Abstract
Mice heterozygous at Aprt (adenine phosphoribosyltransferase) were used as a model to study in vivo loss of heterozygosity (LOH) in normal fibroblasts. Somatic cell variants that exhibited functional loss of the wild-type Aprt in vivo were recovered as APRT-deficient cell colonies after culturing in selection medium containing 2,6-diaminopurine (DAP), an adenine analog that is toxic only to cells with APRT enzyme activity. DAP-resistant (DAPr) fibroblast variants were recovered at a median frequency of 12 × 10−5 from individual ears from progeny of crosses between mouse strains 129/Sv and C3H/HeJ. The frequency of DAPr variants varied greatly among individual ears, suggesting that they preexisted in vivo and arose at various times during development. Polymorphic molecular markers and a cytological marker on the centromere of chromosome 8 made it possible to discriminate between each of six possible mechanistic pathways of LOH. The majority (about 80%) of the DAPr variants were a consequence of mitotic recombination. The prevalence of mitotic recombination in regions proximal to Aprt did not correlate with meiotic map distances. In particular, there was a higher than expected frequency of crossovers within the interval 59 cM to 67 cM. The high spontaneous frequency of Aprt LOH, mediated primarily by mitotic recombination, is fully consistent with our previous results with human peripheral T cells from individuals known to be heterozygous at APRT. Thus, this Aprt heterozygote mouse is a valid model for studying somatic mutagenesis and mitotic recombination in vivo.
Retinoblastoma is a prototype disease for understanding how loss of function of tumor suppressor genes (TSGs) leads to tumor formation. The so-called two-hit (two-mutational events) model explains elegantly the inheritance of genetic predisposition and development of retinal tumors (1). In familial cases, a preexisting RB1 germ-line mutation (the first hit) is inherited, predisposing the retinoblast cells to tumor development by requiring only a second mutational event (the second hit). In sporadic cases, somatic cells lack the predisposing mutation, and a retinoblast cell must acquire two separate RB1 mutations to progress to a tumor. In the two-hit model, the first hit, a rate-limiting step, renders a cell heterozygous or hemizygous at RB1. The second hit, which is frequently referred to as loss of heterozygosity (LOH), leads to the expression of the RB1 mutant phenotype (2). Because the probability of the second mutational event follows a Poisson distribution, the number of tumors and the time at which they arise may vary in heterozygous carriers. Indeed, some carriers (about 5%) may remain tumor free. All somatic cells in Li–Fraumeni syndrome or familial retinoblastoma carry a germ-line mutation in one allele, at TP53 (3) or RB1(4), respectively. When tumors arise, the remaining wild-type allele of TP53 or RB1 is usually either mutated or lost. Concomitant loss of normal allelic function, or functional LOH, leads to unmasking of the recessive phenotypes including deregulation of the cell cycle, perturbation of DNA repair, or alteration of cell–cell communication (5).
Although LOH is well documented in carcinogenesis, it also mediates phenotype expression in some familial autosomal dominant diseases such as polycystic kidney disease (6), and it may serve to generate somatic-cell variants that may have selective advantage during development (2). Although the first mutational event is often a missense mutation or small deletion, subsequent LOH may arise by any of several mechanisms. Detailed characterization of retinoblastoma has identified point mutation, interstitial deletion, gene conversion, mitotic recombination, and chromosome loss/duplication as pathways to LOH at the RB1 locus (4).
Molecular analyses of malignant tumors frequently reveal LOH at multiple known TSG loci, whereas the observation of LOH at other sites suggests locations for undescribed TSGs. These types of correlative data cannot establish whether LOH is causal for the development of a particular tumor or consequential as a result of the genomic instability that frequently accompanies tumorigenesis. By the time LOH is observed in a tumor, cells have undergone several steps of selection for increased growth potential and may have acquired increased genomic instability as a result of functional loss of DNA repair or other genes (7).
To establish a basal frequency of spontaneous LOH in normal cells in vivo, we have developed a model system that uses the selectively neutral marker APRT (encoding adenine phophoribosyltransferase) to circumvent the above limitations inherent in tumor samples. The APRT gene is located on chromosome 16 in humans and on chromosome 8 in mice and can serve as a sensitive reporter of mutagenic events, including LOH, in vivo (8). Analogous to a TSG, two independent allelic mutations are needed to abolish cellular APRT function. Cells lacking APRT are resistant to toxic adenine analogs, such as 2,6-diaminopurine (DAP), allowing their selection in vitro and distinguishing them from their heterozygous and wild-type counterparts. Cells heterozygous at APRT are ideal for study of LOH as they require only a single mutational event to express the selectable recessive phenotype. Previously, we demonstrated multiple pathways leading to APRT deficiency in vitro, including point mutation, interstitial deletion, and mitotic recombination, in an HT1080 human fibrosarcoma cell line, heterozygous at APRT (9, 10). We have also found that the APRT-deficient T-lymphocytes in vivo in human APRT heterozygous subjects were primarily caused by mitotic recombination (11).
