Abstract
The obligate mutualism between yuccas and yucca moths is a major model system for the study of coevolving species interactions. Exploration of the processes that have generated current diversity and associations within this mutualism requires robust phylogenies and timelines for both moths and yuccas. Here we establish a molecular clock for the moths based on mtDNA and use it to estimate the time of major life history events within the yucca moths. Colonization of yuccas had occurred by 41.5 ± 9.8 million years ago (Mya), with rapid life history diversification and the emergence of pollinators within 0–6 My after yucca colonization. A subsequent burst of diversification 3.2 ± 1.8 Mya coincided with evolution of arid habitats in western North America. Derived nonpollinating cheater yucca moths evolved 1.26 ± 0.96 Mya. The estimated age of the moths far predates the host fossil record, but is consistent with suggested host age based on paleobotanical, climatological, biogeographical, and geological data, and a tentative estimation from an rbcL-based molecular clock for yuccas. The moth data are used to establish three alternative scenarios of how the moths and plants have coevolved. They yield specific predictions that can be tested once a robust plant phylogeny becomes available.
Obligate pollination mutualisms such as the yucca-yucca moth and fig-fig wasp associations provide some of the classically cited examples of coevolution (1, 2). In these interactions, the adult insects serve as the exclusive pollinators of their hosts. Their larvae subsequently feed on host seeds, but because many seeds are left intact the interaction carries a net positive effect for the plants. Both plants and insects show obvious coadapted traits, such as specific pollen collection and deposition behaviors in the insects, and structural adaptations that mediate pollinator specificity in the flowers.
Given the strong associations between these organisms, they serve as excellent model systems for exploring many aspects of evolutionary biology, such as the origins of mutualism (3–7), the evolution of virulence (8), evolution of mating systems (9), stability and reversal of obligate mutualism (5, 7, 10, 11), and consequences of specialization on population structure in species (12). Because of this utility for many branches of evolutionary biology, it would be highly desirable to develop a strong phylogenetic framework for the organisms and to establish a timeline for their diversification. This framework would allow for macroevolutionary analyses of the insect-host association, assessing for example the importance of codiversification (13), and the role of intrinsic and extrinsic factors in driving diversification (14).
Here we use molecular data in conjunction with biogeographic and fossil data to develop a phylogeny for the yucca moth family, Prodoxidae, to estimate minimum ages of divergence and determine patterns of diversification within the family. This work builds on an earlier study (15) based on limited molecular data that suggested variable rates of diversification among prodoxid genera. The present results support the following conclusions: (i) an explosive radiation of the yucca moths and evolution of pollination behavior had occurred no later than 40 million years ago (Mya), (ii) a second burst of yucca moth diversification is coincident with Pliocene desertification, (iii) derived nonpollinating cheater yucca moths have co-occurred with pollinators for at least 1.26 ± 0.96 My, and (iv) the radiation of yuccas occurred much earlier than suggested by the sparse fossil record. The emerging picture of early diversification in this obligate mutualism is consistent with a model of pollinator colonization of partly diversified host plant taxa, rather than strictly congruent diversification.
MATERIALS AND METHODS
Phylogenetic reconstruction was based on a 2.1-kb stretch of mtDNA, including most of the cytochrome oxidase I and II subunits and an intervening tRNAlys region (positions 1495–3603 in the Drosophila yakuba genome; ref. 16). The region was PCR-amplified, and both strands were sequenced either manually with Sequenase T7 polymerase or with Amersham Dye-terminator chemistry on an Applied Biosystems 377 automated sequencer following the manufacturer’s protocols (Amersham Pharmacia). All samples used for the analyses are listed in Table 1. For Prodoxidae, all but one genus were included, with a sufficient number of taxa for each genus to provide a stable topology. The unavailable genus Prodoxides from South America, known only from the type series, is basal or near-basal in the family (17), and its omission does not affect the current analyses. For the pollinating yucca moths and the derived nonpollinating cheater yucca moths, all taxa used in ref. 11 were included, resulting in representatives of all major life history categories. Outgroup taxa included single species from the three incurvarioid families (Adelidae, Heliozelidae, Cecidosidae) and four species from the Incurvariidae.
Table 1.
