Abstract
We measured the rate of mutations impairing sporulation ability in Bacillus subtilis as 0.003 in a mutator population, following 6000 generations of strong selection for sporulation that have previously been described. This means that the product of the population size and the functional mutation rate is ∼105, well within the parameter range for which genetic canalization of sporulation ability is expected.
IN a well-adapted population, most mutations are deleterious. Genetic canalization refers to buffering or robustness to the phenotypic effects of mutations. According to theoretical models, genetic canalization may evolve due to selection against the harmful effects of mutations (Wagner 1996; Eshel and Matessi 1998; Rice 1998, 2002; van Nimwegen et al. 1999; Wilke et al. 2001). Alternatively, genetic canalization may evolve as a by-product of selection for environmental canalization (Wagner et al. 1997; Ancel and Fontana 2000; Meiklejohn and Hartl 2002).
In contrast to the recent plethora of theoretical models, canalization has been difficult to study using experimental approaches (Scharloo 1991). Waddington (1942) observed that mutants tend to be more variable than wild-type individuals, at least in multicellular organisms whose phenotypes can be easily scored on an individual basis, such as Drosophila. This observation has been well confirmed since then (Scharloo 1991). This variation, whether revealed by a major mutation or by environmental perturbation, has been shown to depend on preexisting genetic variation (Bateman 1959; Rutherford and Lindquist 1998; True and Lindquist 2000; Queitsch et al. 2002; Dworkin et al. 2003).
The uncovering of variation by a major perturbation seems to imply that wild-type individuals are canalized against this preexisting variation, since it did not affect their phenotype. Hermisson and Wagner (2004) have described a flaw in this logic. According to their metaphor, standing genetic variation is “dust” and canalization is a “rug.” A major mutation shifts the rug, revealing dust (variation) underneath. Modeling certain forms of epistasis using a quantitative genetic approach, Hermisson and Wagner (2004) showed that revealing variation (shifting the rug) can occur without a change in canalization (size of the rug). Mutants can be more variable than wild-type individuals simply because of a shift in the rug, and so this observation does not prove that the system was canalized.
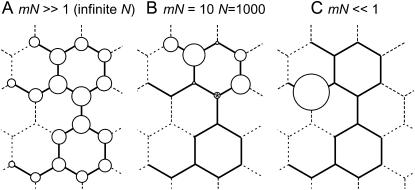
In an alternative metaphor, canalization or the “rug” can be viewed as a neutral network (van Nimwegen et al. 1999; Ancel and Fontana 2000), as illustrated in Figure 1. This is a set of genotypes that all encode the same phenotype, where each connection corresponds to a single mutation, and there is a path connecting any two genotypes that form part of the same neutral network. Other mutations lead to genotypes with different phenotypes, and these fall outside the neutral network. If the mutation rate is sufficiently high, then no single genotype dominates a population, which is better represented by a cloud of genotypes known as a quasi-species (Eigen et al. 1989). The distribution of the cloud is represented by the size of the circles in Figure 1A. Natural selection for genetic canalization can then lead to “survival of the flattest” (Schuster and Swetina 1988; Wilke et al. 2001), where a large neutral network whose genotype has lower fitness may be favored over a smaller neutral network whose genotype has higher fitness.
Figure 1.—
A simple, schematic neutral network. Nodes indicate possible genotypes and the lines between them indicate possible mutations. Mutationally accessible genotypes with the same phenotype, i.e., genotypes lying on the neutral network, are connected by solid lines. The area of the circles shows the relative frequency of genotypes found in a population. (A) The genotype frequencies in an infinite population [mN ≫ 1 (infinite N)] were solved by finding the principal eigenvector of the graph adjacency matrix (van Nimwegen et al. 1999). We see that interior nodes, which are more canalized, have a higher genotype frequency than nodes at the edge of the neutral network. (B) For smaller population sizes, dispersal of genotypes is more random. Frequencies shown were generated by a single simulation of a Wright–Fisher process with N = 1000, m = 0.01, and mN = 10, beginning with all individuals having the starred genotype, and ending after 105 generations. (C) When mN ≪ 1, mutation to new genotypes is much rarer than loss of segregating variation by genetic drift. In both A and B, when a major genetic or environmental upheaval significantly rewires the structure of the neutral network, then the preexisting genetic variation indicated by the variety of circles is revealed. The population shown in A has the maximum adaptive canalization, with the distribution of genotype frequencies optimized rather than random. The population shown in B has a lower level of adaptive canalization, and the population in C has no adaptive canalization. Note that in more realistic scenarios than shown in this schematic, the number of possible sequences may exceed the population size, and so the predicted optimal distribution, such as that shown in A, cannot be realized (Jenkins et al. 2001). Canalization may still be effectively optimized in these cases, however (van Nimwegen et al. 1999; Forster et al. 2006).