To study systematically the frequency with which spontaneous LOH occurs in vivo and the predominant mechanisms that result in LOH, we have produced mice that are Aprt+/neo. These mice have a bacterial neo gene inserted into the third exon of one Aprt allele (8, 12). We found that APRT-deficient mouse fibroblasts arise at high frequency in vivo and that most are derived from mitotic recombination.
MATERIALS AND METHODS
Mice.
The production of the Aprt+/neo mice by gene targeting has been reported (12). Male and female Aprt+/neo or Aprtneo/neo mutant mice of 129/Sv background or 129/C57 mixed background were crossed to wild-type C3H/HeJ to generate Aprt+/neo. Such hybrids are not only heterozygous for many of the flanking simple sequence repeat (SSR) markers, but also heteromorphic at the centromeric region of chromosome 8, with the 129/Sv strain exhibiting a small centromere and C3H/HeJ strain a large centromere (13). The centromeric heteromorphism thus serves as a useful cytological marker for the strain origin of a centromere. By analyzing the SSR markers, cytological markers, and Aprt gene sequence, it is possible to assign each somatic cell variant to a particular LOH pathway.
Preparation of Single Skin Cells.
The mice, aged 2 to 4 mo, were euthanized and each ear was briefly swabbed with 75% ethanol, excised, and rinsed twice in PBS containing kanamycin (100 μg/ml). Each ear was minced carefully in a well of a 24-well plate, into which approximately 0.3 ml collagenase D/dispase neutral protease from Bacillus polymyxa, grade II (4 mg/ml of each in DMEM; Boehringer Mannheim) was added. The minced pieces were treated at 37°C for 45 min, then 1.5 ml DMEM/10% FBS medium was added, followed by overnight incubation. The treated tissues were then dissociated by gentle pipetting. Single cells were obtained by passing the cell suspension through a Cell Strainer (70 μm, Falcon). The harvested cells were pelleted by centrifugation, resuspended in growth medium (DMEM supplemented with 10%FBS/1 × MEM nonessential amino acids/1 × penicillin/streptomycin/5 μg/ml fungizone), and enumerated.
Reconstruction Experiment.
To model the efficiency of in vitro recovery of preexisting APRT-deficient cells from Aprt+/neo mice, reconstruction experiments were conducted in which cells prepared from Aprtneo/neo and Aprt+/neo mice were mixed in the same dish (100 mm) containing selection medium (growth medium plus 50 μg/ml DAP). The medium was changed every 4 d and DAP-resistant (DAPr) colonies were scored on day 12.
Clonal Isolation of DAPr Fibroblast Cells.
About 8 −10 × 105 freshly prepared cells were seeded in a 100-mm culture dish containing 10 ml selection medium. The medium was changed every 4 d, and on day 12 the dish was examined for colonies.
APRT Enzyme Assay.
The cells of DAPr variant clones and controls were washed with PBS and disrupted in 50 μl buffer (50 mM Tris⋅HCl, pH 7.4/5 mM MgCl2). Measurement of APRT activity was as previously described (14).
Immunocytochemistry Assay.
The cells of DAPr colonies were characterized as to type with anitvimentin and anticytokeratin antibodies by using immunohistology kits (Sigma).
Allele-Specific PCR.
Three primers, MA009 (5′-ACA ACC TTC CCT CCT TAC CCT AAC AG-3′), neo4 (5′-TGC CTG CTT GCC GAA TAT CAT GGT-3′), and s2 (5′-ATA AGA CCC TGC CCT TCC TCT ACA CA-3′), were used to amplify the mutant and wild-type alleles. PCR conditions were described elsewhere (8). The DAPr clones were divided into two classes on the basis of the loss (class I) or retention (class II) of the wild-type Aprt (Aprt+), as shown by allele-specific PCR.
PCR Assay of the SSR Loci Flanking Aprt.
DAPr clones of class I were further characterized by PCR amplification of SSR markers along the length of mouse chromosome 8 (15). The markers and their chromosome locations are according to the Mouse Genome Database. PCR primers for SSR markers were purchased from Research Genetics (Huntsville, AL). PCR conditions were described elsewhere (8).
Cytogenetic Studies.
Slides containing metaphase spreads were baked at 65°C for 2–3 hr, treated with 0.25% trypsin, and stained with 5% Giemsa for karyotype analysis. Fluorescence in situ hybridization (FISH) was performed with chromosome 8 painting probes (Applied Genetics, Melbourne, FL) or plasmid pMAprt11 (16). The plasmid pMAprt11 was labeled with a Bionick labeling kit (GIBCO). FISH procedures were as described (10). At least 10 metaphase spreads from each clone were analyzed and photographed with a charge-coupled device camera.
