Abstract
cDNA microarrays and a clustering algorithm were used to identify patterns of gene expression in human mammary epithelial cells growing in culture and in primary human breast tumors. Clusters of coexpressed genes identified through manipulations of mammary epithelial cells in vitro also showed consistent patterns of variation in expression among breast tumor samples. By using immunohistochemistry with antibodies against proteins encoded by a particular gene in a cluster, the identity of the cell type within the tumor specimen that contributed the observed gene expression pattern could be determined. Clusters of genes with coherent expression patterns in cultured cells and in the breast tumors samples could be related to specific features of biological variation among the samples. Two such clusters were found to have patterns that correlated with variation in cell proliferation rates and with activation of the IFN-regulated signal transduction pathway, respectively. Clusters of genes expressed by stromal cells and lymphocytes in the breast tumors also were identified in this analysis. These results support the feasibility and usefulness of this systematic approach to studying variation in gene expression patterns in human cancers as a means to dissect and classify solid tumors.
Many of the new genomic analysis tools offer great promise for classifications of tumors based on variations in gene expression. However, the study of gene expression in primary human breast tumors, as in most solid tumors, is complicated for two major reasons. First, breast tumors consist of many different cell types, including not just the carcinoma cells, but also additional epithelial cell types, stromal cells, adipose cells, endothelial cells, and infiltrating lymphocytes (1). Second, breast carcinoma (BC) cells themselves are morphologically and genetically diverse (2). These features have made the study and classification of human breast tumors difficult.
Recently, cDNA microarrays have been used to identify physiologically relevant gene expression patterns in simple biological samples like yeast cultures (3) and cultures of human fibroblasts (4). cDNA microarrays have been extensively described and simply consist of thousands of different cDNA clones spotted onto known locations on glass microscope slides (5–11); these slides/microarrays then are hybridized with differentially labeled cDNA populations made from the mRNAs of two different samples. The primary data obtained are ratios of fluorescence intensity (red/green, R/G) representing the ratio of concentrations of mRNA molecules that hybridized to each of the cDNAs represented on the array.
As a first step in using cDNA microarrays to identify physiologically relevant gene expression patterns in human breast tumors, in vitro experiments were performed by using specific hormones added to breast epithelial cell cultures. By subjecting cells to different conditions, it was possible to identify “clusters” of genes that showed similar patterns of expression by using the algorithms and software described by Eisen et al. (12). In a first attempt to study tumors, mRNA samples from 13 grossly dissected human breast tumors were compared to the mRNA from cultured human mammary epithelial cells (HMEC). Some of the clusters of genes with distinctive expression patterns identified in vitro also varied substantially in their expression in the breast tumor samples. For some of the clusters of coexpressed genes, expression in the tumor samples appeared to be attributable to other, noncarcinoma cell types, including stromal cells and B lymphocytes.
MATERIALS AND METHODS
cDNA Clones, Microarrays, and Data Analysis.
The 5,531 human cDNA microarrays used in this study were made in collaboration with Synteni, Inc. (now Incyte Pharmaceutical, Inc.). These microarrays were hybridized as described in ref. 4 and scanned and quantitated as described in ref. 3 by using the average of the lower 10% of the pixel intensities for the local background calculation. The cDNA clones used to make these microarrays represent a set of approximately 5,000 genes, which is part of a larger 15,000 gene/clone collection that has been described elsewhere (refs. 4 and 13 and see http://www.nhgri.nih.gov/DIR/LCG/15K/HTML/). Sequence confirmation of this clone set is not completed; but the identities of approximately 80% of the clones that were successfully resequenced were confirmed. Many clones are identified here by either a name or number, which is preceded by the designation SID (which stands for the Stanford identification number); these are clones for which satisfactory confirmation is still lacking. All clones/genes named in the text or in the figures (including I.M.A.G.E. expressed sequence tag numbers) were confirmed by resequencing. Any questions concerning cDNA clones/genes presented here should be directed to C.M.P. at perou@genome.stanford.edu.
Gene-clustering analysis was performed as described in ref. 12. For the cluster diagrams presented in Figs. 1 and 2, the input parameters were to select the subset of genes that had a R/G ratio of 3-fold or higher on at least two or more arrays (Fig. 1) or a R/G ratio of 3-fold or higher on at least three or more arrays (Fig. 2). The primary data tables and other materials are available at http://genome-www.stanford.edu/sbcmp/.
