Abstract
We describe a new reverse-genetics system that allows one to efficiently generate influenza A viruses entirely from cloned cDNAs. Human embryonic kidney cells (293T) were transfected with eight plasmids, each encoding a viral RNA of the A/WSN/33 (H1N1) or A/PR/8/34 (H1N1) virus, flanked by the human RNA polymerase I promoter and the mouse RNA polymerase I terminator—together with plasmids encoding viral nucleoprotein and the PB2, PB1, and PA viral polymerases. This strategy yielded >1 × 103 plaque-forming units (pfu) of virus per ml of supernatant at 48 hr posttransfection. The addition of plasmids expressing all of the remaining viral structural proteins led to a substantial increase in virus production, 3 × 104–5 × 107 pfu/ml. We also used reverse genetics to generate a reassortant virus containing the PB1 gene of the A/PR/8/34 virus, with all other genes representing A/WSN/33. Additional viruses produced by this method had mutations in the PA gene or possessed a foreign epitope in the head of the neuraminidase protein. This efficient system, which does not require helper virus infection, should be useful in viral mutagenesis studies and in the production of vaccines and gene therapy vectors.
The ability to generate infectious RNA viruses from cloned cDNAs has contributed greatly to our biological understanding of these pathogens and, hence, to improved methods of disease control (1). However, this progress had been relatively limited for negative-sense as compared with positive-sense RNA viruses, because neither the genomic viral RNA (vRNA) nor the antigenomic complementary RNA (cRNA) of negative-sense RNA viruses can serve as a direct template for protein synthesis. Rather, the vRNA, after its encapsidation by viral nucleoprotein (NP), must be transcribed into positive-sense mRNA by the viral RNA polymerase complex. Thus, the minimal replication unit is formed by the genomic vRNA complexed with NP and the polymerase proteins. Despite these obstacles, reverse-genetics methods have been established to produce nonsegmented, negative-sense RNA viruses, including rabies virus (2), vesicular stomatitis virus (3, 4), measles virus (5), respiratory syncytial virus (6), Sendai virus (7, 8), rinderpest virus (9), human parainfluenza virus type 3 (10), and simian virus 5 (11).
Generating segmented, negative-sense RNA viruses from cloned cDNAs poses a more formidable challenge, as one must produce a separate vRNA for each gene segment. In one study, Bridgen and Elliott (12) produced a Bunyamwera virus (family Bunyaviridae) from cloned cDNAs encoding three segments of negative-sense vRNA; however, the efficiency of virus recovery was low, and there have been no reports of an engineered Bunyamwera mutant. By contrast, none of the orthomyxoviruses, which contain six (Thogotovirus), seven (influenza C virus), or eight (influenza A and B viruses) segments of negative-sense RNA, have been produced entirely from cloned cDNAs. This lag in progress has been felt most acutely in efforts to control influenza virus infections.
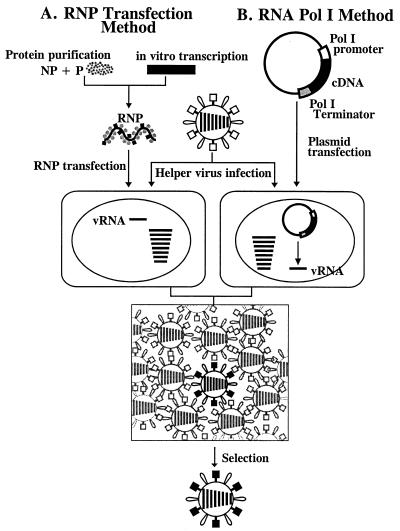
Palese and colleagues (13) pioneered the reverse-genetics, helper virus-dependent system for influenza A virus (Fig. 1A). In their approach, a ribonucleoprotein (RNP) complex is generated by in vitro vRNA synthesis in the presence of purified polymerase and NP proteins and then used to transfect eukaryotic cells. Subsequent infection with influenza A helper virus results in the generation of viruses possessing a gene derived from cloned cDNA. A second method, developed by Neumann et al. (14), is based on the in vivo synthesis of vRNA by RNA polymerase I (Fig. 1B), a cellular enzyme that transcribes ribosomal RNA that lacks both a 5′ cap and a 3′ poly(A) tail. Transfection of cells with a plasmid containing cloned influenza virus cDNAs, flanked by RNA polymerase I promoter and terminator sequences, followed by influenza virus infection, led to the production of transfectant viruses. With both methods, however, transfectants must be selected from a vast background of helper viruses, which requires a strong selection system and complicates the generation of growth-defective viruses.