Sequence Analysis.
An Aprt gene fragment of 2.3 kb was amplified from class II DAPr clones by using primers AMF2 (5′-CCTGGAAAAGCAGGACTGAAA-3′) and MA010 (5′-CACCAAGCAGTTCCTAGTGCT-3′). The PCR conditions were as follows. The template DNA was denatured at 98°C for 6 min, followed by 30 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 2 min. The DNA fragments were excised from the gel and purified with a QIAquick Gel Extraction kit (Qiagen). The purified fragments were either directly sequenced or subcloned with TA cloning kit (Invitrogen) before sequencing. Sequencing was done with a DNA cycle sequencing kit (Amersham Pharmacia).
RESULTS
Recovery of DAP-Resistant Fibroblast Variants.
Reconstruction experiments were performed to model the in vitro recovery of preexisting APRT-deficient cells from Aprt+/neo mouse ears. In one set, different numbers of cells from Aprt+/neo mice and a fixed small number of Aprtneo/neo cells were plated in selection medium. We observed that the recovery [colony-forming efficiency (CFE)] of Aprtneo/neo cells correlated with the number of the Aprt+/neo cells coplated (data not shown). This correlation indicates that cooperative effects outweighed adverse effects, if any, from the crossfeeding of toxic metabolites produced by Aprt+/neo cells (17). In a second set of experiments, different numbers of Aprtneo/neo cells were mixed with a constant number of cells (8 × 105) from Aprt+/neo mice. The CFE of Aprtneo/neo cells in DAP ranged from 1.4% to 1.6% and did not significantly vary with the number of the Aprtneo/neo cells seeded (data not shown). For subsequent selection experiments, we seeded about 1 × 106 cells per 100-mm dish.
The cell suspension obtained after initial enzymatic dissociation of ear skin fragments contains fibroblasts, melanocytes, and keratinocytes. However, only fibroblasts grow and form colonies under culture conditions. To verify that the DAPr colonies were composed of fibroblasts, representative colonies were stained with antibodies to vimentin and cytokeratin. Cells in all of the colonies tested were vimentin positive and cytokeratin negative, and intermediate filaments were visible microscopically (data not shown).
Mitotic Recombination Accounts for the Majority of DAPr Variants.
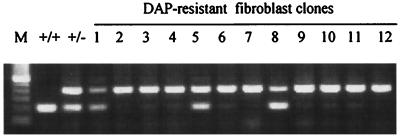
Of 113 clones (from 33 mice) analyzed by allele-specific PCR, 92 (81%) were of class I, exhibiting loss of Aprt+. Allele-specific PCR results from representative clones are shown in Fig. 1.
Figure 1.
Allele-specific PCR of representative DAPr clones. Clones 2, 3, 4, 6, 7, 9, 10, 11, and 12, designated as class I, exhibited physical loss of Aprt+. Clones 1, 5, and 8, designated as class II, retained Aprt+.
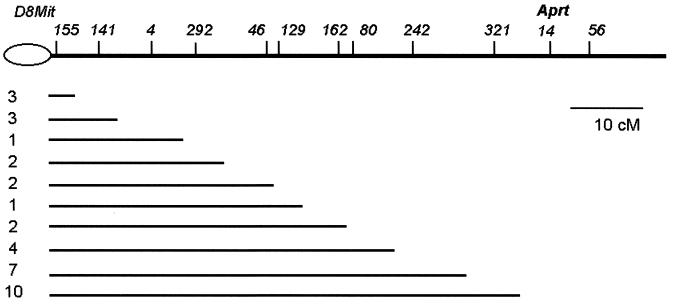
Eighty-four class I variant clones were genotyped further with SSR markers distributed from 1 to 73 cM along the 84 cM genetic length of chromosome 8 to determine the extent of LOH and to provide insight into possible chromosomal mechanisms for Aprt loss. All 84 clones exhibited LOH of large contiguous regions, starting at one or another locus proximal to Aprt, including Aprt, and extending through the most telomeric marker, D8Mit56 (Fig. 2).
Figure 2.
Intervals of LOH in class I clones. The lines correspond to the interval for which the SSR markers remained heterozygous. All markers right to the lines exhibited LOH. The number of independent events in each group is shown on the left. The map positions of the SSR markers are according to Mouse Genome Database [http://www.informatics.jax.org (6/98)].