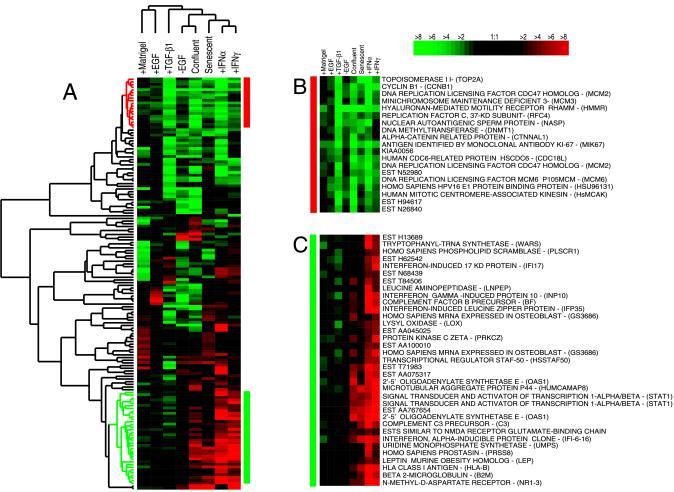
Figure 1.
(A) Cluster diagram of HMEC in vitro experiments. Each column represents a single experiment, and each row represents a single gene. Ratios of gene expression relative to HMEC control samples grown under standard conditions are shown. Green squares represent lower than control levels of gene expression in the experimental samples (ratios less than 1); black squares represent genes equally expressed (ratios near 1); red squares represent higher than control levels of gene expression (ratios greater than 1); gray squares indicate insufficient or missing data. The color saturation reflects the magnitude of the log/ratio [see scale at top right and Fig. 5 (see Supplemental data at www.pnas.org) for the full cluster diagram with all gene names]. (B) Expanded view of the subset of genes whose expression was decreased in association with reduced HMEC proliferation. (C) Expanded view of the IFN-regulated gene cluster. In many instances, multiple independent clones/cDNA representing the same gene were spotted on different locations on these microarrays, and in most cases, these copies usually clustered together, either very near each other or immediately adjacent to each other.
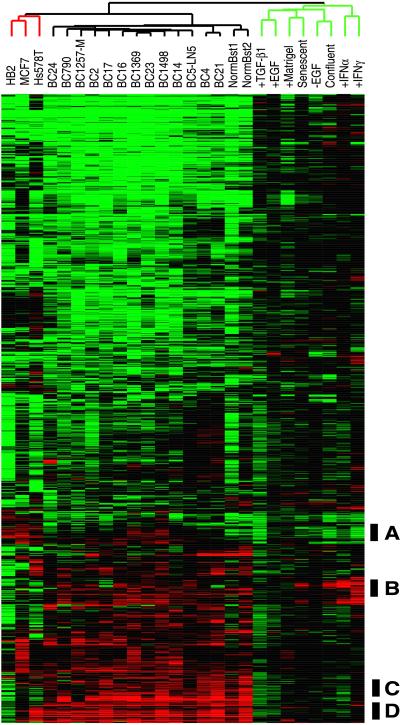
Figure 2.
Overview of the combined in vitro and breast tissue specimen cluster diagram. A scaled-down representation of the 1,247-gene cluster diagram (see Supplemental Fig. 6 at www.pnas.org for the full cluster diagram with all gene names). The black bars show the positions of the clusters discussed in the text: (A) proliferation-associated, (B) IFN-regulated, (C) B lymphocytes, and (D) stromal cells.
Cell Culture.
HMEC were obtained from Clonetics (San Diego)/BioWhittaker and grown in the recommended complete mammary epithelial growth medium. A single HMEC isolate was used for all studies. The control HMEC reference samples used were passage 9–14 cultures that were harvested at 60–80% confluence and had their medium changed 2 days before the mRNA harvest. To determine how much variation in gene expression there was in these independently prepared HMEC samples, two different passage HMEC control samples were compared on a 1,952-gene microarray (data not shown). Of the 1,952 genes analyzed, four genes showed a R/G ratio between 2.3 and 2.0, 14 genes showed a R/G ratio between 0.5 and 0.37, whereas all other genes tested that gave an appreciable signal fell into the R/G ratio range of 2 to 0.5 (2-fold difference or less).
The senescent HMEC cells were a passage-19 culture that showed very little cell division over a 2-week period, with senescence occurring at the M1 stage (14); these senescent cells also contained numerous large vacuoles, which were rarely seen in early to mid passage cells and have been shown previously to occur in senescent fibroblasts, endothelial cells, and HMEC cultures (15–17). For the confluent HMEC sample, the cells were allowed to reach 100% confluence and refed with fresh medium, and mRNA was harvested 2 days later. The epidermal growth factor (EGF) withdrawal HMEC samples were prepared as described in ref. 18. The Matrigel (Becton Dickinson) samples were prepared by applying a thick coating of Matrigel to tissue culture plates according to the manufacturer’s instructions, followed by the seeding of the plates with 2 × 106 HMEC; mRNA then was harvested 24 hr after plating.