Samples used for the analyses
| Species | Locality | GenBank accession numbers |
|---|---|---|
| Adelidae | ||
| Adela trigrapha Powell | USA: Tulare Co, CA | U04880 |
| Heliozelidae | ||
| Coptodisca kalmiella Dietz | USA: Litchfield Co, CT | AF150907 |
| Incurvariidae | ||
| Perthida glyphopa Common | Australia: Perth, WA | AF150908 |
| Vespina quercivora (Davis) | USA: Kern Co, CA | AF150925 |
| Incurvaria masculella (Denis & Schiff) | Sweden: Prov. Småland | AF150926 |
| Paraclemensia acerifoliella (Fitch) | Canada: Ontario, Carleton Co. | AF150927 |
| Cecidosidae | ||
| Cecidoses eremita Curtis | Argentina: Prov. Neuquen | U04881 |
| Prodoxidae | ||
| Greya variabilis Davis & Pellmyr | USA: Clallam Co, WA | AF150909 |
| G. politella (Walsingham) | USA: San Juan Co, CO | U49021 |
| G. solenobiella (Walsingham) | USA: Monterrey Co, CA | AF150910 |
| G. punctiferella (Walsingham) | USA: Clallam Co, WA | AF150911 |
| Lampronia aenescens (Walsingham) | USA: Garfield Co, WA | AF150912 |
| Tetragma gei Davis & Pellmyr | USA: Asotin Co, WA | AF150913 |
| Mesepiola specca Davis | USA: Santa Cruz Co, AZ | U49022 |
| Mesepiola n. sp. | USA: Pima Co, AZ | AF150914 |
| Prodoxus aenescens Riley | USA: Tulare Co, CA | AF150915 |
| P. marginatus Riley | USA: Tulare Co, CA | AF150916 |
| P. coloradensis Riley | USA: Mohave Co, AZ | AF150917 |
| P. y-inversus Riley | USA: Clark Co, NV | AF150918 |
| P. (Agavenema) pallida Davis | USA: Riverside Co, CA | AF150919 |
| P. (Agavenema) n. sp. | USA: San Bernardino Co, CA | AF150920 |
| Parategeticula pollenifera Davis | USA: Santa Cruz Co, AZ | AF150921 |
| P. n. sp. 1 | Mexico: Est. Veracruz | AF150922 |
| P. n. sp. 2 | Mexico: Est. Coahuila | AF150923 |
| P. n. sp. 3 | Mexico: Est. Coahuila | AF150924 |
| Tegeticula m. maculata (Riley) | USA: Tulare Co, CA | U49024 |
| T. m. extranea (Edwards) | USA: Riverside Co, CA | U49023 |
| T. synthetica (Riley) | USA: Clark Co, NV | U49025 |
| T. yuccasella (Riley) ex. Y. torreyi | USA: Brewster Co, TX | U49041 |
| T. yuccasella ex. Y. schidigera | USA: San Bernardino Co, CA | U49039 |
| T. yuccasella ex. Y. baccata | USA: Dona Ana Co, NM | U49026 |
| T. yuccasella ex. Y. intermedia | USA: Torrance Co, NM | U49037 |
| T. yuccasella ex. Y. utahensis | USA: Washington Co, UT | U49042 |
| T. yuccasella ex. Y. elata | USA: Cochise Co, AZ | U49028 |
| T. yuccasella ex. Y. elata (late cheater) | USA: Cochise Co, AZ | U49027 |
| T. yuccasella ex. Y. glauca (late cheater) | USA: Crook Co, WY | U49035 |
| T. yuccasella ex. Y. schidigera (late cheater) | USA: Riverside Co, CA | U49034 |
| T. yuccasella ex. Y. torreyi (late cheater) | USA: Brewster Co, TX | U49049 |
| T. yuccasella ex. Y. filamentosa (early cheater) | USA: Wilson Co, TN | U49039 |
| T. yuccasella ex. Y. glauca (early cheater) | USA: Meade Co, KS | U49033 |
| T. yuccasella ex. Y. intermedia (early cheater) | USA: Valencia Co, NM | U49036 |
| T. yuccasella ex. Y. filamentosa | USA: Wilson Co, TN | U49032 |
| T. yuccasella ex. Y. glauca | USA: Meade Co, KS | U49043 |
Locality and GenBank accession is given for each taxon.