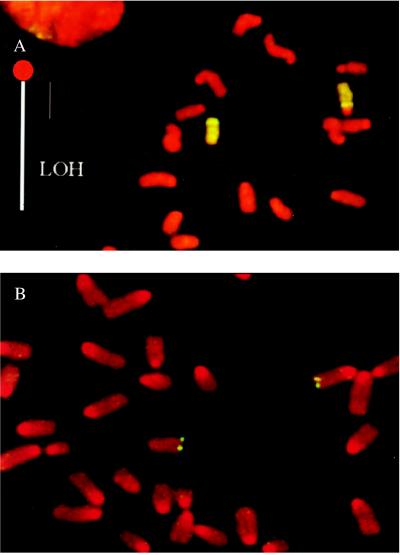
The observation of large contiguous segments of LOH, extending through the most telomeric marker, but not including the centromeric region, excludes chromosome loss/duplication, gene conversion, and interstitial deletion as the primary mechanisms for such class I clones. Chromosome loss/duplication would result in LOH of all loci on chromosome 8, whereas gene conversion or interstitial deletion would not extend LOH as far as the most telomeric marker. Rather, it is most likely that those clones were caused by mitotic recombination, the reciprocal exchange between chromatids of homologues. To confirm this mitotic recombination as a mechanism of LOH, G-banding analysis and whole-chromosome painting were performed on eight clones that exhibited LOH of the terminal 40 to 70 cM (on the meiotic map). We observed that all eight clones contained two normal copies of chromosome 8 in diploid metaphases and four copies in tetraploid metaphases. Importantly, a large centromere characteristic of the C3H/HeJ strain was retained in each metaphase (Fig. 3A), thus providing cytological evidence that the proximal centromeric segment of the chromosome 8 of strain C3H/HeJ (Chr8C3H) was retained.
Figure 3.
Cytogenetic evidence that LOH was caused by mitotic recombination. (A) Whole-chromosome painting of a class I DAPr fibroblast clone. Chromosome regions exhibiting hybridization signal are green, otherwise they stain red. Chromosome 8 of strain C3H/HeJ (Chr8C3H) exhibits a large centromeric region. Although SSR genotyping showed a large interval of LOH (loss of C3H alleles), the homologue with a large centromere, Chr8C3H, exhibited no corresponding terminal deletion. (B) Aprt FISH of a class I DAPr clone. SSR genotyping showed terminal LOH (loss of C3H alleles) beginning at 59 cM, but Aprt hybridization signals were evident on both Chr8129 and Chr8C3H.
FISH with a plasmid containing Aprt (pMAprt11) produced two hybridization signals in most diploid metaphases and four signals in tetraploid metaphases in all five clones examined (Fig. 3B). This presence of Aprt hybridization signals suggests that the Aprt sequences (presumably both Aprtneo) are retained in these clones. It is interesting to note that Aprt hybridization signals were located at or very close to the telomere, even though the meiotic distance of Aprt to the telomere is 17 cM (18). This finding suggests that the physical length from Aprt to the telomere may be much shorter than the genetic length suggests. The loss of distal C3H markers, the normal morphology of both chromosomes 8, and the retention of Aprt sequences, as well as the proximal portion of C3H chromosome 8, unequivocally establish mitotic recombination as the only plausible pathway underlying most, if not all, of the class I variants.
Characterization of Class II Variants.
In class II variants, the full-sized Aprt+ allele from C3H/HeJ was physically retained. To detect possible point mutations in Aprt+, all five exons, three introns (introns 1, 3, and 4), and part of the promoter from 16 clones were sequenced. Point mutations, all in the coding region, were detected in four clones (Table 1). The failure to detect point mutations in the majority of class II variants raises the possibility that mechanisms other than point mutations might be responsible for some class II variants.
Table 1.
Point mutations observed in class II clones
| Clone | Position | Mutation | Change |
|---|---|---|---|
| 129-5LA | 2720 (exon 5) | T → G | Val → Gly |
| 129-7R | 889 (exon 1) | T → A | Leu → Stop |
| E62A | 2233 (exon 3) | T → C | Ile → Ser |
| 30b1 | 873 (exon 1) | A → T | Met → Phe |
To determine whether class II colonies were APRT-deficient, APRT activity of cells from representative colonies was individually measured. Although all class I clones (clones with physical loss of Aprt+) had less than 15% of the activity of the parental Aprt+/neo cells, the APRT activity of class II clones (clones without physical loss of Aprt+) varied, ranging from less than 10% to 90% that of the parental heterozygous cells. Tests for mycoplasma in clones with higher APRT activity were negative, indicating that the APRT activity was not derived from mycoplasma contamination (19). To test the crossresistance of these clones to another more toxic adenine analog, cells from representative clones were incubated in medium containing 2 μg/ml 2-fluoroadenine (FA). Although cells from all of the class I clones grew in FA, cells from class II clones differed in their sensitivity to FA medium. Those with greater APRT activity did not survive FA treatment.
Estimation of the Frequency of DAPr in Individual Ears.