In the framework of neutral network theory, mutational load is given by the number of individuals falling off the neutral network due to mutation. Genetic canalization is a function of the position of the cloud of genotypes on the neutral network. Adaptive canalization occurs when selection moves the cloud toward the center of the network: this reduces the mutational load for a given mutation rate by reducing the probability that a single mutation will lead off the network. Neutral networks also exhibit “neutral” canalization, which is an intrinsic by-product of the genotype–phenotype map, in which not all mutations affect the phenotype. For example, the redundancy of the genetic code shows neutral canalization, while codon bias toward mutationally robust codons might be caused by adaptive canalization (Archetti 2006). These two scenarios are illustrated in Figure 1, A and C, respectively. Observations such as the fact that mutants are more variable than wild-type individuals (Waddington 1942) or that experimental evolution becomes progressively more difficult as a character approaches the adaptive peak (Scharloo 1991) can potentially be explained by neutral canalization alone, leaving it an open question as to whether adaptive genetic canalization occurs.
Theoretical models have been used to predict the conditions under which adaptive genetic canalization is expected. In particular, a cloud of genotypes will shift toward its optimal distribution on a neutral network if and only if mN > 1, where m is the total rate of mutations either staying on or falling off the network and N is the population size (van Nimwegen et al. 1999).
Reinterpreting the results of Hermisson and Wagner (2004) in the context of neutral network theory, variation can be revealed when a set of genotypes occupy a range of locations on the neutral network, as shown in Figure 1, A and B. This is a distinct concept from adaptive canalization, which refers to the shift of the set of genotypes toward their optimal distribution shown in Figure 1A. A major mutation can be interpreted as an “earthquake” that rearranges the neutral networks, effectively rewiring them and moving the current cloud of genotypes to another random location on the new landscape. If only neutral canalization were originally present, then the cloud would move from one random location to another random location, and on average no change in canalization would occur. If, on the other hand, the original system were adaptively genetically canalized, then movement would be from a central, nonrandom position to a random position, and hence a decrease in canalization would occur.
According to the theory of neutral networks, adaptive canalization is optimized when mN ≫ 1 (van Nimwegen et al. 1999), as shown in Figure 1A. When mN ≪ 1, neither adaptive canalization nor segregating variation are present in the system (van Nimwegen et al. 1999), as shown in Figure 1C. For mN not much larger than one, both a limited degree of segregating variation and a limited, nonoptimal degree of canalization are predicted (van Nimwegen et al. 1999), as shown in Figure 1B. Note that adaptive canalization and segregating variation are present under identical conditions on mN. This means that although “shifting the rug” and “shrinking the rug” can be separated conceptually and mathematically (Hermisson and Wagner 2004), in practice they will always occur together, whenever mN > 1.
The product mN is clearly critical to understanding genetic canalization, but both m and N are difficult to estimate in practice. RNA viruses are known to have high values for both m and N, and in agreement with this, genetic canalization has been observed for RNA viruses (Montville et al. 2005). Genetic canalization has also been inferred from microRNA sequences in a range of eukaryotes with smaller effective population sizes (Borenstein and Ruppin 2006). It is, however, difficult to rule out selection for environmental canalization (e.g., thermodynamic stability) rather than direct selection for genetic canalization as the cause of observed genetic canalization (Wagner et al. 1997; Ancel and Fontana 2000; Meiklejohn and Hartl 2002).
Since complex biological systems are highly modular, in general the mutation rate m should be interpreted as the mutation rate of one module, rather than of the genome as a whole. Here we estimate the rate of mutations leading to deterioration in sporulation ability in Bacillus subtilis to estimate the strength of selection for genetic canalization.