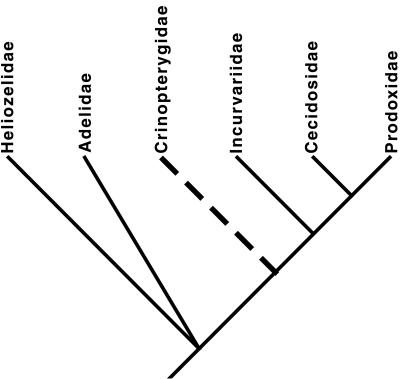
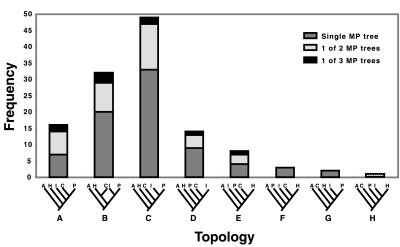
Relationships of the incurvarioid families have been established from morphological studies (17) (Fig. 1). Initial parsimony, neighbor-joining, and likelihood analyses of the complete COI-COII data set supported this phylogeny, except that the single available cecidosid, Cecidoses eremita, clustered with the basal Heliozelidae in all analyses. This finding also held true in an analysis based on amino acids for the COI and COII regions of the data. We hypothesized that this result was caused by an accelerated substitution rate in these lineages, leading to long branch attraction. This hypothesis was tested by using a parametric bootstrap procedure (18). Parameters for a five-taxon tree were estimated by assuming the topology in Fig. 1 and used as input data for the simulation program seq-gen, version 1.1 (19). After removing all third codon sites, maximum-likelihood estimates of base frequencies, transition bias (Tr/Ti), the distribution of substitution rates (approximation of the gamma parameter assuming four rate categories), and branch lengths were calculated on a tree constrained to have the topology presented in Fig. 1. Average stem node-to-tip branch lengths were estimated for Incurvariidae and Prodoxidae, giving single length estimates for each branch in the constraint tree (Fig. 2). By using the estimated parameters, 100 five-taxon data sets with sequence lengths equal to the analyzed data (1,182 bp) were generated under the evolutionary model of Hasegawa et al. (20) and Yang (21) and analyzed by using the unweighted parsimony algorithm in paup* (22). When the resulting most-parsimonious (MP) trees were tallied by topology, it became clear that the long branch leading to C. eremita was in fact causing problems. The true input topology was recovered only from 16% of the simulated data sets (Fig. 3). For this reason, we removed the C. eremita sequence from further analyses.
Figure 1.
Proposed phylogeny for the families of Incurvarioidea, based on morphological data (17). Dashed line indicates that taxon was unavailable for DNA analysis, and placement is based on morphology alone.
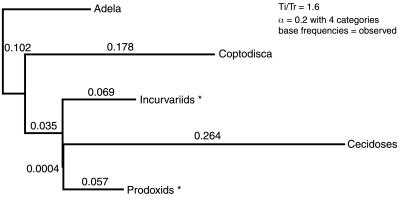
Figure 2.
Input parameters (topology, branch lengths, transition bias, and gamma parameter) used to generate simulated sequence data sets for the parametric bootstrap analysis. Branch lengths leading to prodoxids and incurvariids are averages for all sampled (see text).
Figure 3.
Frequency of parametric bootstrap replicates from which each topology was recovered as a single MP tree, one of two MP trees, or one of three MP trees. The true topology (lane A) was only recovered from 10 of the 100 simulated data sets. A = Adelidae, H = Heliozelidae, I = Incurvariidae, C = Cecidosidae, P = Prodoxidae.
After removing the Cecidoses sequence, topologies were estimated with paup* (22) by using unweighted parsimony, maximum likelihood, and neighbor joining. The maximum-likelihood and neighbor-joining trees were estimated assuming the nucleotide substitution model of Hasegawa et al. (20). Bootstrap support values for each node were estimated in the parsimony and neighbor-joining analyses. Given the low level of sequence divergence among members of the Tegeticula yuccasella complex, all codon positions were used for initial estimates of the topology.
Likelihood ratio tests were used to establish the best-supported model of molecular evolution for estimating minimum ages for the key life history traits. Rate heterogeneity across sites and among branches was tested. The substitution model of Hasegawa et al. (20) was assumed. As with the simulation experiment, third codon positions were dropped from the analysis to avoid the effects of substantial saturation. Standard likelihood ratio tests were applied in which the null hypothesis is a special case of the alternative hypothesis (23, 24). For example, the model that specifies a single substitution rate for all branches in the phylogeny is a special case of the model that estimates a separate rate parameter for each branch. In such cases, where the null hypothesis is nested within the alternative, two times the negative log of the likelihood ratio statistic:
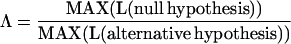 |
approximates the χ2 distribution with the degrees of freedom equal to the difference in the number of parameters estimated under the null and alternative hypotheses (24).
