, genomic regions from which these cDNAs are transcribed may not exhibit sufficient nucleotide identity (>98%) to be classified as NIPs. In addition, CPs can be identified only in genes for which at least four ESTs have been captured.
To address these limitations and to identify more NIPs in the maize genome, we endeavored to locate CPs within version 3.1 of our MAGIs (Fu et al. 2005), which consists of 114,173 contigs. Because MAGIs include introns, the selection of MAGI-derived NIPs is even more stringent than for EST-based NIPs. A total of 15,375 MAGIs contain at least four overlapping clones and are therefore CP competent; 289 of these competent contigs exhibit at least one CP.
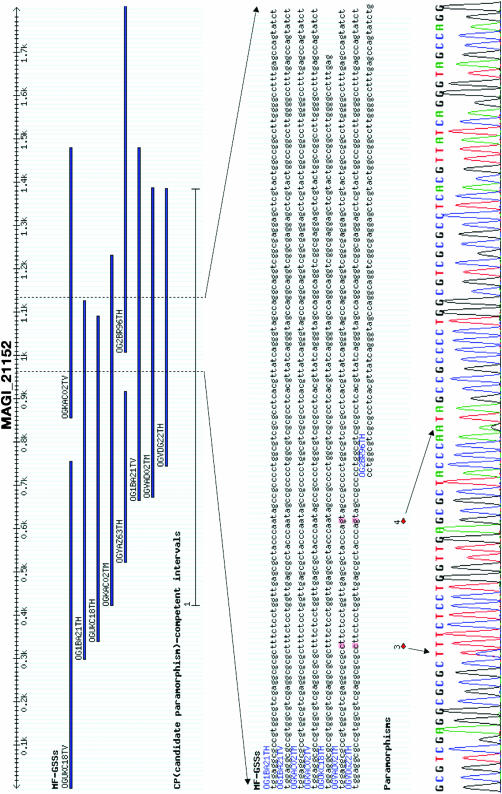
Primer pairs that flank CP sites for 280 of the 289 candidate MAGIs were designed, of which 231 amplified a single band from B73 genomic DNA. Sequence analyses of these amplicons validated a total of 258 paramorphisms (PMs) in 116 PM-containing MAGIs (Figure 2; see also supplemental Figure 1 at http://www.genetics.org/supplemental/) via a strategy identical to that used to validate NIPs identified from EST contigs. In several cases, primer pairs appeared to amplify multiple amplicons as evidenced by numerous multiple peaks in the sequence trace files. This suggests that a somewhat more distant paralog was also being amplified. Although at least one CP site was confirmed in these cases, to be conservative, these MAGIs were not included in subsequent analyses and calculations.
Figure 2.—
An example of a validated NIP (MAGI_21152). The membership and layout of MF GSSs, a CP-competent interval (∼900 bp), and the trace file for a 150-bp subinterval of the CP-competent interval (the bottom chromatograph) are shown relative to the two paramorphisms highlighted.
Expression of NIPs:
Evidence for the expression of each of the 116 PM-containing MAGIs was sought via EST alignments, FGENESH predictions, and BLASTX results (materials and methods; Figure 2). The 84 PM-containing MAGIs for which evidence of gene expression was obtained were deemed to be NIPs (see supplemental Table 1 at http://www.genetics.org/supplemental/). These 84 NIPs contain a total of 170 validated paramorphic sites, which are located in both coding and noncoding regions.
Of the 44 NIPs that could be assigned functions via significant BLASTX matches, 10 are predicted kinases and 3 are predicted transcription factors and/or contain a zinc-finger domain. The remaining 31 NIPs are involved in a wide variety of biochemical pathways (e.g., metabolism, nitrogen utilization, and DNA methylation). We therefore conclude that NIPs are not restricted to a limited number of biological functions.
Frequency of NIPs:
The experiments described above identified 84 genic MAGIs that contain one or more paramorphisms and are therefore classified as NIPs. Of the 15,375 CP-competent MAGIs, 12,012 appear to be genes on the basis of their lack of similarity to transposons and evidence of expression. The CP-competent intervals associated with the 84 validated NIPs exhibit ≥98% nucleotide identity, include both coding and noncoding sequences, and can be as long as 2.6 kb (supplemental Figure 2 at http://www.genetics.org/supplemental/). Because <80% (231/289) of the CP-containing MAGIs were analyzed, we conservatively estimate that 0.9% [84/(12,012 × 0.8)] of the genes in this assembly have a NIP.