Figure 1.
Schematic diagram of established reverse-genetics systems. In the RNP transfection method (A), purified NP and polymerase proteins are assembled into RNPs with the use of in vitro-synthesized vRNA. Cells are transfected with RNPs, followed by helper virus infection. In the RNA polymerase I method (B), a plasmid containing the RNA polymerase I promoter, a cDNA encoding the vRNA to be rescued, and the RNA polymerase I terminator are transfected into cells. Intracellular transcription by RNA polymerase I yields synthetic vRNA, which is packaged into progeny virus particles upon infection with helper virus. With both methods, transfectant viruses (i.e., those containing RNA derived from cloned cDNA) are selected from the helper virus population.
We report here the generation of influenza A viruses entirely from cloned cDNAs. The reverse-genetics approach we describe is highly efficient and can be used to introduce mutations into any gene segment and to develop influenza virus-based gene delivery systems.
MATERIALS AND METHODS
Cells and Viruses.
293T human embryonic kidney cells and Madin–Darby canine kidney cells (MDCK) were maintained in DMEM supplemented with 10% FCS and in MEM containing 5% newborn calf serum, respectively. The 293T cell line is a derivative of 293, into which the gene for the temperature-sensitive simian virus 40 T antigen has been inserted. This line produces replication-competent T antigen in large amounts at 37°C (15). All cells were maintained at 37°C in 5% CO2. Influenza viruses A/WSN/33 (H1N1) and A/PR/8/34 (H1N1) were propagated in 10-day-old eggs.
Construction of Plasmids.
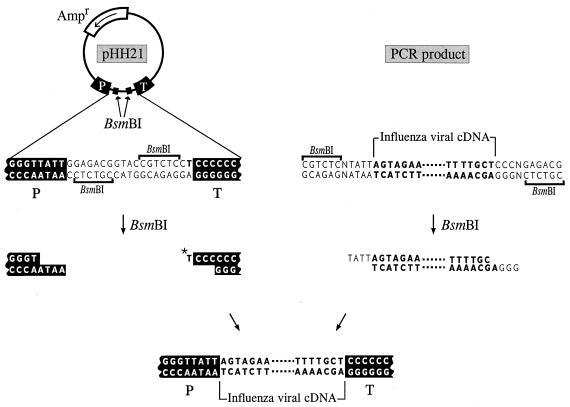
To generate RNA polymerase I constructs, we cloned cDNAs derived from A/WSN/33 or A/PR/8/34 viral RNA between the promoter and terminator sequences of RNA polymerase I. Briefly, the cloned cDNAs were amplified by PCR with primers containing BsmBI sites, digested with BsmBI, and cloned into the BsmBI sites of the pHH21 vector that contains the human RNA polymerase I promoter and the mouse RNA polymerase I terminator, separated by BsmBI sites (Fig. 2). The PB2, PB1, PA, hemagglutinin (HA), NP, neuraminidase (NA), M, and NS genes of the A/WSN/33 strain were PCR-amplified by use of the following plasmids: pSCWPB2, pGW-PB1, and pSCWPA (all obtained from Debi Nayak at the University of California, Los Angeles) and pWH17, pWNP152, pT3WNA15 (16), pGT3WM, and pWNS1, respectively. The PB1 gene of influenza A/PR/8/34 virus was amplified by using pcDNA774(PB1) (17) as a template. The sequences of the primers will be provided on request. To ensure that the genes were free of unwanted mutations, we sequenced PCR-derived fragments with an autosequencer (Applied Biosystems) according to the protocol recommended by the manufacturer. The cDNAs encoding the HA, NP, NA, and M1 genes of A/WSN/33 virus were cloned as described (18) and subcloned into the eukaryotic expression vector pCAGGS/MCS (controlled by the chicken β-actin promoter) (19), resulting in pEWSN-HA, pCAGGS-WSN-NP0/14, pCAGGS-WNA15, and pCAGGS-WSN-M1–2/1, respectively. The M2 and NS2 genes from the A/PR/8/34 virus were amplified by PCR and cloned into pCAGGS/MCS, yielding pEP24c and pCA-NS2. Finally, pcDNA774(PB1), pcDNA762(PB2), and pcDNA787(PA) were used to express the PB2, PB1, and PA proteins under control of the cytomegalovirus promoter, as reported elsewhere (17).