Finally, boundary ages were estimated for key events in the evolutionary history within the Prodoxidae. The topology shown in Fig. 4 was assumed. At each node, we used the two-cluster test of Takezaki et al. (26) to test the null hypothesis of equal substitution rates for the two clades above. When the null hypothesis was not rejected, the “height” of the node was estimated as half the average genetic distance between pairs of taxa sampled from the separate clades above the node (26). The relative age of the node then was estimated as quotient of the height of node of interest divided by the height of the reference node that defines the split between Incurvariidae and Prodoxidae+Cecidosidae (Fig. 1). Standard errors on the age estimates were estimated from the variance in this ratio (27). Covariances between node heights were estimated as described by Ayala et al. (28). Finally, the minimum age of each node was estimated as the relative age multiplied by 95 Mya, the minimum age of the split between the Incurvariidae and Prodoxidae+Cecidosidae clades.
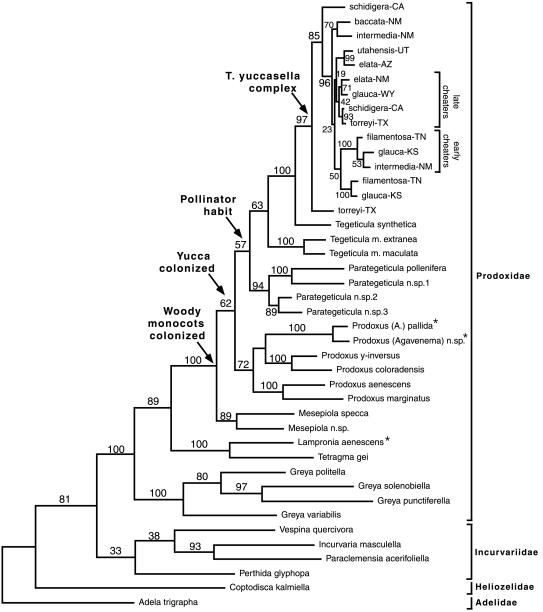
Figure 4.
Phylogeny for Prodoxidae and three outgroup families; shown is one of 10 MP trees, with neighbor-joining and maximum-likelihood trees having identical topology. Numbers are bootstrap values based on 100 iterations. Moth families are given on right; names for unnamed taxa within the yuccasella complex are those given in ref. 11 and are based on host yucca and state (all of which are described in ref. 25). The terms early and late cheaters reflect use in ref. 11 and refer to the host phenological stage attacked. Asterisks identify taxa that were excluded in age estimations caused by significant rate deviations in two-cluster tests. Arrows indicate latest origin of four named traits to be explored.
RESULTS AND DISCUSSION
Establishment of Topology.
Phylogenetic analysis of the entire COI-COII data set resulted in 10 MP trees that differed only in the placement of taxa within the yuccasella complex. One of the 10 MP trees was recovered in the neighbor-joining analysis of the full data set, assuming no rate variation among sites (Fig. 4). The maximum-likelihood tree recovered under the single rate model differed in that Vespina was pulled out of the Incurvariidae clade and placed below it. Both the maximum likelihood and MP/neighbor joining trees are consistent with an earlier phylogeny (15), including far fewer taxa, except that the basal genera within Prodoxidae (Greya and Lampronia+Tetragma) have switched positions. This difference holds up in all analyses of the current data. More importantly, inferences concerning yucca moth diversification are unaffected by alternative topological resolutions of the basal prodoxid genera.
Testing for Rate Variation Among Sites and Branches.
Under the assumption that the tree in Fig. 4 represents the true topology, the maximum-likelihood estimate of the gamma-distribution parameter was 0.18. The likelihood for the model including rate heterogeneity was significantly better than that of the single rate model (−2 log Λ = 3,406, P ≪ 0.01). The estimated value of gamma was very robust, remaining unchanged given alternative resolution of poorly supported nodes within the topology. The observed rate heterogeneity across sites was accounted for in the age estimations.
The likelihood ratio test for rate variation among branches was run, assuming the topology shown in Fig. 4 and rate variation among sites. The molecular clock assumption (no rate variation among branches) was rejected (−2 log Λ = 134, P = ≪ 0.01). Given this result, we turned to the distance-based tests of Takezaki et al. (26). The two-cluster test identified rate variation between clades at two nodes. The node-to-tip test identified taxa within these nodes as having significantly higher (two Prodoxus species) or lower (Lampronia) than average substitution rates. Therefore, these three taxa were not included in the age estimation analyses.