Both members of many NIP families are expressed:
Forty-six NIPs that contained at least one exon or putative exon (materials and methods) were selected for analysis. Touchdown PCR was performed using both genomic DNA and pooled cDNA isolated from various tissues and organs of the inbred line B73. A total of 29 NIPs yielded a single band from both PCR reactions, of which 25 could be confirmed to be derived from the target NIP via sequencing. As shown in Table 1, these sequencing experiments provided evidence that both members of 20 NIP families (80%; 20/25) are expressed (materials and methods). For the remaining 5 NIPs (20%; 5/25), only one copy could be shown to be expressed. This is, however, a highly conservative assay for the expression because only a portion of the transcriptome was sampled. We conclude that both members of at least four-fifths of NIP families are expressed.
TABLE 1.
NIP pairs for which RT–PCR validated expression of both members
| Paramorphisms
|
|||
|---|---|---|---|
| MAGI ID | Annotationa | Position | Haplotypeb |
| A. NIPs with EST support | |||
| 33361 | Class III peroxidase 70 precursor | 2571, 2586 | G … T |
| A … C | |||
| 43016 | Putative proteosome subunit | 825 | C |
| G | |||
| 53926 | Putative cytochrome P450 | 2594, 2635 | T … G |
| C … A | |||
| 58637 | Putative membrane related | 651 | C |
| T | |||
| 65202 | Hypothetical protein | 3315 | G |
| A | |||
| 80184 | Receptor-like kinase-like | 1442 | T |
| G | |||
| 86866 | Putative acyltransferase | 1669 | C |
| T | |||
| 89568 | NA | 1085 | C |
| T | |||
| 97955 | Putative nitrate reductase apoenzyme | 1684 | C |
| G | |||
| 100946 | Putative trehalose-6-phosphate synthase/phosphatase | 715 | A |
| G | |||
| B. NIPs with only FGENESH support | |||
| 21152 | Putative strictosidine synthase | 904, 909, 975, 999 | G … A … T … A |
| A … G … C … G | |||
| 36788 | NA | 1176 | T |
| C | |||
| 45574 | Protein kinase | 1448 | C |
| T | |||
| 67751 | Putative S-receptor kinase | 1048, 1087, 1122 | T … G …G |
| C … A … C | |||
| 85672 | NA | 330 | A |
| C | |||
| 89009 | Pentatricopeptide repeat-containing | 862 | C |
| T | |||
| 98934 | Putative cytochrome P450 | 652, 660, 785 | C … T … A |
| G … C … T | |||
| 95980 | AKIN β1-like protein | 2126 | T |
| C | |||
| 101406 | Terpene syntase 5 related | 1263 | T |
| A | |||
BLASTX search against UniRef protein database using e−10 as E-value cutoff.
The presence of “. . .” between paramorphisms indicates that sites are not adjacent.
Members of many NIP families exhibit differential expression:
Ten NIP families in which both members were expressed were further analyzed using RNA samples extracted from 16 different developmental stages of various tissues and organs. Members of 8 (80%) of these 10 NIP families were differentially expressed in at least one RNA sample (Table 2). We conclude that the members of many expressed NIP families are differentially expressed.
TABLE 2.