Figure 2.
Schematic diagram of the generation of RNA polymerase I constructs. cDNAs derived from influenza virus were amplified by PCR, digested with BsmBI, and cloned into the BsmBI sites of the pHH21 vector (34), which contains the human RNA polymerase I promoter (P) and the mouse RNA polymerase I terminator (T). The thymidine nucleotide upstream of the terminator sequence (*T) represents the 3′ end of the influenza viral RNA. Influenza A virus sequences are shown in boldface letters.
Generation of Infectious Influenza Particles.
293T cells (1 × 106) were transfected with a maximum of 17 plasmids in different amounts (as described in Results), with the use of Trans IT LT-1 (Panvera, Madison, WI) according to the manufacturer’s instructions. Briefly, DNA and transfection reagent were mixed (2 μl of Trans IT LT-1 per μg of DNA), incubated at room temperature for 45 min, and added to the cells. Six hours later, the DNA–transfection reagent mixture was replaced by Opti-MEM (GIBCO/BRL) containing 0.3% BSA and 0.01% FCS. At different times after transfection, we harvested viruses from the supernatant and titrated them in MDCK cells. Because helper virus was not required by this procedure, we analyzed the recovered transfectant viruses without plaque purification.
Determination of the Percentage of Plasmid-Transfected Cells Producing Viruses.
Twenty-four hours after transfection, 293T cells were dispersed with 0.02% EDTA into single cells. The cell suspension then was diluted 10-fold and transferred to confluent monolayers of MDCK cells in 24-well plates. Viruses were detected by the hemagglutination assay.
Immunostaining Assay.
Nine hours after infection with influenza virus, cells were washed twice with PBS and fixed with 3.7% paraformaldehyde (in PBS) for 20 min at room temperature. Next, they were treated with 0.1% Triton X-100 and processed as described earlier (20).
RESULTS
Generation of Infectious Virus by Plasmid-Driven Expression of Viral RNA Segments, Three Polymerase Subunits, and NP Protein.