Individual hormones were added to 60–70% confluent HMEC cultures. In each case, the concentrations of hormone used were based on previously published studies (19, 20), with the medium removed and replaced with complete mammary epithelial growth medium (MEGM) that contained either 7 ng/ml transforming growth factor β-1 (TGF-β1) (R & D Systems), 500 units/ml universal type I IFN-α (RDI, Flanders, NJ), or 33 ng/ml IFN-γ (RDI) followed by a 24-hr incubation at 37°C. MCF7 cells (J. Weinstein, National Cancer Institute) and Hs578T cells (American Type Culture Collection) were grown in RPMI + 10% FCS + penicillin/streptomycin to 70–80% confluence. HB2 cells (21) were obtained from H. S. Wiley (University of Utah) and grown in complete MEGM medium.
mRNA Isolation from Cells and Breast Tumors.
Cultured cells were harvested by scrapping and mRNA was prepared by using an Invitrogen FastTrack 2.0 mRNA Isolation Kit and protocol. All of the breast tumor samples used in this study were pieces of the primary tumor except for BC5-LN5, which was a BC-filled lymph node, and BC1257-M, which was a BC metastasis to an ovary. After surgical resection, the breast tumors were dissected and a piece(s) was quickly frozen in liquid nitrogen and stored at −80°C. A frozen tumor specimen then was cut into small pieces and immediately placed into 12 ml of TRIzol Reagent (GIBCO/BRL). The tumor sample in TRIzol was homogenized by using a PowerGen 125 Tissue Homogenizer (Fisher Scientific), and total RNA was isolated by using the TRIzol reagent protocol. Tumor mRNA was isolated by using the above-mentioned Invitrogen kit and protocol. The normal breast samples used here were obtained from CLONTECH and were pools of six (NorBst1) or two (NorBst2) whole normal breasts.
Histology and Immunohistochemistry.
Immunohistochemistry was performed on paraffin-embedded sections from all tumors in this study by using either a STAT1 mAb (Santa Cruz Biotechnology) or a Ki-67 antibody (Immunotech, Westbrook, ME) following the protocol described in ref. 22. Antibody reactivity was detected by using diaminobenzidine, and each section was counterstained with hematoxylin to visualize tumor morphology.
RESULTS
To begin to define patterns of gene expression relevant to mammary epithelial cell biology, a HMEC line growing in vitro was subjected to a set of experimental perturbations. These included (i) addition of TGF-β1 for 24 hr, (ii) withdrawal of EGF for 2 days, (iii) withdrawal of EGF for 2 days followed by the addition of EGF for 90 min, (iv) addition of IFN-α for 24 hr, (v) addition of IFN-γ for 24 hr, (vi) response to 100% confluence, (vii) response to senescence, and (viii) growth on Matrigel for 24 hr. The mRNAs from these experimental cultures each were labeled with the Cy5/red fluorescent nucleotide. All but one of the experiments presented in this paper (i.e., the EGF addition experiment that used the –EGF sample as the Cy3-labeled sample) used the same green reference sample, which was a Cy3-labeled cDNA sample prepared from mRNA taken from 60–80% confluent, passage 9–14, normally growing HMEC cultures.
The data from this study were analyzed and displayed as described (12). Briefly, a hierarchical clustering algorithm produces a table of results wherein the elements/cDNAs of the array (representing specific genes) are grouped together based on similarities in their patterns of gene expression. The same algorithm is applied to cluster the experimental samples (i.e., cell lines and tumors) according to the similarities in their overall patterns of gene expression. The data tables, thus ordered, are presented graphically as colored images. Along the vertical axis, the genes analyzed are arranged as ordered by the clustering algorithm, so that the genes with the most similar patterns of expression are placed adjacent to each other. Along the horizontal axis, experimental samples are similarly arranged such that those with the most similar patterns of expression across all genes are placed adjacent to each other. The color of each cell/square in this tabular image represents the measured expression ratio of each gene in question. The color saturation is also directly proportional to the magnitude of the measured gene expression ratio with the brightest red squares having the highest R/G ratio (i.e., >8-fold difference), the brightest green squares having the lowest R/G ratio, black squares indicating a ratio of approximately 1, and gray squares indicating insufficient data quality.