Spores were isolated after 6000 generations of experimental evolution in which selection for growth was interrupted by 892 episodes during which sporulation in inducing medium was necessary for survival (Maughan and Nicholson 2004). These evolved spores were diluted and plated on sporulation-inducing medium (Schaeffer et al. 1965) and 7244 colonies were counted after ∼16 hr of growth. During sporulation, wild-type colonies produce a brown pigment while sporulation mutants do not (Piggot and Coote 1976), resulting in a relatively simple screen for mutant colonies deficient in sporulation. To determine the number of sporulation mutants, the 7244 colonies were examined under a dissecting microscope ∼3–4 days after plating (which allows ample time for sporulation to be completed) to look for complete lack of pigment production throughout the colony, indicating that a mutation affecting sporulation ability had occurred in the first one or two generations of growth. The 3- to 4-day incubation period was also important for ensuring that sporulation phenotypes have a genetic basis and are not environmentally induced. This is because all wild-type colonies will sporulate after 3–4 days, regardless of minor environmental fluctuations.
A total of 44 colonies lacked the pigment, suggesting that they were sporulation mutants, and were examined using phase-contrast microscopy to determine the proportion of spores vs. vegetative cells within the colony. Of the 44 colonies that lacked pigment, 12 were reduced (<50% spores) and 10 were severely reduced (<1–2% spores) in the number of spores produced within the colony when compared to a wild-type colony (∼70–80% spores). These experimental values were used to calculate the total functional mutation rate as 22/7244 = 0.003. This functional mutation rate represents the frequency of individuals falling off the sporulation neutral network due to mutation.
Note that the population used to measure the functional mutation rate had spontaneously acquired a mutator phenotype over the course of experimental evolution. Using fluctuation tests, we measured the per base pair/per generation point mutation rate in this population to be 3.73 × 10−8, which is ∼10 times greater than that of the ancestral B. subtilis strain (Maughan et al. 2006).
Using this point mutation rate, we can also calculate a second, indirect estimate of the functional mutation rate using the following logic. It is known that at least 210 genes are involved in making a functional spore (Piggot and Losick 2002). The average size of a bacterial gene is 1000 bp, and thus there is a mutational target of ∼210,000 bp. Of course, not all of these sites, when mutated, would be deleterious to sporulation ability. If we assume that one-third of sites, when mutated, are deleterious to some aspect of sporulation, then we have a 70,000-bp target. Multiplying this by the point mutation rate, 3.73 × 10−8, gives a functional mutation rate of 0.0026, a value remarkably close to the experimentally determined value of 0.003. Note that both are most likely underestimates: the direct measure includes only mutants unable to form a spore, and not cells mutant for the ability to germinate and for spore characteristics, such as environmental resistance properties, while the indirect estimate neglects indels and other nonpoint mutations, and also potentially underestimates the number of genes. On the other hand, note that we have measured the point mutation rate during a cycle that encompasses germination, growth, sporulation, and storage portions of the life cycle, rather than the mutation rate during a single generation of normal growth and division. It is possible that mutations also occurred during germination and sporulation of this strain, making our direct measure an overestimate.
We have measured only those mutations that fall off the neutral network. The mutation rate m relevant to the neutral network model includes mutations that stay on the neutral network (van Nimwegen et al. 1999), and therefore must be higher. Our measure is of the genetic load L resulting from mutational trait loss and serves as a lower bound on m.
The structure of the neutral network is normally considered fixed over the course of evolution, although the precise manifestation of the genetic architecture, such as the epistatic effects of particular mutations, may depend on the spread of genotypes within the neutral network at a particular point in time. In an alternative view, genetic canalization may evolve in a more radical sense by providing sufficient mutational pressure to favor a completely new genetic architecture, for example, through gene duplication (Proulx and Phillips 2005). The maximum strength of selection for this to occur is equal to the mutation load L (Proulx and Phillips 2005), and so, by standard population genetic reasoning, this form of genetic canalization requires LN > 1 rather than mN > 1. Note also that for the specific neutral network simulated by van Nimwegen et al. (1999), mN ≥ 500 was found to be necessary and sufficient for optimal canalization, although lower levels of adaptive canalization were seen in the full range mN > 1. It remains possible, however, that larger and/or more realistic neutral networks may have larger cutoffs and that LN > 1 may be a more reliable condition for adaptive canalization to occur.
To estimate the effective population size, Ne, overnight growth densities, dilution ratios, and the number of generations of bacterial growth per day were measured at various time points during the 6000-generation evolution experiment (Maughan et al. 2006). These data were inserted into equation 15 from Wahl and Gerrish (2001), which takes into account fluctuations in population size, giving us Ne = 3 × 107.