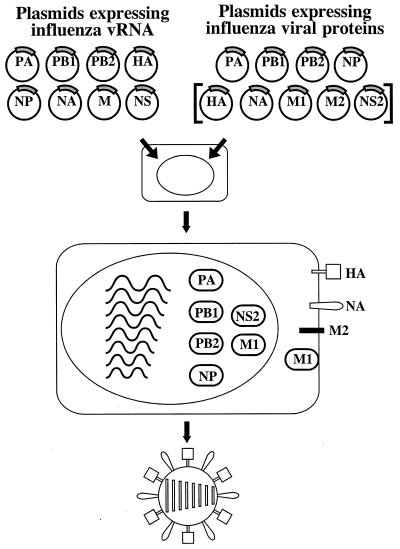
Although transfection of cells with a mixture of RNPs extracted from purified virions results in infectious influenza particles (unpublished data), this strategy is not likely to be efficient when used with eight different in vitro generated RNPs. To produce infectious influenza viruses entirely from cDNAs, we attempted to generate the eight viral RNPs in vivo. Thus, we constructed plasmids that contain cDNAs for the full-length viral RNAs of the A/WSN/33 virus, flanked by the human RNA polymerase I promoter and the mouse RNA polymerase I terminator. In principle, transfection of these eight plasmids into eukaryotic cells should result in the synthesis of all eight influenza vRNAs. The PB2, PB1, PA, and NP proteins, generated by cotransfection of protein expression plasmids, then should assemble the vRNAs into functional vRNPs that are replicated and transcribed, ultimately forming infectious influenza viruses (Fig. 3). To test this prediction, we transfected 1 × 106 293T cells with protein expression plasmids [1 μg of pcDNA762(PB2), 1 μg of pcDNA774(PB1), 0.1 μg of pcDNA787(PA), and 1 μg of pCAGGS-WSN-NP0/14] and 1 μg of each of the following RNA polymerase I plasmids: pPolI-WSN-PB2, pPolI-WSN-PB1, pPolI-WSN-PA, pPolI-WSN-HA, pPolI-WSN-NP, pPolI-WSN-NA, pPolI-WSN-M, and pPolI-WSN-NS. The decision to use a reduced amount of pcDNA787(PA) was based on previous observations (21) and our preliminary data on the optimal conditions for the generation of virus-like particles (VLPs) (unpublished data). Twenty-four hours after transfection of 293T cells, we found 7 × 103 pfu of virus per ml of supernatant (Experiment 1; Table 1), demonstrating the capacity of reverse genetics to produce influenza A virus entirely from plasmids.
Figure 3.
Reverse-genetics method for generating segmented, negative-sense RNA viruses entirely from cloned cDNA. Plasmids containing the RNA polymerase I promoter, a cDNA for each of the eight viral RNA segments, and the RNA polymerase I terminator are transfected into cells together with protein expression plasmids. Although infectious viruses can be generated with plasmids expressing PA, PB1, PB2, and NP, expression of all remaining structural proteins (shown in brackets) increases the efficiency of virus production (see text).
Table 1.
Plasmid sets used to produce influenza virus from cloned cDNA*
| Experiment
|
||||||||
|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |
| RNA polymerase I plasmids for† | ||||||||
| PB1 | + | + | − | − | − | − | − | − |
| PR8-PB1 | − | − | + | + | + | + | + | + |
| PB2 | + | + | + | + | + | + | + | + |
| PA | + | + | + | + | + | + | + | + |
| HA | + | + | + | + | + | + | + | + |
| NP | + | + | + | + | + | + | + | + |
| NA | + | + | + | + | + | + | + | + |
| M | + | + | + | + | + | + | + | + |
| NS | + | + | + | + | + | + | + | + |
| Protein expression plasmids for | ||||||||
| PB1 | + | + | + | + | − | + | + | + |
| PB2 | + | + | + | + | + | − | + | + |
| PA | + | + | + | + | + | + | − | + |
| NP | + | + | + | + | + | + | + | − |
| HA | − | + | − | + | + | + | + | + |
| NA | − | + | − | + | + | + | + | + |
| M1 | − | + | − | + | + | + | + | + |
| M2 | − | + | − | + | + | + | + | + |
| NS2 | − | + | − | + | + | + | + | + |
| Virus titer, pfu/ml | 7 × 103 | 7 × 103 | 1 × 103 | 3 × 104 | 0 | 0 | 0 | 0 |
293T cells were transfected with the indicated plasmids. Twenty-four (Experiments 1 and 2) or 48 hr (Experiments 3–8) later, the virus titer in the supernatant was determined in MDCK cells.
Unless otherwise indicated, plasmids were constructed with cDNAs representing the RNAs of A/WSN/33 virus.
Efficiency of Influenza Virus Production with Coexpression of All Viral Structural Proteins.