Age Estimation Within the Prodoxidae.
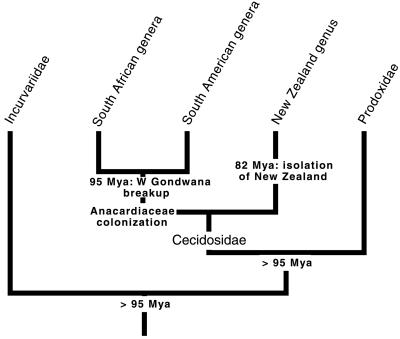
We proceeded to estimate ages for nodes of particular significance in prodoxid life history evolution. Both biogeographic and fossil data support a minimum age of 95 My for the prodoxid stemgroup (Fig. 5). Cecidosid moths combine a classical Gondwanan distribution with very low dispersal ability (29). Six genera within the family are gall-makers on Anacardiaceae, and they make up separate monophyletic groups in South America and southern Africa (30). Their shared host utilization suggests an origin before breakup of West Gondwana, which was definitive for all but very able dispersers 95–100 Mya (31). This date substantially predates the proposed 70-My age of the Anacardiaceae (32, 33), but is consistent with other biogeographic evidence (34). Further support for old age of the Cecidosidae is a recently discovered, unnamed genus (35) endemic to New Zealand, now regarded as basal within the family (R. Hoare and E.S. Nielsen, personal communication), indicating family origin before the isolation of New Zealand, approximately 82 Mya (36). Cecidosid diversification thus indicates a conservative age of >95 My for the prodoxid-cecidosid divergence. Because the Cecidosidae has evolved at a faster rate than the other families, however, it cannot be used for calibration. Instead we applied the 95-My estimate to the next lower node, between the Incurvariidae and Prodoxidae+ Cecidosidae clades.
Figure 5.
Phylogeny for Incurvariidae, Cecidosidae, and Prodoxidae, indicating key geological and biological events, and biogeographic benchmark dates used to infer minimum age of stemgroup Prodoxidae.
The fossil record is scant, but consistent with the age calculations based on biogeographic data. There are no prodoxid fossils, but 97-My-old leaf fossils with highly characteristic mines of the more derived group Ditrysia provide indirect evidence that the incurvarioid stemgroup was present at that time (37, 38), whereas a proposed 110- to 120-My-old incurvarioid wing (39) cannot be exclusively tied to the superfamily (40).
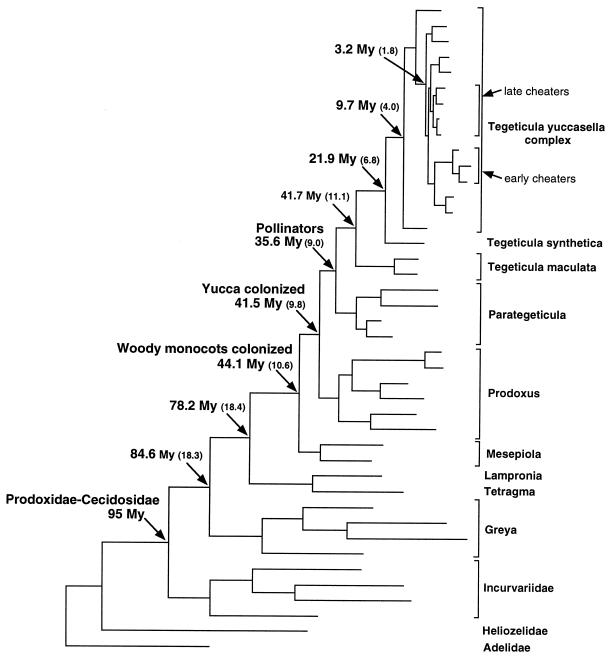
After age estimation for the family, we calculated minimum ages for nodes associated with major evolutionary events within the Prodoxidae (Fig. 6). Basal genera are confined to humid and semiarid habitats and feed on a variety of dicotyledonous families. Coincident with the invasion of arid habitats by Prodoxidae, woody monocots were colonized no later than 44.1 Mya, with extant members of the basal genus Mesepiola feeding on seeds of Nolinaceae. A very short internode to the Prodoxus+pollinator node indicates rapid colonization of the yuccas by 41.5 Mya. The pollinating genera Parategeticula and Tegeticula originated very closely in time 35–41 Mya. The estimated age of the node separating these two genera is younger than the basal node within Tegeticula, but not significantly so. In fact, all three genera feeding on Yucca arose so quickly that their ages overlap in the analysis. The distinct feeding and life history characteristics exhibited by Prodoxus, Parategeticula, and Tegeticula seem to have arisen in rapid succession shortly after the colonization of woody monocots (Fig. 6). Parsimonious trait reconstruction on the phylogeny places the colonization of woody monocots on the internode between the Lampronia+Tetragma and Mesepiola branches. and is consistent with a late burst of lineages quickly diversifying into all extant life histories. This reconstruction is consistent with the hypothesis that a burst of diversification occurred as the woody monocots were colonized, yielding all of the extant life histories observed in the monocot feeding prodoxids. Alternative scenarios of earlier colonization of the woody monocots require an early period of limited cladogenesis followed by explosive radiation, or consistent rate of cladogenesis with subsequent extinction of basal monocot feeders.