Taking the product of the experimentally measured functional mutation rate and effective population size, we see that 105 = LNe < mNe in this mutator population. There are of course inaccuracies in this estimation: in particular, Ne is an overestimate since it neglects the effect of background selection and selective sweeps, and L is an underestimate, since it includes only mutants unable to form a spore, and not cells mutant for spore resistance characteristics and the ability to germinate. Nevertheless, this value is easily high enough, allowing a wide margin of error, for there to be substantial selection for higher canalization (van Nimwegen et al. 1999; Proulx and Phillips 2005). This is particularly true, given that the population has been under extremely strong selection for sporulation ability; i.e., cells that did not sporulate once every seven generations would die, for a total of 892 rounds of strong selection for 100% sporulation (Maughan and Nicholson 2004).
Although it has been found that certain aspects of the sporulation phenotype are evolvable (Voigt et al. 2005), previous work has found no evidence for an increased frequency of sporulation after 6000 generations of strong selection for it (Maughan and Nicholson 2004). Taken together, these results suggest that not only did the frequency of sporulation not increase, but also the mutational load associated with loss of sporulation ability did not evolve to attain the lower level of LN ∼1 that one would expect.
These observations suggest that the genetic architecture is already constrained and unable to canalize further. Another way of looking at this is that the distribution of genotypes on the neutral network has already reached the optimum, corresponding to that found in an infinite population (van Nimwegen et al. 1999). Sporulation might already be adaptively canalized to some extent, but whether or not this is the case, constraint prevents further canalization, despite the fact that selection for further canalization exists. This implies that the system has been unable to “add nodes” or to change by other means the underlying genetic architecture of the neutral network over the course of 6000 generations of strong selection. Note that even in nonmutator strains, we still expect LNe = 104, and hence are still in the parameter range in which further genetic canalization is expected.
In conclusion, we have found a strikingly high functional mutation rate leading to loss of sporulation ability despite 6000 generations of strong selection for sporulation. This shows that B. subtilis are in the parameter range LN ≫ 1 where adaptive genetic canalization of sporulation is expected, but suggests that they have reached a limit that genetically constrains further evolution of genetic canalization. Whether such genetic constraints, which likely vary for different phenotypes in different organisms, commonly prevent the further evolution of genetic canalization in other systems remains to be determined.
Acknowledgments
We thank Günter Wagner and Joachim Hermisson for helpful discussions. J.M. was supported by the BIO5 Institute at the University of Arizona and NIH grant GM076041. H.M. was supported by funds from the Department of Ecology and Evolutionary Biology at the University of Arizona.
References
- Ancel, L. W., and W. Fontana, 2000. Plasticity, evolvability, and modularity in RNA. J. Exp. Zool. 288: 242–283. [DOI] [PubMed] [Google Scholar]
- Archetti, M., 2006. Genetic robustness and selection at the protein level for synonymous codons. J. Evol. Biol. 19: 353–365. [DOI] [PubMed] [Google Scholar]
- Bateman, K. G., 1959. The genetic assimilation of four venation phenocopies. J. Genet. 56: 443–474. [DOI] [PubMed] [Google Scholar]
- Borenstein, E., and E. Ruppin, 2006. Direct evolution of genetic robustness in microRNA. Proc. Natl. Acad. Sci. USA 103: 6593–6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin, I., A. Palsson, K. Birdsall and G. Gibson, 2003. Evidence that Egfr contributes to cryptic genetic variation for photoreceptor determination in natural populations of Drosophila melanogaster. Curr. Biol. 13: 1888–1893. [DOI] [PubMed] [Google Scholar]
- Eigen, M., J. McCaskill and P. Schuster, 1989. The molecular quasi-species. Adv. Chem. Phys. 75: 149–263. [Google Scholar]