Fig. 1A shows the single cluster diagram produced by the set of HMEC in vitro experiments (see also Fig. 5, which is published as supplemental data to this article on the PNAS web site, www.pnas.org). Six of the eight experimental treatments tested here caused a significant reduction in the proliferation rate of these cultures, which is reflected by the variation in expression of a set of genes highlighted in the uppermost portion of Fig. 1A. This set of genes included many genes involved in the progression through the cell cycle and included the human homologue of the yeast CDC47 gene (MCM2), MCM3, MCM6, cyclin B1, and the proliferation-associated antigen Ki-67 (Fig. 1B). The repression of cell-cycle transit and DNA replication is consistent with the inhibition of proliferation that is known to occur in HMEC upon the addition of TGF-β1 and IFN-γ and the withdrawal of EGF (18, 23, 24). These results show that the expression level of this set of genes was reduced by a diverse set of growth inhibitory pathways and suggests that this pattern may be linked to cellular proliferation.
Two important processes of many primary human epithelial cells in culture are a reduction in cell proliferation caused by contact inhibition or replicative senescence (25, 26). The cluster of genes that showed reduced expression after treatment with TGF-β1, IFN-γ, or IFN-α, or after withdrawal of EGF, showed a similar reduction in transcript levels at confluence and senescence (Fig. 1B).
A striking feature of the gene expression patterns seen in the confluent and senescent samples is apparent at the bottom of Fig. 1A. Under both of these conditions numerous IFN-regulated genes, including (2′-5′) oligoadenylate synthetase E, IFN-induced 17KD protein, and STAT1 were induced (Fig. 1C) (27–29). The induction of these IFN-regulated genes is known to occur via the JAK/STAT pathway, which when activated, results in the phosphorylation of some of the STAT family member proteins (30); the phosphorylated STAT protein(s) then translocates into the nucleus where it directly activates the transcription of target genes, including the STAT1 gene itself (29). Numerous transcriptional targets of this pathway, as well as one of the central regulators (i.e., STAT1) all were coordinately expressed in this “IFN-regulated” cluster. The response of this set of genes to IFN-α and IFN-γ was also very similar to the genes’ response to confluence or senescence (Fig. 1 A and C). A simple interpretation of these results is that the induction of these IFN-regulated genes in the confluent and senescent cells was the result of the activation of STAT1. The activating STAT1 signal, at confluence and senescence, however, remains unknown.
In addition to the patterns present in Fig. 1, there were other patterns to be seen in the primary data tables (which are available at http://genome-www.stanford.edu/sbcmp/ in text/tab delimited format). Although all of these patterns can be seen in the primary data, many were not represented in Fig. 1 because we selected for the subset of genes whose transcript levels varied 3-fold or more in at least two experiments. Hence, any gene whose expression varied by less than this magnitude, or was highly expressed in only a single experiment, would not have been included.
Our goal is to use cDNA microarrays as a tool to understand and classify tumors on the basis of their global patterns of gene expression. To this end, several human breast tumors, tissues, and cell lines were compared, each in turn, to the same HMEC control samples used for the experiments presented in Fig. 1. Thirteen grade II-III, grossly dissected, infiltrating ductal carcinomas were collected, and mRNA was prepared from each. In addition, two normal breast samples (pools of six and two whole normal breasts) also were tested. Finally, three immortal breast-derived cell lines were studied (MCF7, ref. 31; HB2, ref. 21; and Hs578T, ref.32). A single clustered image was generated for the 1,247 genes that varied 3-fold or more in three or more of the 26 samples/experiments (Fig. 2 and Fig. 6, which is published as supplemental data to this article on the PNAS web site, www.pnas.org). The experimental sample dendrogram above the cluster diagram shows the identity of each column/experiment, with the branching pattern and length of the branches conveying a measure of the relatedness between the samples (12).
The microarrays used in this study did not contain all of the genes known to be important for breast cancer biology; for example, the estrogen receptor and HER2/NEU were notably absent. Nevertheless, there were still too many clusters of coexpressed genes with obvious relevance to breast cancer biology to discuss here in detail. We therefore will discuss only a few clusters of genes (indicated by the black bars in Fig. 2) that we considered to be particularly significant. All of the other clusters, which included a cluster of coexpressed ribosomal protein genes, a cluster of EGF-responsive genes, and a cluster of genes that were highly expressed in normal breast, can be found in Fig. 6 (see Supplemental data). We recognize as well that important features remain to be found by further exploration of this large data set (approximately 140,000 independent data points).