Expression patterns of NIPs in the B73 inbred line
| cDNA sample no.a
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAGI ID | Paramorphism sites | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 |
| 21152 | G … A … T … A | +b | NDc | + | + | + | + | + | ND | + | + | + | + | + | −b | + | + | + |
| A … G … C … G | + | ND | + | + | + | + | + | ND | + | + | − | − | + | + | + | + | + | |
| 43016 | G | + | + | + | + | + | + | ND | + | + | + | + | + | + | + | + | + | ND |
| C | + | + | + | + | + | + | ND | + | + | + | + | + | + | + | + | + | ND | |
| 53926 | T … G | + | + | − | − | − | − | + | − | + | + | ND | − | − | − | + | + | − |
| C … A | + | + | − | − | − | − | + | − | + | + | ND | − | + | − | + | + | + | |
| 65202 | G | + | − | − | + | + | + | − | + | + | + | ND | + | ND | + | ND | + | − |
| A | + | + | − | + | + | + | + | + | + | + | ND | + | ND | + | ND | + | + | |
| 67751 | T … G … G | + | − | − | − | − | − | − | − | − | + | + | − | − | + | − | − | − |
| C … A … C | + | + | − | − | − | − | − | − | − | + | + | − | − | + | + | − | − | |
| 80184 | T | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + |
| G | + | + | + | + | + | + | + | + | + | − | + | + | + | + | + | + | + | |
| 86866 | C | + | + | − | − | + | − | − | − | + | + | + | + | ND | − | + | + | + |
| T | + | + | − | − | + | − | − | − | + | − | − | − | ND | − | − | − | + | |
| 89568 | C | + | − | − | − | − | − | + | − | − | − | + | − | − | − | + | − | − |
| T | + | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| 97955 | C | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + |
| G | + | + | − | + | + | + | + | + | + | + | + | + | − | + | + | + | + | |
| 100946 | G | + | + | − | − | ND | ND | + | − | + | − | + | ND | − | − | + | + | + |
| A | + | − | − | − | ND | ND | − | − | + | − | − | ND | − | − | − | + | − | |
cDNA samples: 1, pooled cDNA; 2, 14DAPL shoot; 3, 59DAPL root; 4, 59DAPL husk; 5, 65DAPL husk; 6, 79DAPL husk; 7, 59DAPL unpollinated ear; 8, 65DAPL unpollinated ear; 9, 79DAPL unpollinated ear; 10, 59DAPL tassel; 11, 65DAPL tassel; 12, 79DAPL unpollinated silk; 13, 1 DAP silk; 14, 1DAP kernel; 15, 5DAP kernel; 16, 15DAP kernel; 17, mature pollen. (DAPL, days after planting; DAP, days after pollination.)
+, RT–PCR product present; −, RT–PCR product not present.
ND, no data for sequencing result.
Genomic organization of maize NIPs:
To begin to define the molecular events that give rise to NIPs, it would be useful to know the relative positions of members of NIP families within the maize genome. These experiments were conducted by using PCR primers that flank paramorphisms to amplify genomic DNA from the inbreds B73 and Mo17 and the IBM RIs derived from a cross between B73 and Mo17.
Most of the 84 NIP primer pairs could amplify both B73 and Mo17 and the resulting amplicons from these two inbreds were the same size at the resolution afforded by gel electrophoresis. However, B73 genomic DNA but not Mo17 was amplified when 14 of the primer pairs were used in PCR. This indicates either that the corresponding Mo17 NIPs exhibit a high degree of sequence or structural polymorphism relative to the B73 NIPs from which the PCR primers were designed or that the Mo17 genome does not contain the corresponding NIP, a result that would extend the violations of genomic colinearity among maize inbreds initially observed by Fu and Dooner (2002) and extended by others (Brunner et al. 2005; Lai et al. 2005; Lal and Hannah 2005). Using the PCR primers that amplify B73 NIPs but not Mo17 to genotype the IBM RIs, it was possible to determine the positions of the members of all 14 NIP families relative to each other (materials and methods). The members of 7 and 2 NIP families were tightly and loosely linked, respectively (see supplemental Table 1 at http://www.genetics.org/supplemental/). The members of an additional 5 NIP families were unlinked genetically.
Arabidopsis NIPs:
Although Arabidopsis has a much smaller genome than maize, it is also thought to have undergone an ancient polyploidization event (Vision et al. 2000). To compare the relative rates of NIPs in these two model plants, we sought EST-based NIPs in Arabidopsis using the Columbia ecotype. Of the 33 initial EST clusters analyzed that contained at least one statistically significant CP, 7 were found to have already been reported to be transcribed from two or more copies in the Arabidopsis genome; however, the inclusion of introns for all seven of these genes results in <98% identity. A total of 117 CPs were tested in 24 of the 26 novel Arabidopsis NIPs using primer pairs that successfully amplified a single band of DNA from Columbia genomic template (25 primer pairs total); 100 were definitively established as false positives. The remaining 17 putative CP sites could not be verified as negative due to low-quality sequence reads. Hence, there is no evidence that any of the Arabidopsis EST clusters surveyed here represent novel collapsed paralogs.
To confirm this observation, we located NIPs among all 68 low-copy Arabidopsis gene pairs that have rates of synonymous substitution (Ks) that are <2% (Lynch and Conery 2000; Moore and Purugganan 2003). Only 39 pairs meet the NIP criteria and are annotated as distinct loci (materials and methods), which is consistent with the EST result. Of these NIP families, 28 are located <10 cM apart (materials and methods). Of the remaining 11 NIP families, 9 of these are located on different chromosomes.