Although expression of the viral NP and polymerase proteins is sufficient for the plasmid-driven generation of influenza viruses, we sought to improve on this method. In previous studies, the expression of all influenza virus structural proteins (PB2, PB1, PA, HA, NP, NA, M1, M2, and NS2) resulted in VLPs that contained an artificial vRNA encoding a reporter chloramphenicol-acetyltransferase gene (21). We therefore reasoned that the availability of the entire complement of structural proteins, instead of only those required for viral RNA replication and transcription, might improve the efficiency of virus production. To this end, we transfected 293T cells with optimal amounts of viral protein expression plasmids (as judged by VLP production; unpublished data): 1 μg of pcDNA762(PB2) and pcDNA774(PB1); 0.1 μg of pcDNA787(PA); 1 μg of pEWSN-HA, pCAGGS-WSN-NP0/14, and pCAGGS-WNA15; 2 μg of pCAGGS-WSN-M1–2/1; 0.3 μg of pCA-NS2; and 0.03 μg of pEP24c (for M2), together with 1 μg of each RNA polymerase I plasmid (Experiment 2; Table 1). A second set of cells was transfected with the same set of RNA polymerase I plasmids, with the exception of the PB1 gene, for which we substituted pPolI-PR/8/34-PB1 in an effort to generate a reassortant virus, together with plasmids expressing only PA, PB1, PB2, and NP (Experiment 3; Table 1) or those expressing all the influenza structural proteins (Experiment 4; Table 1). In one experiment, yields of WSN virus did not differ appreciably at 24 hr (Experiments 1 and 2; Table 1) posttransfection. However, in other experiments, we found more than a 10-fold increase in yields of WSN virus (Experiments 1 and 2; Table 2) and the virus with PR/8/34-PB1 (Experiments 3 and 4; Table 1) when all the influenza viral structural proteins were provided. Negative controls, which lacked one of the plasmids for the expression of PA, PB1, PB2, or NP proteins, did not yield any virus (Experiments 5–8; Table 1). Thus, expression of all influenza A virus structural proteins appreciably improved the efficiency of our reverse-genetics method.
Table 2.
Kinetics of virus production after plasmid transfection into 293T cells
| Hours after plasmid transfection | Virus titers in culture supernatant, pfu/ml
|
|||
|---|---|---|---|---|
| Experiment
| ||||
| 1 | 2 | 3 | 4 | |
| 12 | 0 | 0 | ND | 0 |
| 18 | 0 | 3 × 10 | ND | 0 |
| 24 | 2 × 102 | 8 × 103 | 2 × 103 | 6 × 103 |
| 30 | 4 × 103 | 1 × 105 | 5 × 104 | 9 × 104 |
| 36 | 5 × 104 | 2 × 106 | >1 × 105 | 7 × 105 |
| 42 | 9 × 105 | 1 × 107 | >1 × 106 | 5 × 106 |
| 48 | 7 × 106 | 5 × 107 | >1 × 106 | 1 × 107 |
293T cells were transfected with eight RNA polymerase I plasmids encoding A/WSN/33 virus genes (Experiments 1 and 2) or with these plasmids except the PB1 gene, which was derived from A/PR/8/34 virus (Experiments 3 and 4), and only PA, PB1, PB2, and NP (Experiment 1) or nine (Experiments 2–4) protein expression plasmids as described in the text. At different time points, we titrated virus in the culture supernatant in MDCK cells. ND, not determined.
Next, we determined the kinetics of virus production after transfection of cells with the set of plasmids used to generate WSN virus (Experiments 1 and 2; Table 2) or the virus with the A/PR/8/34-PB1 gene (Experiments 3 and 4; Table 2). In three of four experiments, virus was first detected at 24 hr after transfection. The titer measured at that time, >103 pfu/ml (with all nine structural proteins), had increased to >106 pfu/ml by 48 hr after transfection (Table 2). To estimate the percentage of plasmid-transfected cells that were producing viruses, we treated 293T cells with EDTA (0.02%) at 24 hr after transfection to disperse the cells and then performed limiting dilution studies. In this experiment, no free virus was found in the culture supernatant at this time point. The results indicated that 1 in 102.8–103.3 cells was generating infectious virus particles. Although this number of virus-producing cells may appear low considering the high transfection efficiency of 293T cells (>90%; ref. 23), this is likely because not all of the cells are transfected with an optimal amount of plasmids for virus recovery.