When 1 × 104 cells from Aprt+/neo mice were cultured in the presence of feeder cells in drug-free medium, the average colony-forming efficiency was 1.6% (Table 2), similar to that obtained in reconstruction experiments. The frequency of DAPr clones varied greatly between individual ears, from less than 1.4 × 10−5 (in 1515R) to more than 32 × 10−5 (in 1505R), with a median frequency of 12 × 10−5. The frequency even differed dramatically between two ears from the same animal (e.g., animals 1515 and 1512). The number of DAPr variants per 55,000 colony-forming units did not follow a Poisson distribution (P < 0.0001).
Table 2.
Frequency of DAPr skin fibroblasts in ears of 129 × C3H hybrid mice
| Mouse | Ear | Total no. cells (×106) | CFE % | CFU (×104) | No. DAPr colonies | Frequency (×10−5) |
|---|---|---|---|---|---|---|
| 1505 | L | 4.3 | 1.5 | 6.5 | 7 | 10.9 |
| R | 4.5 | 1.1 | 5 | 16 | 32.3 | |
| 1518 | L | 3.4 | 1.7 | 5.8 | 7 | 12.1 |
| R | 3.6 | 1.2 | 4.3 | 5 | 11.6 | |
| 1521 | L | 3.2 | 1.6 | 5.1 | 7 | 13.7 |
| R | 4.3 | 1.5 | 6.5 | 16 | 24.8 | |
| 1515 | L | 3.7 | 1.8 | 6.7 | 14 | 21 |
| R | 4.1 | 1.8 | 7.4 | 0 | 0 | |
| 1519 | L | 3.3 | 2.3 | 7.6 | 3 | 4 |
| R | 2.9 | 1.4 | 4.1 | 11 | 27.1 | |
| 1512 | L | 2.6 | 1.5 | 3.9 | 7 | 17.9 |
| R | 3.2 | 2.3 | 7.4 | 0 | 0 | |
| 1534 | L | 2.8 | 1.4 | 3.8 | 0 | 0 |
| R | 2.9 | 1.3 | 3.9 | 2 | 5.1 | |
| Mean | 3.5 | 1.6 | 5.5 | 6.8 | 12.9 | |
| Median = 11.9 |
The CFE was estimated by plating 1 × 104 cells in a 100-mm dish (in duplicate for each ear) containing growth medium and 8 to 10 × 105 feeder cells. About 1 × 106 cells were plated in selection medium to recover DAPr colonies. L, left ear; R, right ear. CFU, colony-forming unit; CFE, colony-forming efficiency.
We hypothesize that the great variation in the number of DAPr variants between individual ears was caused by the stochastic nature of Aprt LOH in both temporal and spatial terms. For example, when LOH occurs early in development of a particular ear and cellular descendents of the variant cell remain in situ, a relatively higher frequency of the DAPr cell variants would be anticipated from that ear. In this regard, the spatial clustering of DAPr fibroblasts in an intact ear would be analogous to the wing spot in Drosophila (20) or coat color spot in Wei heterozygous mice (21). We tested this hypothesis by comparing chromosomal intervals of LOH in different colonies obtained from the same ear. If DAPr ear cells were all derived from one progenitor cell, then individual colonies from the same ear would exhibit the same interval of LOH. Analysis of chromosome 8 SSR markers supports our hypothesis, because crossovers in clones from the same ear tend to lie in the same interval (Table 3). For example, seven of the eight clones from two ears of mouse 28 had crossovers in the 3-cM interval between D8Mit46 and D8Mit129 and five of six clones from ear 44L had crossovers in the 6-cM interval between D8Mit155 and D8Mit141. Thus, there was probably clustering in situ of recombinant sib cells, and the size of a cluster is likely related to the time during ear development when recombination occurred.
Table 3.
Distribution of LOH endpoints in class I colonies recovered from individual ears
| Locus | Map distances | Identification
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 11R | 28LR | 42L | 44L | 44R | 1521R | 1515L | 7015L | 1505R | ||
| D8Mit155 | ||||||||||
| 5 | 5 | |||||||||
| –141 | ||||||||||
| 8 | 1 | 3 | ||||||||
| –4 | ||||||||||
| –46 | ||||||||||
| 3 | 7 | |||||||||
| –129 | ||||||||||
| –162 | ||||||||||
| 4 | 3 | |||||||||
| –80 | ||||||||||
| 6 | 1 | 1 | ||||||||
| –242 | ||||||||||
| 12 | 3 | 7 | 3 | |||||||
| –321 | ||||||||||
| 8 | 4 | 4 | 4 | |||||||
| Aprt | ||||||||||
| Total no. of colonies | 5 | 8 | 3 | 6 | 3 | 7 | 7 | 3 | 4 | |
The interval in which mitotic crossover occurred for each colony was determined by testing for LOH of a series of syntenic SSR markers. Colonies from the same ear and with the same interval of LOH were considered to be a single event for calculation purpose. L, left ear; R, right ear; LR, two ears combined. Meiotic map distances (in cM) between markers are shown in the second column.
Distribution of Mitotic Crossovers.