DISCUSSION
The maize genome contains a high frequency of NIPs:
Plant genomes contain large numbers of paralogs, many of which are tandemly arrayed (Sun et al. 2001; Yuan et al. 2002; Messing et al. 2004). In addition, maize contains a substantial degree of intraspecies diversity for gene content (Fu and Dooner 2002). At least some of the intraspecific violations of genetic colinearity are due to “hitchhiking” gene fragments that have been duplicated by active transposons (Brunner et al. 2005; Lai et al. 2005; Lal and Hannah 2005; Morgante et al. 2005). Potentially, these duplications of genic sequences have significant evolutionary implications. The extent to which these duplications are functional is, however, under debate (Juretic et al. 2005).
It has previously been reported that several pairs of NIPs are expressed. These include the genetically unlinked ciszog1 and ciszog2 genes (Swigonova et al. 2005), the tightly linked p1 and p2 genes (Zhang et al. 2000), and the locally duplicated zein seed storage protein gene families that exhibit 98% identity (Song et al. 2001). This study demonstrates that most NIPs are expressed and that individual members of many NIP families exhibit differential expression patterns. Given their high degree of sequence identity, it likely that these different expression patterns are controlled by sequence variation outside the NIPs or differing epigenetic states, including local chromatin structure. Taken together, this study provides the first conclusive evidence that substantial numbers of hypomethylated duplications have successfully diversified their expression profiles and may therefore have unique functional roles.
Origins of NIPs:
Following duplication, gene pairs would be expected to decay into NIPs. Although transposons can “capture” gene sequences and duplicate them via transposition, Mu elements do not preferentially insert at genetically linked sites (Lisch et al. 1995). It is therefore unlikely that Pack-MULEs (Jiang et al. 2004) would be able to generate the large proportion of genetically linked NIPs observed in this study. Similarly, unless Helitrons (Lal et al. 2003; Brunner et al. 2005; Lai et al. 2005; Lal and Hannah 2005; Morgante et al. 2005) preferentially insert in nearby locations, tandemly arrayed NIPs are unlikely to have arisen via the action of Helitrons. We therefore consider several alternative mechanisms that could generate NIPs.
Unequal recombination between repetitive sequences that flank genes can generate gene duplications (Babcock et al. 2003). In humans, such processes are thought to be responsible for ∼30% of the recent segmental duplications (Zhou and Mishra 2005). Unequal recombination occurs between the long terminal repeats of rice retrotransposons (Ma et al. 2004; Ma and Bennetzen 2006). Tandem gene duplications generated via this mechanism would be flanked by repeats of high identity. An ∼10-kb segment of BAC clone ZMMBBb0483G05 deposited in GenBank (accession no. AC157776) by the McCombie laboratory contains two pairs of tandemly duplicated NIPs; each pair of NIPs exhibits >99.5% identity. Significantly, conserved repeats (as defined by the Iowa State University MAGI Cereal Repeat Database 3.1; Fu et al. 2005) are located between and flanking the duplications. The positioning of these repeats is consistent with duplication via unequal pairing between the repeats.
More exotic mechanisms of NIP generation are also possible. For example, break-induced replication at stalled replication forks could stimulate the production of segmental duplications (Figure 3A, iii) and rearrangements in regions of genomic instability (Koszul et al. 2004; Zhou and Mishra 2005). Gene conversion or similar mechanisms may have also homogenized diverged paralogs. Because many of the characterized maize gene conversion events have conversion tracts >1 kb (reviewed by Yandeau-Nelson et al. 2005), it is possible that this mechanism could generate NIPs. In support of this hypothesis, we have recently observed that the duplicate gl8 genes (gl8a and gl8b), which reside on syntenic regions of different chromosomes and therefore presumably originated during the ancient allotetraploidization event, exhibit a degree of nucleotide identity (96%; Dietrich et al. 2005) that is substantially higher than the 80–90% identity expected for ancient paralogs (Blanc and Wolfe 2004). Because tandemly arrayed paralogs undergo frequent recombination (Yandeau-Nelson et al. 2006), gene conversion can also maintain a high degree of nucleotide identity between them (Zhang and Peterson 2005).