Figure 6.
Phylogeny for Prodoxidae as in Fig. 4, showing estimated minimum ages (± SE in parentheses) for select nodes. Taxa are given on the right, with nested brackets within the yuccasella complex indicating cheater yucca moth clades. Prodoxid hosts are given at far right.
After the early rapid diversification of prodoxids on Yucca, a subsequent explosive radiation occurred within the T. yuccasella complex 3.2 ± 1.8 Mya, giving rise to all pollinator lineages on capsular-fruited yuccas and the nonpollinating cheater yucca moths so quickly that the exact topology among major branches cannot be established from data presented here. However, additional data on nucleotide variation among host specific populations show definitively that there have been at least two independent losses of pollinating behavior in the T. yuccasella complex (unpublished work). By using maximum sequence divergence within the two independent cheater lineages, we find that two losses of pollination behavior in these two lineages is simultaneous given the coarse resolution of our age estimates. The early and late cheater lineages identified by Pellmyr et al. (11) arose 1.26 ± 0.96 and 1.26 ± 0.95 Mya, respectively.
Implications for Early Stages of the Mutualism.
The availability of a timeline for diversification of the yucca moths makes possible the erection of explicit predictions for patterns of host diversification under different models of how this plant-moth association has evolved (25, 41). Specific models that generate different predictions include strict cospeciation, synchronous diversification without cospeciation, and asynchronous diversification without cospeciation. In a strict cospeciation model, which is the most constrained model, parallel topologies are predicted between the yuccas and the moths, and diversification rates also should be synchronous. This is the predicted outcome if host shifts are very unlikely, and diversification strongly linked between the taxa. This model already can be rejected (4, 11). If moths can colonize new hosts, but still respond evolutionarily to host diversification, synchronous patterns of diversification are predicted for the moths and their hosts, but their topologies are not predicted to be congruent. In the least constrained model, where host switching is common and factors driving diversification of yuccas and yucca moths are unlinked, no correlations are expected in either relative rates or timing of plant and pollinator diversification.
Information about phylogeny and history of diversification of yuccas is still limited (42), preventing strong tests of the above predictions, but some inferences about the history of the association can be drawn. The estimated Eocene age of the association between yuccas and prodoxids provides indirect evidence of far older age of Yucca than previously documented. The macrofossil record (excluding Pleistocene subfossils; ref. 43) is limited to a 14-My-old vegetative fragment sharing one synapomorphy with extant Yucca (44). Applying a molecular clock to rbcL data, Eguiarte (45) proposed that Agavaceae and Nolinaceae diversified about 47 Mya, a date slightly older than that inferred here for basal diversification of prodoxids onto these families. Geologically, this period coincides with the onset of uplifting of western North America (46–48), which led to widespread development of semiarid habitats where extant yuccas are constituent taxa. Alternatively, Axelrod (49) speculated that Yucca may have evolved as early as late Cretaceous. Although a proposed Eocene origin would not exclude simultaneous diversification of moths and plants, a Cretaceous origin would indicate that early yuccas persisted without moths as pollinators, and that the yucca-yucca moth association originated as moths colonized extant hosts rather than through parallel diversification.
The second explosive radiation of yucca moths, within the yuccasella complex, occurred 3.2 ± 1.8 Mya. The moths derived from this radiation were primarily those of capsular-fruited yuccas, which occur in the northern portion of the Yucca geographic range. The onset of this radiation coincided with rapid aridification (50), leading to true deserts and treeless steppes, thus creating or extending the primary extant habitats for the capsular yuccas and their moth associates. There is currently neither fossil nor phylogenetic information for the history of Yucca within this Pliocene window. Rigorous tests of the predictions above must await a robust plant phylogeny.