Although the crossovers in class I clones from the same ear tended to fall into the same interval, the crossovers from different animals were more randomly distributed, from the most centromeric locus, D8Mit155 (1 cM) to Aprt (67 cM). However, nearly one-third (28/84) of the class I variants have breakpoints in the 8-cM interval between 59 cM (D8Mit321) and 67 cM (Aprt) (P < 0.001). The clustering of crossovers within this interval was not caused by overrepresentation of sib clones from a few ears. When all of the putative sib clones were scored as only a single event for each ear (Fig. 2), there were still more than the expected number of crossovers in this interval than predicted from the meiotic map distance (P < 0.007). The elevated frequency of mitotic crossovers in this region suggests that either it contains a hot spot for mitotic recombination or the meiotic linkage map is contracted because of suppression of meiotic crossover.
DISCUSSION
Aprt Heterozygous Mice as Models for Study of in Vivo LOH.
The Aprt+/neo mouse has several advantages for the study of LOH in vivo. First, the reporter gene is an endogenous gene at its resident locus, near the telomere of chromosome 8. It is worth noting that human APRT is located near the telomere of 16q. Second, the Aprtneo and wild-type alleles can be easily distinguished. Third, by appropriate crosses, interstrain hybrid Aprt+/neo mice can be generated with heterozygous marker loci along the length of the chromosome and heteromorphic cytogenetic markers. To our knowledge, this is the first mouse assay that allows both the quantitative and molecular characterization of in vivo LOH in a solid tissue. Spontaneous and induced frequencies of in vivo LOH have been reported for murine small intestine in Dlb-1 heterozygous mice (22–24), but the histochemical approach used did not allow a molecular characterization of the mutational events. Interestingly, the frequency of Aprt LOH that we observed in mouse ear fibroblasts is quite similar to that of human T cells, ≈10−4 (11), though it is about 10-fold higher than that reported for mouse T cells (25)
Combined molecular and cytogenetic analyses enabled us to demonstrate that mitotic recombination is the predominant mechanism for LOH of chromosome 8 in vivo in fibroblasts of 129 × C3H and 129/C57 × C3H hybrids. Of all possible pathways to LOH other than mitotic recombination, only point mutation was identified. It remains to be determined whether epigenetic inactivation is responsible for some of the class II variants. Epigenetic inactivation of Aprt has been reported in cultured embryonal carcinoma cells (26). Variants caused by chromosome loss/reduplication, gene conversion, or interstitial deletion are probably very rare in normal cells in vivo, though chromosome loss/duplication and interstitial deletion are not uncommon in embryonal carcinoma cells (27)
The Recovered Apro LOH Variants Preexisted in Vivo.
Studies in which recessive mutant cells are induced by a relatively short mutagen treatment suggest that significant APRT enzyme activity persists for at least 42 hr after loss of allelic function (28). Thus, tissue-derived cells that survive DAP selection immediately after harvest must have undergone mutation at Aprt or lost APRT activity in vivo. There is additional evidence in support of the in vivo origin of the DAPr variants. First, the distribution of the number of DAPr variants between individual ears is compatible only with their preexistence in vivo. If they had been induced by DAP, a Poisson distribution of the variants in individual ears would be expected (variance ≅ mean), and the numbers for each ear would be more similar. On the other hand, if they preexist in vivo, we would expect a greater variation (variance > mean) between individual ears, reminiscent of the distribution of bacterial mutants in the Luria–Delbruck fluctuation assay (29). DAPr fibroblasts apparently differ with respect to the developmental stage at which they arose. For example, if a cell variant arose just 12 hr before the animal was killed, it might not have time to divide, and it would probably be killed by DAP because of residual APRT activity. However, had the mutational event occurred early in development, the mutant progenitor cell would have undergone clonal expansion and would give rise to many colonies when cells are harvested and placed under DAP selection.
The observation that DAPr clones from an individual ear are more likely to be identical also supports their in vivo origin. Analysis of SSR markers showed that most of the mitotic recombinants isolated from one ear, though from different culture dishes, tended to have the same interval of allelic loss. For instance, in ear 44L, five of the six mutants have a crossover in an interval of 6 cM, between D8Mit155 and D8Mit141, whereas in the other ear from the same animal, 44R, all three mutants analyzed had a crossover between D8Mit162 and D8Mit80. Thus, it is unlikely that these latter variants were independently induced in vitro. There is no reason to presume that DAP would induce the same variants in cultures from each ear. Rather, we would expect a spectrum of variants from each ear if they were induced by DAP in vitro.
Mitotic Recombination in Vivo in Normal Fibroblasts.