Recovery of Influenza Virus Containing the FLAG Epitope in the NA Protein.
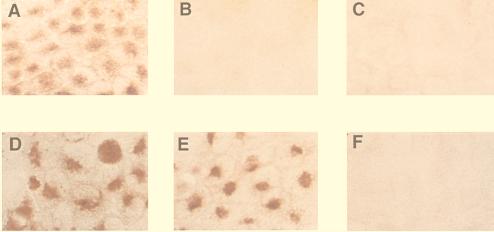
To verify that the new reverse-genetics system allowed the introduction of mutations into the genome of influenza A viruses, we attempted to generate a virus containing a Flag epitope (16) in the NA protein. 293T cells were transfected with an RNA polymerase I plasmid (pPolI-WSN-NA/FL79) that contained a cDNA encoding the NA protein and a Flag epitope at the bottom of the protein’s head, together with the required RNA polymerase I and protein expression plasmids. To confirm that the recovered virus (PR8-WSN-FL79) did, in fact, express the NA-Flag protein, we performed immunostaining assays of cells infected with PR8-WSN-FL79 or A/WSN/33 wild-type virus. A mAb to the Flag epitope detected cells infected with PR8-WSN-FL79, but not those infected with wild-type virus (Fig. 4). Recovery of the PR8-WSN-FL79 virus was as efficient as that for the untagged wild-type virus (data not shown). These results indicate that the new reverse-genetics system allows one to introduce mutations into the influenza A virus genome.
Figure 4.
Detection of the Flag epitope in cells infected with a transfectant virus. Antibody staining was used to identify the NA in MDCK cells infected with either PR8-WSN-FL79 (A and D) or A/WSN/33 wild-type virus (B and E) or on mock-infected MDCK cells (C and F). Nine hours after infection, cells were fixed with paraformaldehyde, treated with Triton X-100, and incubated with either anti-Flag (A–C) or anti-WSN NA (D–F) mAbs. Intensive Golgi staining (red) is apparent in positive samples (A, D, and E).
Generation of Infectious Influenza Virus Containing Mutations in the PA Gene.
To test further the utility of our new reverse-genetics system, we attempted to produce viruses possessing mutations in the PA gene. Previously, it was not possible to modify this gene by reverse genetics, because of the lack of a reliable selection system. The two silent mutations that we introduced created new recognition sequences for restriction endonucleases (Bsp120I at position 846 or PvuII at position 1284 of the cRNA), allowing us to distinguish between the wild-type and mutated PA genes. We recovered the transfectant viruses, PA-T846C and PA-A1284C, by using the method described above. The recovered transfectant viruses were biologically cloned by two consecutive limiting dilutions. To verify that the recovered viruses indeed were transfectants with mutations in the PA gene, we produced cDNA for the PA gene by reverse transcriptase–PCR. As shown in Fig. 5, PA-T846C and PA-A1284C viruses had the expected mutations within the PA gene, as demonstrated by the presence of the newly introduced restriction sites. PCR of the same viral samples and primers without the reverse transcription step failed to produce any products (data not shown), indicating that the PA cDNA indeed was originated from vRNA instead of the plasmid used to generate the viruses. These results illustrate how viruses with mutated genes can be produced and recovered without the use of helper viruses.
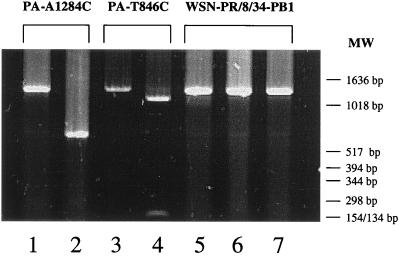
Figure 5.
Recovery of PA mutants. The PA gene of each virus was amplified by reverse transcriptase–PCR with primers that yield a 1,226-bp fragment (position 677-1903 of the mRNA; lanes 1, 3, and 5), which then was digested with the restriction enzyme Bsp120I (at position 846 of the mRNA; lanes 4 and 7) or PvuII (at position 1284 of the mRNA; lanes 2 and 6). The presence of Bsp120I or PvuII sites in the PCR products yielded either 169- and 1,057-bp or 607- and 619-bp fragments, respectively. MW, molecular weight markers.