Comparison with Other Obligate Mutualisms.
Obligate mutualisms have attracted particular interest in terms of their stability and endurance (10, 51). The presence of pollinating yucca moths for at least 40 My can be compared against available data for another obligate pollination mutualism, between fig wasps and figs. A fossil fig wasp of Oligocene age is known (52), but the presence of fig fruits from the Eocene onward (53) suggest that this mutualism has persisted as long as the yucca-yucca moth association. Furthermore, many associations between specific yuccas and yucca moths have persisted since the evolution of derived cheater yucca moths, which dramatically increased the cost to the plants (11), suggesting that these mutualisms retain evolutionary stability over long time spans.
Acknowledgments
We thank J. Donahue, T. Friedlander, M. Gentili, J.-F. Landry, R. Leuschner, Z. Mazanec, N. Ryrholm, and D. Wagner for moth samples and E.A. Herre for references to fossil fig wasps. Collecting permits were provided by Big Bend National Park and the Texas Department of Parks and Wildlife. Work was funded by National Science Foundation Grants 95–09056 and 95–28072 and a grant from the National Geographic Society.
ABBREVIATIONS
- Mya
million years ago
- MP
most parsimonious
Footnotes
References
- 1.Thompson J N. The Coevolutionary Process. Chicago: Univ. of Chicago; 1994. [Google Scholar]
- 2.Fleming T H, Holland J N. Oecologia. 1998;114:368–375. doi: 10.1007/s004420050459. [DOI] [PubMed] [Google Scholar]
- 3.Bronstein J L. Q Rev Biol. 1994;69:31–51. [Google Scholar]
- 4.Bogler D J, Neff J L, Simpson B B. Proc Natl Acad Sci USA. 1995;92:6864–6867. doi: 10.1073/pnas.92.15.6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pellmyr O, Huth C J. Nature (London) 1994;372:257–260. [Google Scholar]
- 6.Pellmyr O, Thompson J N, Brown J M, Harrison R G. Am Nat. 1996;148:827–847. [Google Scholar]
- 7.Richter K S, Weis A E. Nature (London) 1995;376:557–558. [Google Scholar]
- 8.Herre E A. Science. 1993;259:1442–1445. doi: 10.1126/science.259.5100.1442. [DOI] [PubMed] [Google Scholar]
- 9.Herre E A. Nature (London) 1987;329:627–629. [Google Scholar]
- 10.Bull J J, Rice W R. J Theor Biol. 1991;149:63–74. doi: 10.1016/s0022-5193(05)80072-4. [DOI] [PubMed] [Google Scholar]
- 11.Pellmyr O, Leebens-Mack J, Huth C J. Nature (London) 1996;380:155–156. doi: 10.1038/380155a0. [DOI] [PubMed] [Google Scholar]
- 12.Nason J D, Herre E A, Hamrick J L. Nature (London) 1998;391:685–687. [Google Scholar]
- 13.Farrell B D. Science. 1998;281:555–559. doi: 10.1126/science.281.5376.555. [DOI] [PubMed] [Google Scholar]
- 14.Becerra J X. Science. 1997;276:253–256. doi: 10.1126/science.276.5310.253. [DOI] [PubMed] [Google Scholar]
- 15.Brown J M, Pellmyr O, Thompson J N, Harrison R G. Ann Entomol Soc Am. 1994;87:795–802. [Google Scholar]
- 16.Clary D O, Wolstenholme D R. J Mol Evol. 1985;22:252–271. doi: 10.1007/BF02099755. [DOI] [PubMed] [Google Scholar]
- 17.Nielsen E S, Davis D R. Syst Entomol. 1985;10:307–322. [Google Scholar]
- 18.Huelsenbeck J P, Hillis D M, Jones R. In: Molecular Zoology: Advances, Strategies and Protocols. Ferris J D, Palumbi S R, editors. New York: Wiley; 1996. pp. 19–45. [Google Scholar]
- 19.Rambaut A, Grassly N C. Comput Appl Biosci. 1997;13:235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]
- 20.Hasegawa M, Kishino H, Yano T. J Mol Evol. 1985;21:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- 21.Yang Z. J Mol Evol. 1994;39:306–314. doi: 10.1007/BF00160154. [DOI] [PubMed] [Google Scholar]