Molecular and cytogenetic characterization of DAPr fibroblast cell variants in 129 × C3H hybrid mice revealed that about 80% were segregants of mitotic recombination. Our current findings are highly consistent with those we made with T cells from human APRT heterozygotes, where LOH mutants were observed at a frequency of 1 × 10−4 and were predominantly caused by mitotic recombination (11). Mitotic recombination has also been suggested to occur in vivo in mouse T cells (25, 30). Because T lymphocyte precursors normally undergo T cell receptor recombination, it was important to exclude the possibility that the high level of mitotic recombination observed in T cells is a consequence of their V(D)J recombinase activity. We have now demonstrated, with combined molecular and cytogenetic evidence, that segregants of mitotic recombination occur in vivo at a high frequency in normal fibroblasts from hybrids of inbred mouse strains, and that they account for the majority of the APRT-deficient variants. This observation suggests that mitotic recombination is a common mechanism for generating homozygous somatic cell variants in vivo.
Mitotic vs. Meiotic Crossover.
There were more mitotic crossovers than expected from the meiotic linkage map in the region from 59 cM to 67 cM. Several factors may account for this discrepancy. One is that the meiotic map distance does not reflect the actual physical distance. It has been observed that chiasmata are not evenly distributed along the chromosomes, with the telomeric regions usually having more chiasmata (31–34). According to the meiotic map, Aprt (67 cM) should be located about one-fifth the distance of the total chromosome length (84 cM) from the telomere; however, its hybridization signal was consistently observed near the telomere of chromosome 8. It is likely that the physical distance from Aprt to the telomere is shorter than that predicted by the meiotic map. This would be consistent with the observation that a region adjacent to a meiotic crossover hot spot would be suppressed for a second crossover because of chiasma interference (34). A crossover distal to Aprt would likely suppress another crossover in the region immediately proximal to Aprt, resulting in a contracted map distance in the region proximal to Aprt. Another factor is the possibility of hot or cold spots in which mitotic crossover is enhanced or suppressed. It has been reported that deletion of a 37-bp region in a mouse retrotransposon strongly suppresses recombinational activity in mouse cells in culture (35). Furthermore, hypervariable minisatellite DNA sequences were shown to stimulate homologous recombination in cultured human cells (36). The presence of mouse retrotransposons and minisatellite DNA sequences throughout the mouse genome could act as favorable niches for mitotic recombination. Lastly, because of their different transcriptional activities, meiotic cells and somatic cells have unique chromatin structures, which could affect the capacity to undergo meiotic and/or mitotic recombination at certain chromosomal loci.
Acknowledgments
This work is dedicated to Professor Frank H. Ruddle on the occasion of his 70th birthday and was supported by National Institutes of Health grants DK38185 and ES05652.
ABBREVIATIONS
- TSG
tumor-suppressor gene
- Aprt
adenine phosphoribosyltransferase
- LOH
loss of heterozygosity
- DAP
2,6-diaminopurine
- DAPr
DAP resistant
- SSR
simple sequence repeat
- FISH
fluorescence in situ hybridization
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Knudson A G. Proc Natl Acad Sci USA. 1971;68:820–823. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tischfield J A. Am J Hum Genet. 1997;61:995–999. doi: 10.1086/301617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malkin D, Li F P, Strong L C, Fraumeni J F, Nelson C E, Kim D H, Kassel J, Gryka M A, Bischoff F Z, Tainsky M A, et al. Science. 1990;250:1233–1238. doi: 10.1126/science.1978757. [DOI] [PubMed] [Google Scholar]
- 4.Cavenee W K, Dryja T P, Philips R A, Benedict W F, Godbout R, Gallie B L, Murphree A L, Strong L C, White R L. Nature (London) 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 5.Kinzler K W, Vogelstein B. Cell. 1996;87:159–170. doi: 10.1016/s0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 6.Qian F, Germino G G. Am J Hum Genet. 1997;61:1000–1005. doi: 10.1086/301618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loeb L A. Cancer Res. 1991;51:3075–3079. [PubMed] [Google Scholar]