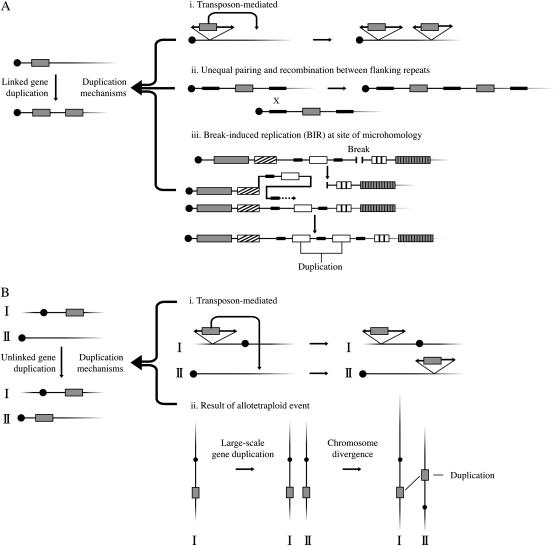
Figure 3.—
Mechanisms of gene duplication for (A) genetically linked (i–iii) and (B) genetically unlinked (i–iii) NIPs. Unequal pairing between flanking repeats (A, ii) can occur between homologs or sister chromatids, but probably at a lower rate. Transposon-mediated duplication can generate genetically tightly linked (A, i) and unlinked (B, i) NIPs. Unlinked NIPs could reside on separate chromosomes as depicted in (B, i) or could be at least 50 cM apart on the same chromosome. (B) Genetically unlinked NIPs are shown on two separate chromosomes (I and II). Unlinked NIPs can result from duplications of entire chromosomes (B, ii) or large segments of chromosomes that subsequently diverge (i.e., chromosomal rearrangements and gene loss or gain). Unlinked NIPs might also be generated by chromosomal rearrangements between duplicates that were originally genetically linked. Both linked and unlinked gene duplications might also occur by currently uncharacterized mechanisms. Boxes, thick lines, and solid circles represent genes, nongenic repeats, and centromeres, respectively.
While it is not currently possible to identify the mechanism by which a given NIP pair was generated, it is likely that multiple mechanisms are involved. It may be possible to decipher these mechanisms once the maize genome sequence has been completed by locating the specific sequence signatures that are associated with each duplication mechanism (Figures 3 and 4).
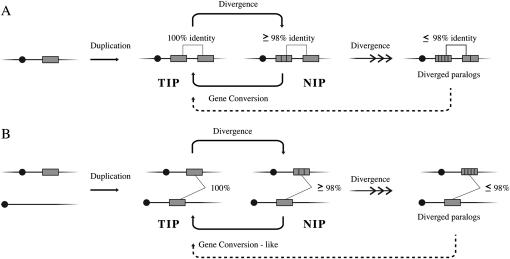
Figure 4.—
A proposed mechanism for the evolution of gene duplications and the generation of NIPs and totally identical paralogs (TIPs). Genetically linked (A) and unlinked (B) duplication events generate TIPs that can diverge over time to produce NIPs. NIPs can be homogenized back into TIPs via nonallelic gene conversion or can further diverge. More diverged paralogs might also be homogenized into TIPs, but likely at a lower rate (dashed line). Shaded boxes represent genes and vertical lines within the boxes represent paramorphisms.
Why does maize have more NIPs than Arabidopsis?
We conservatively estimate that the maize genome contains at least 500 NIPs. In contrast, we identified <10% of this number of NIPs in the Arabidopsis genome (N = 39). This is true even though the Arabidopsis genome contains Helitrons (Kapitonov and Jurka 2001), which duplicate genes in maize (Brunner et al. 2005; Lai et al. 2005; Lal and Hannah 2005; Morgante et al. 2005).
The frequency of NIPs within a species depends on the rates of four parameters: the rate and timing of initial duplication events, the rate at which NIPs decay (mutation rate), and the rates of gene loss and gene conversion. Hence, the lower frequency of NIPs in Arabidopsis as compared to maize could be a consequence of a lower rate of gene duplication. Alternatively, if gene conversion is a dominant mechanism for gene duplication, the fact that only ∼12.6–16.6% of Arabidopsis genes are members of tandemly arrayed gene families (Zhang and Gaut 2003) as compared to ∼35% of maize genes (Messing et al. 2004) may contribute to the observed differences in NIP content between these species.