- 22.Swofford D. paup* 4.0b2. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 23.Goldman N. J Mol Evol. 1993;36:182–198. doi: 10.1007/BF00166252. [DOI] [PubMed] [Google Scholar]
- 24.Huelsenbeck J P, Rannala B, Yang Z H. Evolution. 1997;51:410–419. doi: 10.1111/j.1558-5646.1997.tb02428.x. [DOI] [PubMed] [Google Scholar]
- 25.Pellmyr O. Syst Entomol. 1999;24:243–270. [Google Scholar]
- 26.Takezaki N, Rzhetsky A, Nei M. Mol Biol Evol. 1995;12:823–833. doi: 10.1093/oxfordjournals.molbev.a040259. [DOI] [PubMed] [Google Scholar]
- 27.Lynch M, Walsh B. Genetic Analysis of Quantitative Traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 28.Ayala F J, Rzhetsky A, Ayala F J. Proc Natl Acad Sci USA. 1998;95:606–611. doi: 10.1073/pnas.95.2.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Becker V O. Polsk P Entomol. 1977;47:79–86. [Google Scholar]
- 30.Nielsen E S. J. Res. Lepid. 1985. Suppl. 1, 1–16. [Google Scholar]
- 31.Pitman W S, III, Cande S, LaBrecque J, Pindell J. In: Biological Relationships Between Africa and South America. Goldblatt P, editor. New Haven: Yale Univ. Press; 1993. pp. 15–34. [Google Scholar]
- 32.Cronquist A. An Integrated System of Classification of the Flowering Plants. New York: Columbia Univ. Press; 1981. [Google Scholar]
- 33.Moran N A, Munson M A, Baumann P, Ishikawa H. Proc R Soc London B. 1993;253:167–171. [Google Scholar]
- 34.Raven P H, Axelrod D I. Ann Mo Bot Gard. 1974;61:539–673. [Google Scholar]
- 35.Dugdale J S. N Z J Zool. 1989;16:277–281. [Google Scholar]
- 36.Cooper R A, Millener P R. Trends Ecol Evol. 1993;8:429–433. doi: 10.1016/0169-5347(93)90004-9. [DOI] [PubMed] [Google Scholar]
- 37.Labandeira C C, Dilcher D L, Davis D R, Wagner D L. Proc Natl Acad Sci USA. 1994;91:12278–12282. doi: 10.1073/pnas.91.25.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kristensen N P. Mem Mus Nat Hist Nat. 1997;173:253–271. [Google Scholar]
- 39.Whalley P. Ann Transv Mus. 1977;31:70–89. [Google Scholar]
- 40.Kristensen N P, Skalski A W. In: Lepidoptera: Moths and Butterflies 1, Handbook of Zoology. Kristensen N P, editor. Berlin: de Gruyter; 1999. pp. 7–25. [Google Scholar]
- 41.Page R D M. Syst Biol. 1996;45:151–167. [Google Scholar]
- 42.Bogler D J, Simpson B B. Am J Bot. 1996;83:1225–1235. [Google Scholar]
- 43.Betancourt J L, Van Devender T R, Martin P S, editors. Packrat Middens: Late Quarternary Environments of the Arid West. Tucson: Univ. of Arizona Press; 1990. [Google Scholar]
- 44.Tidwell W D, Parker L R. Rev Palaeobot Palyn. 1990;62:79–95. [Google Scholar]
- 45.Eguiarte L E. Bol Soc Bot Méx. 1995;56:45–56. [Google Scholar]
- 46.Raymo M E, Ruddiman W F. Nature (London) 1992;359:117–122. [Google Scholar]
- 47.Wolfe J A, Schorn H E, Forest C E, Molnar P. Science. 1997;276:1672–1675. [Google Scholar]
- 48.Kerr R E. Science. 1997;275:1564–1565. [Google Scholar]
- 49.Axelrod D L. Occ Pap Calif Acad Sci. 1979;132:1–174. [Google Scholar]
- 50.Webb S D. Annu Rev Ecol Syst. 1977;8:355–380. [Google Scholar]
- 51.Addicott J F, Bronstein J, Kjellberg F. In: Genetics, Evolution, and Coordination of Insect Life Cycles. Gilbert F, editor. London: Springer; 1990. pp. 143–161. [Google Scholar]
- 52.Brues C T. Bull Mus Comp Zool. 1910;54:1–126. [Google Scholar]
- 53.Collinson M E. In: Evolution, Systematics, and Fossil History of the Hamamelidae. Crane P R, Blackmore S, editors. Vol. 2. Oxford: Clarendon; 1989. pp. 319–339. [Google Scholar]