- 8.Stambrook P J, Shao C, Stockelman M, Boivin G, Engle S J, Tischfield J A. Environ Mol Mutagen. 1996;28:471–482. doi: 10.1002/(SICI)1098-2280(1996)28:4<471::AID-EM25>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 9.Zhu Y, Stambrook P J, Tischfield J A. Mol Carcinogen. 1993;8:138–144. doi: 10.1002/mc.2940080304. [DOI] [PubMed] [Google Scholar]
- 10.Shao C, Gupta P K, Sun Y, Sahota A, Tischfield J A. Cytogenet Cell Genet. 1996;75:216–221. doi: 10.1159/000134486. [DOI] [PubMed] [Google Scholar]
- 11.Gupta P K, Sahota A, Bye S, Boyadjiev S, Shao C, O’Neill P, Albertini R J, Stambrook P J, Tischfield J A. Cancer Res. 1997;57:1188–1193. [PubMed] [Google Scholar]
- 12.Engle S J, Stockelman M G, Chen J, Boivin G, Yum M N, Davis P M, Ying M Y, Sahota A, Simmonds M A, Stambrook P J, et al. Proc Natl Acad Sci USA. 1996;93:5307–5312. doi: 10.1073/pnas.93.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dev V G, Miller D A, Miller O J. Genetics. 1973;75:663–670. doi: 10.1093/genetics/75.4.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sahota A, Chen J, Behzadian M A, Ravindra R, Takeuchi H, Stambrook P J, Tischfield J A. Am J Hum Genet. 1991;48:983–989. [PMC free article] [PubMed] [Google Scholar]
- 15.Dietrich W F, Miller J C, Steen R, Merchant M A, Damron-Boles D, Husain Z, Dredge R D, Daly M J, Ingalls K A, O’Connor T J, et al. Nature (London) 1996;380:149–152. doi: 10.1038/380149a0. [DOI] [PubMed] [Google Scholar]
- 16.Sikela J M, Khan S A, Feliciano E, Trill J, Tischfield J A, Stambrook P J. Gene. 1983;22:219–228. doi: 10.1016/0378-1119(83)90106-3. [DOI] [PubMed] [Google Scholar]
- 17.Dickerman L H, Tischfield J A. Mutat Res. 1978;49:83–94. doi: 10.1016/0027-5107(78)90080-5. [DOI] [PubMed] [Google Scholar]
- 18.Ceci J D, Mills K A. Mamm Genome. 1998;8:s160–179. doi: 10.1007/s003359900653. [DOI] [PubMed] [Google Scholar]
- 19.Stanbridge E J, Tischfield J A, Schneider E L. Nature (London) 1975;256:329–321. doi: 10.1038/256329a0. [DOI] [PubMed] [Google Scholar]
- 20.Ramel C, Magnusson J. Mutat Res. 1992;267:221–227. doi: 10.1016/0027-5107(92)90066-b. [DOI] [PubMed] [Google Scholar]
- 21.Panthier J J, Guenet J L, Condamine H, Jacob F. Genetics. 1990;125:175–182. doi: 10.1093/genetics/125.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winton D J, Blount M A, Ponder B A J. Nature (London) 1988;333:463–466. doi: 10.1038/333463a0. [DOI] [PubMed] [Google Scholar]
- 23.Tao K S, Heddle J A. Mutagenesis. 1994;9:187–191. doi: 10.1093/mutage/9.3.187. [DOI] [PubMed] [Google Scholar]
- 24.Clarke A R, Howard L A, Harrison D J, Winton D J. Oncogene. 1997;14:2015–2018. doi: 10.1038/sj.onc.1201040. [DOI] [PubMed] [Google Scholar]
- 25.Van Sloun P P H, Wijnhoven S W P, Kool H J M, Slater R, Weeda G, van Zeeland A A, Lohman P H M, Vrieling H. Nucleic Acids Res. 1998;26:4888–4894. doi: 10.1093/nar/26.21.4888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper G E, Khattar N H, Bishop P L, Turker M S. Somatic Cell Mol Genet. 1992;18:215–225. doi: 10.1007/BF01233858. [DOI] [PubMed] [Google Scholar]
- 27.Turker M S, Pieretti M, Kumar S. Mutat Res. 1997;374:201–208. doi: 10.1016/s0027-5107(96)00230-8. [DOI] [PubMed] [Google Scholar]
- 28.Chu E H, Malling H V. Proc Natl Acad Sci USA. 1968;61:1306–1312. doi: 10.1073/pnas.61.4.1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luria S E, Delbruck M. Genetics. 1943;28:491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dempsey J L, Odagiri Y, Morley A A. Mutat Res. 1993;285:45–51. doi: 10.1016/0027-5107(93)90050-p. [DOI] [PubMed] [Google Scholar]
- 31.Polani P E. Chromosoma. 1972;36:343–374. doi: 10.1007/BF00336793. [DOI] [PubMed] [Google Scholar]
- 32.Maudlin I, Evans E P. Chromosoma. 1980;80:49–56. doi: 10.1007/BF00327565. [DOI] [PubMed] [Google Scholar]
- 33.Ashley T, Cacheiro N L A, Russell L B, Ward D C. Chromosoma. 1993;102:112–120. doi: 10.1007/BF00356028. [DOI] [PubMed] [Google Scholar]
- 34.Lawrie N M, Tease C, Hulten M A. Chromosoma. 1995;104:308–314. doi: 10.1007/BF00352262. [DOI] [PubMed] [Google Scholar]
- 35.Edelman W, Kroger B, Goller M, Horak I. Cell. 1989;57:937–946. doi: 10.1016/0092-8674(89)90332-2. [DOI] [PubMed] [Google Scholar]
- 36.Wahls W P, Wallace L J, Moore P D. Cell. 1990;60:95–103. doi: 10.1016/0092-8674(90)90719-u. [DOI] [PubMed] [Google Scholar]