The “proliferation cluster” described in Fig. 1 was recapitulated in this larger experiment (Figs. 3A, 5, and 6). In addition to the previously observed parallel between expression of these genes in vitro and cell proliferation, we observed that this set of genes was highly expressed in the three rapidly growing immortal cell lines, and most significantly, highly expressed in a subset of tumor specimens. In the tumors, the level of expression of this proliferation-associated cluster showed a range of expression from low levels in BC24 and both normal breast samples (bright green) to high expression levels in BC23 (bright red).
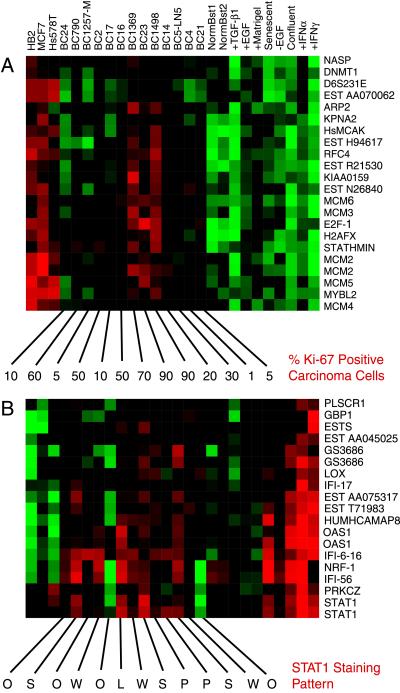
Figure 3.
Expanded view of two gene clusters taken from the 1,247-gene cluster diagram. (A) A portion of the proliferation-associated cluster. The numbers below each breast tumor’s column show the percentage of carcinoma cells in each specimen that stained positive for the Ki-67 antigen. (B) Expanded view of the IFN-regulated gene cluster. The letters below each breast tumor’s column identify the STAT1 staining pattern seen, with O representing no STAT1 staining (BC17 and Fig. 4A), W representing weak STAT1 staining, S representing strong staining (BC23 and Fig. 4C), P representing peripheral tumor cell nest staining (BC14 and Fig. 4E), and L representing staining of lymphocytes/histiocytes only (BC16 and Fig. 4G).
A classic histopathological marker that is associated with cell proliferation is the Ki-67 antigen, which is a large protein of unknown function that is expressed at relatively high levels in proliferating cells and at much lower levels in quiescent cells (33–35). To determine whether there was a relationship between the expression levels of genes in the proliferation cluster and the Ki-67 assessment, the percentage of Ki-67-positive carcinoma cells in each tumor section was determined (Figs. 3A and 4 B, D, F, and H for representative Ki-67 stains). The three tumor specimens that showed the highest level of expression of the genes in the proliferation cluster contained 70% or greater Ki-67-positive carcinoma cells whereas the three tumor specimens with the lowest average expression of these genes showed only 5–10% Ki-67-positive carcinoma cells. Thus, although the correlations in detail are imperfect, there is general agreement between the expression level of the genes in the proliferation cluster and a conventional assessment of proliferation of carcinoma cells.
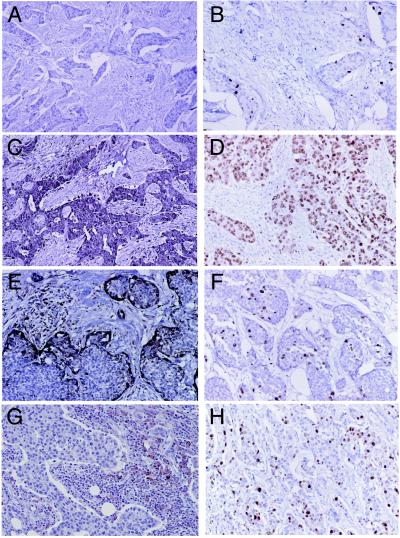
Figure 4.
Immunohistochemical stains of four breast tumor specimens for the STAT1 protein (A, C, E, and G) or for the Ki-67 protein (B, D, F, and H). (A and B) Tumor BC17. (C and D) Tumor BC23. (E and F) Tumor BC14. (G and H) Tumor BC16. Magnification: approximately ×200.
Breast tumor specimens contain other diverse cell types in addition to the carcinoma cells (1). We therefore expected to find gene expression patterns that were contributed by these nonepithelial cell types. Before discussing these data, it first should be noted that the use of a single cell line (HMEC) as a reference made quantitative measurements of gene expression impossible for all those genes not expressed at all in the HMEC reference. In these cases, the HMEC reference sample’s signal intensity at these spots was very close to zero/background, whereas the experimental sample’s intensity gave a significant value. The net result is that the observed ratio is not an accurate measure of the difference between the two samples, but is instead seen as an arbitrarily large ratio in favor of the experimental sample. Therefore, only qualitative measurements could be made on these nonepithelial cell type-specific spots/genes. Nevertheless, this qualitative assessment still permitted the identification of gene expression patterns that were contributed by at least two nonepithelial cell types.