NIPs and genetic markers:
NIPs can complicate the development of SNP-based genetic markers. This is because an apparent “SNP” identified via comparisons of ESTs or shotgun sequences from two inbreds may represent a paramorphism rather than a true SNP. Unlike SNPs, paramorphisms will not necessarily exhibit Mendelian segregation; therefore, it may not be possible to convert them into informative genetic markers. Indeed, such an explanation has been invoked to explain the inability to convert a fraction of human “SNPs” into genetic markers (Fredman et al. 2004).
Evolutionary implications of NIPs:
An individual diploid genome can contain at most two alleles of a given locus. NIPs provide a mechanism for a maize plant to include more than two “alleles” of a given gene within its genome and the differential expression of members within a NIP family can increase the plasticity of the transcriptome. Hence, the genetic diversity provided by NIPs may contribute to the environmental stability of maize. NIPs may also serve as a reservoir of genetic variability upon which selection can act because recombination between highly similar paralogs can generate new “alleles” that condition novel phenotypes (Zhang and Peterson 2005). Finally, the existence of multiple copies of a given sequence (i.e., NIPs) increases the probability of recovering rare favorable mutations. As such, NIPs may have facilitated the domestication of maize and may contribute to the continuing success of long-term selection experiments in closed maize populations (Laurie et al. 2004) and maize breeding in general.
Acknowledgments
We thank Amy Nienaber and Elizabeth Hahn for technical assistance with the NIP detection experiments in maize and Arabidopsis, respectively; Karthik Viswanathan and Mu Zhang for computational support; Hsin D. Chen and Josh Shendelman for genetic mapping; Sang-Duck Seo for assistance in figure design; and An-Ping Hsia for helpful comments. Scott Emrich was supported in part by a National Science Foundation (NSF) Integrative Graduate Education and Research Traineeship fellowship (award no. DGE-9972653). This research was funded in part by competitive grants from the NSF Plant Genome Program (DBI-9975868 and DBI-0321711); additional support was provided by the Hatch Act and State of Iowa funds.
References
- Babcock, M., A. Pavlicek, E. Spiteri, C. D. Kashork, I. Ioshikhes et al., 2003. Shuffling of genes within low-copy repeats on 22q11 (LCR22) by Alu-mediated recombination events during evolution. Genome Res. 13: 2519–2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey, J. A., Z. Gu, R. A. Clark, K. Reinert, R. V. Samonet et al., 2002. Recent segmental duplications in the human genome. Science 297: 1003–1007. [DOI] [PubMed] [Google Scholar]
- Blanc, G., and K. H. Wolfe, 2004. Widespread paleopolyploidy in model plant species inferred from age distributions of duplicate genes. Plant Cell 16: 1667–1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner, S., K. Fengler, M. Morgante, S. Tingey and A. Rafalski, 2005. Evolution of DNA sequence nonhomologies among maize inbreds. Plant Cell 17: 343–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung, J., X. Estivill, R. Khaja, J. R. Macdonald, K. Lau et al., 2003. Genome-wide detection of segmental duplications and potential assembly errors in the human genome sequence. Genome Biol. 4: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, C., F. Cui, M. Packila, J. Li, D. Ashlock et al., 2002. Maize Mu transposons are targeted to the 5′ untranslated region of the gl8 gene and sequences flanking Mu target-site duplications exhibit nonrandom nucleotide composition throughout the genome. Genetics 160: 697–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich, C. R., M. A. Perera, M. D. Yandeau-Nelson, R. B. Meeley, B. J. Nikolau et al., 2005. Characterization of two gl8 paralogs reveals that the 3-ketoacyl reductase component of fatty acid elongase is essential for maize (Zea mays L.) development. Plant J. 42: 844–861. [DOI] [PubMed] [Google Scholar]
- Durand, D., 2003. Vertebrate evolution: doubling and shuffling with a full deck. Trends Genet. 19: 2–5. [DOI] [PubMed] [Google Scholar]
- Emrich, S. J., S. Aluru, Y. Fu, T.-J. Wen, M. Narayanan et al., 2004. A strategy for assembling the maize (Zea mays L.) genome. Bioinformatics 20: 140–147. [DOI] [PubMed] [Google Scholar]
- Fredman, D., S. J. White, S. Potter, E. E. Eichler, J. T. Den Dunnen et al., 2004. Complex SNP-related sequence variation in segmental genome duplications. Nat. Genet. 36: 861–866. [DOI] [PubMed] [Google Scholar]