DISCUSSION
The reverse-genetics systems described in this report allows one to efficiently produce influenza A viruses entirely from cloned cDNAs. Bridgen and Elliott (12) also used reverse genetics to generate a Bunyamwera virus (Bunyaviridae family), but it contains only three segments of negative-sense RNA, and the efficiency of its production was low, 102 pfu/107 cells. Although the virus yields differed among the experiments, we consistently found 103–107 pfu/106 cells for influenza virus, which contains eight segments. There are several explanations for the high efficiency of our reverse-genetics system. Instead of producing RNPs in vitro (22), we generated them in vivo through intracellular synthesis of vRNAs by using RNA polymerase I and through plasmid-driven expression of the viral polymerase proteins and NP. Also, the use of 293T cells, which are readily transfected with plasmids (23), ensured that a large population of cells received all of the plasmids needed for virus production. In addition, we produced vRNA by using cellular RNA polymerase I, which is among the most abundantly expressed enzymes in growing cells. These features likely contributed to the overall efficiency of our system. In nonsegmented negative-strand RNA viruses, the use of plus-sense RNA (i.e., cRNA) improved recovery of infectious viruses (2) by avoiding the possible annealing of (−)vRNA with vast amounts of the (+)mRNAs produced from the plasmids for protein expression. A similar approach (i.e., generation of cRNA instead of vRNA) might even improve the high efficiency of influenza virus recovery described in this study.
Previously established reverse-genetics systems (13, 14, 22, 24) require helper-virus infection and, therefore, selection methods that permit a small number of transfectants to be retrieved from a vast number of helper viruses. Such strategies have been employed to generate influenza viruses that possess one of the following cDNA-derived genes: PB2 (25), HA (26, 27), NP (28), NA (13), M (29, 30), and NS (31). Most of the selection methods, except for those applicable to the HA and NA genes, rely on growth temperature, host-range restriction, or drug sensitivity, thus limiting the utility of reverse genetics for functional analysis of the gene products. Even with the HA and NA genes, for which reliable, antibody-driven selection systems are available, it is difficult to produce viruses with prominent growth defects. By contrast, the reverse-genetics system we describe does not require helper virus and permits one to generate transfectants with mutations in any gene segment or with severe growth defects. This advantage is demonstrated in Fig. 5, which shows the recovery of transfectant viruses with an artificially mutated PA gene. Having the technology to introduce any viable mutation into the influenza A virus genome will enable investigators to address a number of long-standing issues, such as the nature of regulatory sequences in nontranslated regions of the viral genome, structure–function relationships of viral proteins, and the molecular basis of host-range restriction and viral pathogenicity.
Although inactivated influenza vaccines are available, their efficacy is suboptimal partly because of their limited ability to elicit local IgA and cytotoxic T cell responses. Clinical trials of cold-adapted live influenza vaccines now under way suggest that such vaccines are optimally attenuated, so that they will not cause influenza symptoms but will still induce protective immunity (reviewed in ref. 32). However, preliminary results indicate that these live virus vaccines will not be significantly more effective than the best inactivated vaccine (reviewed in ref. 32), leaving room for further improvement. One possibility would be to modify a cold-adapted vaccine with the reverse-genetics system established in this report. Alternatively, one could start from scratch by using reverse genetics to produce a “master” influenza A strain with multiple attenuating mutations in the genes that encode internal proteins. The most intriguing application of our reverse-genetics system may lie in the rapid production of attenuated live-virus vaccines in cases of suspected pandemics involving new HA or NA subtypes of influenza virus.
This new reverse-genetics system will likely enhance the use of influenza viruses as vaccine vectors. The viruses can be engineered to express foreign proteins or immunogenic epitopes in addition to the influenza viral proteins. One could, for example, generate viruses with foreign proteins as a ninth segment, as described (31), and use them as live vaccines. Not only do influenza viruses stimulate strong cell-mediated and humoral immune responses, but they also afford a wide array of virion surface HA and NA proteins (e.g., 15 HA and 9 NA subtypes and their epidemic variants), allowing repeated immunization of the same target population.