A cluster of genes marked at the bottom of Fig. 2 shows an expression pattern likely to have been contributed by B lymphocytes. This cluster included three different Ig genes (lambda heavy, lambda light, and mu chains) and the generic leukocyte antigen CD45 (see Fig. 6). Tumor sections were stained for CD20, a B-lymphocyte cell surface marker (36); the results showed that the tumors that were positive for Ig mRNAs as determined by cDNA microarray analysis also contained CD20 positive cells (data not shown).
A second nontumor cell type that may be contributing a unique gene expression pattern was tumor-associated stromal cells. A distinctive cluster of genes that included the extracellular matrix proteins collagen type Iα and biglycan were expressed at high levels in all of the tumor specimens, and only in Hs578T among all of the cell lines (a carcinosarcoma-derived cell line that has stromal cell characteristics; ref. 32) (see Fig. 6).
The IFN-regulated gene cluster that originally was identified in vitro also was expressed coordinately in many of the tumors (Fig. 3B). This cluster of genes was highly expressed in some of the tumors (BC23), moderately expressed in others (BC14), and apparently silent in others (BC17). To identify the cell type(s) that was contributing the IFN-regulated pattern, paraffin-embedded tumor sections were stained with antibodies specific for the STAT1 protein. This protein was chosen because it is a required component of the IFN-α signal transduction pathway (20) and was expressed coordinately with the other genes in this cluster. Immunohistochemical staining of tumor BC17, which expressed the genes in this cluster at a very low level, showed no reactivity for the STAT1 protein in any cell type, including lymphocytes (Fig. 4A). In contrast, tumor BC23, which had high STAT1 mRNA levels, showed strong and homogenous staining of all tumor cells and many lymphocytes (Fig. 4C). Specimen BC14 showed a unique pattern of STAT1 staining that could be characterized as a positive staining of some lymphocytes and the most peripheral tumor cells of some tumor cell nests, with the tumor cells in the center of most nests showing little STAT1 reactivity (Fig. 4E). Finally, a fourth pattern was seen in which only some of the lymphocytes/histiocytes stained positive for STAT1 whereas the carcinoma cells did not (BC16 and Fig. 4G).
DISCUSSION
Variation in gene expression reflects important aspects of biological variation in cancers. Systematic characterization of expression patterns associated with specific cell types, and in response to specific physiological and pathological perturbations, provides a framework for interpreting the biological significance of the expression patterns observed in each tumor. In this study we found features of gene expression patterns in breast cancers that could be related to (i) a complex physiological property (e.g., proliferation), (ii) the activity of specific signaling pathways (e.g., the IFN-regulated pathway), and (iii) the cellular composition of the tumors (e.g., the presence of stromal cells and B lymphocytes).
One of our main goals is to use cDNA microarrays to classify breast tumors into categories based on shared gene expression patterns. The tumor specimens analyzed here could readily be classified based on at least two different gene-expression parameters: expression levels of the genes in the proliferation-associated cluster and the IFN-regulated cluster. A much larger clinical study will no doubt be required to determine whether expression levels of these, or other sets of genes, can be used as prognostic indicators. Diverse additional features of variation in gene expression patterns also were seen among the tumor samples (see primary data tables at http://genome-www.stanford.edu/sbcmp and Fig. 6); all of these gene expression features may provide the basis for a more precise molecular taxonomy of breast cancers.
A cluster of IFN-regulated genes was highly expressed in HMEC under three circumstances: addition of IFN, senescence, and confluence. The latter two suggest that there may be circumstances that activate expression of these genes other than the presence of IFN. Similarly, we found multiple circumstances in which these genes were expressed in the tumor specimens, some of which may not involve IFNs and may resemble confluence and/or senescence (BC23). The role of the STAT family of proteins in breast cell biology is extensive and complex, with numerous STAT proteins playing important roles (37–39). Watson and Miller (40) have demonstrated the presence of high levels of STAT1 protein in some primary breast tumor nuclear extracts, whereas others have documented high levels of both STAT1 and STAT3 proteins in primary breast tumors (41). We, too, see high levels of STAT3 protein in many of the same tumors that express high levels of STAT1 mRNA and protein (data not shown). It is not clear at this time what effects the high levels of STAT1 and STAT3 proteins are having on the carcinogenic process, but it is clear that a subset of BCs express the STAT1 protein at relatively high levels, which appears to have resulted in the induction of a known set of IFN-regulated genes.