- Fu, H., and H. Dooner, 2002. Intraspecific violation of genetic colinearity and its implications in maize. Proc. Natl. Acad. Sci. USA 99: 9573–9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., A.-P. Hsia, L. Guo and P. S. Schnable, 2004. Types and frequencies of sequencing errors in methyl-filtered and high C0t maize genome survey sequences. Plant Physiol. 135: 2040–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., S. J. Emrich, L. Guo, T.-J. Wen, D. Ashlock et al., 2005. Quality assessment of maize assembled genomic islands (MAGIs) and large-scale experimental verification of predicted genes. Proc. Natl. Acad. Sci. USA 102: 12282–12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu, Y., T. J. Wen, Y. I. Ronin, H. D. Chen, L. Guo et al., 2006. Genetic dissection of intermated recombinant inbred lines using a new genetic map of maize. Genetics 174: 1671–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaut, B. S., and J. F. Doebley, 1997. DNA sequence evidence for the segmental allotetraploid origin of maize. Proc. Natl. Acad. Sci. USA 94: 6809–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, X., and A. Madan, 1999. CAP3: a DNA sequence assembly program. Genome Res. 9: 868–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N., Z. Bao, X. Zhang, S. R. Eddy and S. R. Wessler, 2004. Pack-MULE transposable elements mediate gene evolution in plants. Nature 431: 569–573. [DOI] [PubMed] [Google Scholar]
- Juretic, N., D. R. Hoen, M. L. Huynh, P. M. Harrison and T. E. Bureau, 2005. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 15: 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyanaraman, A., S. Aluru, S. Kothari and V. Brendel, 2003. Efficient clustering of large EST data sets on parallel computers. Nucleic Acids Res. 31: 2963–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov, V. V., and J. Jurka, 2001. Rolling-circle transposons in eukaryotes. Proc. Natl. Acad. Sci. USA 98: 8714–8719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koszul, R., S. Caburet, B. Dujon and G. Fischer, 2004. Eucaryotic genome evolution through the spontaneous duplication of large chromosomal segments. EMBO J. 23: 234–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai, J., Y. Li, J. Messing and H. K. Dooner, 2005. Gene movement by Helitron transposons contributes to the haplotype variability of maize. Proc. Natl. Acad. Sci. USA 102: 9068–9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal, S. K., M. J. Giroux, V. Brendel, C. E. Vallejos and L. C. Hannah, 2003. The maize genome contains a Helitron insertion. Plant Cell 15: 381–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal, S. K., and L.C. Hannah, 2005. Helitrons contribute to the lack of gene colinearity observed in modern maize inbreds. Proc. Natl. Acad. Sci. USA 102: 9993–9994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie, C. C., S. D. Chasalow, J. R. Ledeaux, R. McCarroll, D. Bush et al., 2004. The genetic architecture of response to long-term artificial selection for oil concentration in the maize kernel. Genetics 168: 2141–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M., N. Sharopova, W. D. Beavis, D. Grant, M. Katt et al., 2002. Expanding the genetic map of maize with the intermated B73 × Mo17 (IBM) population. Plant Mol. Biol. 48: 453–461. [DOI] [PubMed] [Google Scholar]
- Lisch, D., P. Chomet and M. Freeling, 1995. Genetic characterization of the Mutator system in maize: behavior and regulation of Mu transposons in a minimal line. Genetics 139: 1777–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, M., and J. S. Conery, 2000. The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- Ma, J., and J. L. Bennetzen, 2006. Recombination, rearrangement, reshuffling, and divergence in a centromeric region of rice. Proc. Natl. Acad. Sci. USA 103: 383–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, J., K. M. Devos and J. L. Bennetzen, 2004. Analyses of LTR-retrotransposon structures reveal recent and rapid genomic DNA loss in rice. Genome Res. 14: 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messing, J., A. K. Bharti, W. M. Karlowski, H. Gundlach, H. R. Kim et al., 2004. Sequence composition and genome organization of maize. Proc. Natl. Acad. Sci. USA 101: 14349–14354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R. C., and M. D. Purugganan, 2003. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 100: 15682–15687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, R. C., and M. D. Purugganan, 2005. The evolutionary dynamics of plant duplicate genes. Curr. Opin. Plant Biol. 8: 122–128. [DOI] [PubMed] [Google Scholar]
- Morgante, M., S. Brunner, G. Pea, K. Fengler, A. Zuccolo et al., 2005. Gene duplication and exon shuffling by Helitron-like transposons generate intraspecies diversity in maize. Nat. Genet. 37: 997–1002. [DOI] [PubMed] [Google Scholar]
- Ohno, S., 1970. Evolution by Gene Duplication. Springer-Verlag, New York.