- Eshel, I., and C. Matessi, 1998. Canalization, genetic assimilation and preadaptation: a quantitative genetic model. Genetics 149: 2119–2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forster, R., C. Adami and C. O. Wilke, 2006. Selection for mutational robustness in finite populations. J. Theor. Biol. 243: 181. [DOI] [PubMed] [Google Scholar]
- Hermisson, J., and G. P. Wagner, 2004. The population genetic theory of hidden variation and genetic robustness. Genetics 168: 2271–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins, G. M., M. Worobey, C. H. Woelk and E. C. Holmes, 2001. Evidence for the non-quasispecies evolution of RNA viruses. Mol. Biol. Evol. 18: 987–994. [DOI] [PubMed] [Google Scholar]
- Maughan, H., and W. L. Nicholson, 2004. Stochastic processes influence stationary-phase decisions in Bacillus subtilis. J. Bacteriol. 186: 2212–2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maughan, H., V. Callicotte, A. Hancock, C. W. Birky, W. L. Nicholson et al., 2006. The population genetics of phenotypic deterioration in experimental populations of Bacillus subtilis. Evolution 60: 686–695. [PubMed] [Google Scholar]
- Meiklejohn, C. D., and D. L. Hartl, 2002. A single mode of canalization. Trends Ecol. Evol. 17: 468–473. [Google Scholar]
- Montville, R., R. Froissart, S. K. Remold, O. Tenaillon and P. E. Turner, 2005. Evolution of mutational robustness in an RNA virus. PLoS Biol. 3: 1939–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot, P. J., and J. G. Coote, 1976. Genetic aspects of bacterial endospore formation. Bacteriol. Rev. 40: 908–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piggot, P. J., and R. Losick, 2002. Sporulation genes and intercompartmental regulation, pp. 483–517 in Bacillus subtilis and Its Closest Relatives: From Genes to Cells, edited by A. L. Sonenshien, J. A. Hoch and R. Losick. American Society of Microbiology, Washington, DC.
- Proulx, S. R., and P. C. Phillips, 2005. The opportunity for canalization and the evolution of genetic networks. Am. Nat. 165: 147–162. [DOI] [PubMed] [Google Scholar]
- Queitsch, C., T. A. Sangster and S. Lindquist, 2002. Hsp90 as a capacitor of phenotypic variation. Nature 417: 618–624. [DOI] [PubMed] [Google Scholar]
- Rice, S. H., 1998. The evolution of canalization and the breaking of von Baer's laws: modeling the evolution of development with epistasis. Evolution 52: 647–656. [DOI] [PubMed] [Google Scholar]
- Rice, S. H., 2002. A general population genetic theory for the evolution of developmental interactions. Proc. Natl. Acad. Sci. USA 99: 15518–15523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutherford, S. L., and S. Lindquist, 1998. Hsp90 as a capacitor for morphological evolution. Nature 396: 336–342. [DOI] [PubMed] [Google Scholar]
- Schaeffer, P., J. Millet and J. P. Aubert, 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. USA 54: 704–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharloo, W., 1991. Canalization: genetic and developmental aspects. Annu. Rev. Ecol. Syst. 22: 65–93. [Google Scholar]
- Schuster, P., and J. Swetina, 1988. Stationary mutant distributions and evolutionary optimization. Bull. Math. Biol. 50: 635. [DOI] [PubMed] [Google Scholar]
- True, H. L., and S. L. Lindquist, 2000. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature 407: 477–483. [DOI] [PubMed] [Google Scholar]
- van Nimwegen, E., J. P. Crutchfield and M. Huynen, 1999. Neutral evolution of mutational robustness. Proc. Natl. Acad. Sci. USA 96: 9716–9720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt, C. A., D. M. Wolf and A. P. Arkin, 2005. The Bacillus subtilis sin operon: an evolvable network motif. Genetics 169: 1187–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington, C. H., 1942. Canalization of development and the inheritance of acquired characters. Nature 150: 563–565. [Google Scholar]
- Wagner, A., 1996. Does evolutionary plasticity evolve? Evolution 50: 1008–1023. [DOI] [PubMed] [Google Scholar]
- Wagner, G. P., G. Booth and H. Bagheri-Chaichian, 1997. A population genetic theory of canalization. Evolution 51: 329–347. [DOI] [PubMed] [Google Scholar]
- Wahl, L. M., and P. J. Gerrish, 2001. The probability that beneficial mutations are lost in populations with periodic bottlenecks. Evolution 55: 2606–2610. [DOI] [PubMed] [Google Scholar]
- Wilke, C. O., J. L. Wang, C. Ofria, R. E. Lenski and C. Adami, 2001. Evolution of digital organisms at high mutation rates leads to survival of the flattest. Nature 412: 331–333. [DOI] [PubMed] [Google Scholar]