Breast cancers are complex and highly variable in their histology. It is not a priori evident that measurements of gene expression based on total mRNA isolated from such a complex tissue can be interpreted in terms of the properties of specific cells (e.g., the carcinoma cells). It therefore is noteworthy that discrete gene expression patterns could be identified within a complex mixture of cell types, and that many of these patterns then could be assigned to specific cell types, either by using prior knowledge about the cell type-specific expression of genes within a cluster (e.g., Igs) or by immunohistochemistry using either classical reagents (e.g., Ki-67 antibodies) or antibodies specific for products of genes contained within a cluster (e.g., STAT1). By identifying characteristic clusters of coexpressed genes and then by using antibodies directed against a subset of these genes for immunohistochemistry, archived tumor samples with extensive clinical data can be made accessible to insights derived from gene expression analyses. We therefore have reason for optimism that clustering analysis of cDNA microarray data can be used generally to study and interpret variation in gene expression in tumors, without prior separation of the constituent cell types.
Supplementary Material
Acknowledgments
We thank William Gerald and Larry Norton for discussions and tumor specimens, Peter Nagy for tumor procurement, H.S. Wiley for the m225 antibody and HB2 cell line, Lee and Len Hertzenberg for the use of their tissue culture facility, and members of the P.O.B. and D.B. labs for discussions. This work was supported by a grant from the National Cancer Institute (National Institutes of Health CA 77097) and the Howard Hughes Medical Institute. C.M.P. is a SmithKline Beecham Pharmaceuticals Fellow of the Life Sciences Research Foundation. M.B.E. is an Alfred E. Sloan Foundation Postdoctoral Fellow in Computational Molecular Biology, and D.T.R. is a Walter and Idun Berry Fellow. P.O.B. is an Associate Investigator of the Howard Hughes Medical Institute.
ABBREVIATIONS
- HMEC
human mammary epithelial cells
- EGF
epidermal growth factor
- TGF-β1
transforming growth factor β-1
- BC
breast carcinoma
- R/G
red/green
References
- 1.Ronnov-Jessen L, Petersen O W, Bissell M J. Physiol Rev. 1996;76:69–125. doi: 10.1152/physrev.1996.76.1.69. [DOI] [PubMed] [Google Scholar]
- 2.Tavassoli F A, Schnitt S J. Pathology of the Breast. New York: Elsevier; 1992. [Google Scholar]
- 3.DeRisi J L, Iyer V R, Brown P O. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- 4.Iyer V R, Eisen M B, Ross D T, Schuler G, Moore T, Lee J C F, Trent J M, Staudt L M, Hudson J, Jr, Boguski M S, et al. Science. 1999;283:83–87. doi: 10.1126/science.283.5398.83. [DOI] [PubMed] [Google Scholar]
- 5.Schena M, Shalon D, Davis R W, Brown P O. Science. 1995;270:467–470. doi: 10.1126/science.270.5235.467. [DOI] [PubMed] [Google Scholar]
- 6.Schena M, Shalon D, Heller R, Chai A, Brown P O, Davis R W. Proc Natl Acad Sci USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shalon D, Smith S J, Brown P O. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 8.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 9.Chu S, DeRisi J, Eisen M, Mulholland J, Botstein D, Brown P O, Herskowitz I. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 10.Brown P O, Botstein D. Nat Genet. 1999;21:33–37. doi: 10.1038/4462. [DOI] [PubMed] [Google Scholar]
- 11.Alizadeh A, Eisen M, Botstein D, Brown P O, Staudt L M. J Clin Immunol. 1998;18:373–379. doi: 10.1023/a:1023293621057. [DOI] [PubMed] [Google Scholar]
- 12.Eisen M B, Spellman P T, Brown P O, Botstein D. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ermolaeva O, Rastogi M, Pruitt K D, Schuler G D, Bittner M L, Chen Y, Simon R, Meltzer P, Trent J M, Boguski M S. Nat Genet. 1998;20:19–23. doi: 10.1038/1670. [DOI] [PubMed] [Google Scholar]