- Palmer, L. E., P. D. Rabinowicz, A. L. O'Shaughnessy, V. S. Balija, L. U. Nascimento et al., 2003. Maize genome sequencing by methylation filtration. Science 302: 2115–2117. [DOI] [PubMed] [Google Scholar]
- Qiu, F., L. Guo, T.-J. Wen, F. Liu, D. A. Ashlock et al., 2003. DNA sequence-based “bar-codes” for tracking the origins of ESTs from a maize cDNA library constructed using multiple mRNA sources. Plant Physiol. 133: 475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter, T. E., T. J. Pryor, J. L. Bennetzen and S. H. Hulbert, 1995. New rust resistance specificities associated with recombination in the Rp1 complex in maize. Genetics 141: 373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins, T. P., E. L. Walker, J. L. Kermicle, M. Alleman and S. L. Dellaporta, 1991. Meiotic instability of the R-r complex arising from displaced intragenic exchange and intrachromosomal rearrangement. Genetics 129: 271–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen, S., and H. J. Skaletsky, 2000. Primer3 on the WWW for general users and for biologist programmers, pp. 365–386 in Bioinformatics Methods and Protocols: Methods in Molecular Biology, edited by S. Misener and S. A. Krawetz. Humana Press, Totowa, NJ. [DOI] [PubMed]
- She, X., Z. Jiang, R. A. Clark, G. Liu, Z. Cheng et al., 2004. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature 431: 927–930. [DOI] [PubMed] [Google Scholar]
- Song, R., V. Llaca, E. Linton and J. Messing, 2001. Sequence, regulation, and evolution of the maize 22-kD alpha zein gene family. Genome Res. 11: 1817–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Q., N. C. Collins, M. Ayliffe, S. M. Smith, J. Drake et al., 2001. Recombination between paralogues at the rp1 rust resistance locus in maize. Genetics 158: 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigonova, Z., J. L. Bennetzen and J. Messing, 2005. Structure and evolution of the r/b chromosomal regions in rice, maize and sorghum. Genetics 169: 891–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vision, T. J., D. G. Brown and S. D. Tanksley, 2000. The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117. [DOI] [PubMed] [Google Scholar]
- Whitelaw, C. A., W. B. Barbazuk, G. Pertea, A. P. Chan, F. Cheung et al., 2003. Enrichment of gene-coding sequences in maize by genome filtration. Science 302: 2118–2120. [DOI] [PubMed] [Google Scholar]
- Yandeau-Nelson, M. D., Q. Zhou, H. Yao, X. Xu, B. J. Nikolau et al., 2005. MuDR transposase increases the frequency of meiotic crossovers in the vicinity of a Mu insertion in the maize a1 gene. Genetics 169: 917–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yandeau-Nelson, M. D., X. Yiji, L. Jin, M. G. Neuffer and P. S. Schnable, 2006. Unequal sister chromatid and homolog recombination at a tandem duplication of the a1 locus in maize. Genetics 173: 2211–2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan, Q., J. Hill, J. Hsiao, K. Moffat, S. Ouyang et al., 2002. Genome sequencing of a 239-kb region of rice chromosome 10L reveals a high frequency of gene duplication and a large chloroplast DNA insertion. Mol. Genet. Genomics 267: 713–720. [DOI] [PubMed] [Google Scholar]
- Zhang, F., and T. Peterson, 2005. Comparisons of maize pericarp color1 alleles reveal paralogous gene recombination and an organ-specific enhancer region. Plant Cell 17: 903–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., and B. S. Gaut, 2003. Does recombination shape the distribution and evolution of tandemly arrayed genes (TAGs) in the Arabidopsis thaliana genome? Genome Res. 13: 2533–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, P., S. Chopra and T. Peterson, 2000. A segmental gene duplication generated differentially expressed myb-homologous genes in maize. Plant Cell 12: 2311–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, Y., and B. Mishra, 2005. Quantifying the mechanisms for segmental duplications in mammalian genomes by statistical analysis and modeling. Proc. Natl. Acad. Sci. USA 102: 4051–4056. [DOI] [PMC free article] [PubMed] [Google Scholar]