- 14.Foster S A, Galloway D A. Oncogene. 1996;12:1773–1779. [PubMed] [Google Scholar]
- 15.Goldstein S, Moerman E J, Porter K. Exp Cell Res. 1984;154:101–111. doi: 10.1016/0014-4827(84)90671-2. [DOI] [PubMed] [Google Scholar]
- 16.Piotrowicz R S, Weber L A, Hickey E, Levin E G. FASEB J. 1995;9:1079–1084. doi: 10.1096/fasebj.9.11.7649407. [DOI] [PubMed] [Google Scholar]
- 17.Stampfer M R, Yaswen P. Transformation of Human Epithelial Cells: Molecular and Oncogenetic Mechanisms. Boca Raton, FL: CRC; 1992. [Google Scholar]
- 18.Stampfer M R, Pan C H, Hosoda J, Bartholomew J, Mendelsohn J, Yaswen P. Exp Cell Res. 1993;208:175–188. doi: 10.1006/excr.1993.1236. [DOI] [PubMed] [Google Scholar]
- 19.Stampfer M R, Bodnar A, Garbe J, Wong M, Pan A, Villeponteau B, Yaswen P. Mol Biol Cell. 1997;8:2391–2405. doi: 10.1091/mbc.8.12.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bromberg J F, Horvath C M, Wen Z, Schreiber R D, Darnell J E., Jr Proc Natl Acad Sci USA. 1996;93:7673–7678. doi: 10.1073/pnas.93.15.7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berdichevsky F, Alford D, D’Souza B, Taylor-Papadimitriou J. J Cell Sci. 1994;107:3557–3568. doi: 10.1242/jcs.107.12.3557. [DOI] [PubMed] [Google Scholar]
- 22.Bindl J M, Warnke R A. Am J Clin Pathol. 1986;85:490–493. doi: 10.1093/ajcp/85.4.490. [DOI] [PubMed] [Google Scholar]
- 23.Stampfer M R, Yaswen P, Alhadeff M, Hosoda J. J Cell Physiol. 1993;155:210–221. doi: 10.1002/jcp.1041550127. [DOI] [PubMed] [Google Scholar]
- 24.Harvat B L, Jetten A M. Cell Growth Differ. 1996;7:289–300. [PubMed] [Google Scholar]
- 25.Abercrombie M. Nature (London) 1979;281:259–262. doi: 10.1038/281259a0. [DOI] [PubMed] [Google Scholar]
- 26.Hayflick L. Exp Cell Res. 1965;37:614–636. doi: 10.1016/0014-4827(65)90211-9. [DOI] [PubMed] [Google Scholar]
- 27.Benech P, Mory Y, Revel M, Chebath J. EMBO J. 1985;4:2249–2256. doi: 10.1002/j.1460-2075.1985.tb03922.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blomstrom D C, Fahey D, Kutny R, Korant B D, Knight E., Jr J Biol Chem. 1986;261:8811–8816. [PubMed] [Google Scholar]
- 29.Lehtonen A, Matikainen S, Julkunen I. J Immunol. 1997;159:794–803. [PubMed] [Google Scholar]
- 30.Darnell J E., Jr Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 31.Levenson A S, Jordan V C. Cancer Res. 1997;57:3071–3078. [PubMed] [Google Scholar]
- 32.Hackett A J, Smith H S, Springer E L, Owens R B, Nelson-Rees W A, Riggs J L, Gardner M B. J Natl Cancer Inst. 1977;58:1795–1806. doi: 10.1093/jnci/58.6.1795. [DOI] [PubMed] [Google Scholar]
- 33.Gerdes J. Semin Cancer Biol. 1990;1:199–206. [PubMed] [Google Scholar]
- 34.Goodson W H, 3rd, Moore D H, 2nd, Ljung B M, Chew K, Florendo C, Mayall B, Smith H S, Waldman F M. Breast Cancer Res Treat. 1998;49:155–164. doi: 10.1023/a:1005926228093. [DOI] [PubMed] [Google Scholar]
- 35.Schluter C, Duchrow M, Wohlenberg C, Becker M H, Key G, Flad H D, Gerdes J. J Cell Biol. 1993;123:513–522. doi: 10.1083/jcb.123.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stamenkovic I, Seed B. J Exp Med. 1988;167:1975–1980. doi: 10.1084/jem.167.6.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu X, Robinson G W, Wagner K U, Garrett L, Wynshaw-Boris A, Hennighausen L. Genes Dev. 1997;11:179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 38.Udy G B, Towers R P, Snell R G, Wilkins R J, Park S H, Ram P A, Waxman D J, Davey H W. Proc Natl Acad Sci USA. 1997;94:7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith P D, Crompton M R. Biochem J. 1998;331:381–385. doi: 10.1042/bj3310381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson C J, Miller W R. Br J Cancer. 1995;71:840–844. doi: 10.1038/bjc.1995.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garcia R, Jove R. J Biomed Sci. 1998;5:79–85. doi: 10.1007/BF02258360. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.