Influenza VLPs possessing an artificial vRNA encoding a reporter gene have been produced by expressing viral structural proteins and vRNA with the vaccinia-T7 polymerase system (21). We recently improved on this method by generating VLPs entirely from plasmids (unpublished data). Thus, using reverse genetics, one can now generate VLPs containing vRNAs that encode proteins required for vRNA transcription and replication (i.e., PA, PB1, PB2, and NP), as well as vRNAs encoding proteins of interest. Such VLPs could be useful gene-delivery vehicles. Importantly, their lack of genes encoding viral structural proteins would ensure that infectious viruses would not be produced after VLP-gene therapy. Because the influenza virus genome is not integrated into host chromosome, the VLP system would be suitable for gene therapy in situations requiring only short-term transduction of cells (e.g., for cancer treatment). In contrast to adenovirus vectors (33), influenza VLPs could contain both HA and NA variants, allowing repeated treatment of target populations.
The family Orthomyxoviridae comprises influenza A, B, and C viruses, as well as the recently classified Thogotovirus. We suggest that our strategy for generating infectious influenza A viruses entirely from cloned cDNAs would apply to any orthomyxovirus and perhaps to other segmented, negative-sense RNA viruses as well (e.g., Bunyaviridae, Arenaviridae). The ability to manipulate the viral genome without technical limitations has profound implications for the study of viral life cycles and their regulation, the function of viral proteins, and the molecular mechanisms of viral pathogenicity.
Acknowledgments
We thank Krisna Wells and Martha McGregor for excellent technical assistance and John Gilbert for editing the manuscript. We are also grateful to Dr. Debi Nayak for WSN polymerase plasmids, Dr. A. James Cooley for immunostaining pictures, and Yuko Kawaoka for illustrations. Automated sequencing was performed at the University of Wisconsin–Biotechnology Center. Support for this work came from National Institute of Allergy and Infectious Diseases Public Health Service research grants.
ABBREVIATIONS
- cRNA
complementary RNA
- MDCK
Madin–Darby canine kidney
- HA
hemagglutinin
- NA
neuraminidase
- NP
nucleoprotein
- pfu
plaque-forming units
- RNP
ribonucleoprotein complex
- VLP
virus-like particle
- vRNA
viral RNA
Footnotes
A Commentary on this article begins on page 8804.
References
- 1.Palese P, Zheng H, Engelhardt O G, Pleschka S, Garcia-Sastre A. Proc Natl Acad Sci USA. 1996;93:11354–11358. doi: 10.1073/pnas.93.21.11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schnell M J, Mebatsion T, Conzelmann K K. EMBO J. 1994;13:4195–4203. doi: 10.1002/j.1460-2075.1994.tb06739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lawson N D, Stillman E A, Whitt M A, Rose J K. Proc Natl Acad Sci USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Whelan S P, Ball L A, Barr J N, Wertz G T. Proc Natl Acad Sci USA. 1995;92:8388–8392. doi: 10.1073/pnas.92.18.8388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dotsch C, Christiansen G, Billeter M A. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins P L, Hill M G, Camargo E, Grosfeld H, Chanock R M, Murphy B R. Proc Natl Acad Sci USA. 1995;92:11563–11567. doi: 10.1073/pnas.92.25.11563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcin D, Pelet T, Calain P, Roux L, Curran J, Kolakofsky D. EMBO J. 1995;14:6087–6094. doi: 10.1002/j.1460-2075.1995.tb00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato A, Sakai Y, Shioda T, Kondo T, Nakanishi M, Nagai Y. Genes Cells. 1996;1:569–579. doi: 10.1046/j.1365-2443.1996.d01-261.x. [DOI] [PubMed] [Google Scholar]
- 9.Baron M D, Barrett T. J Virol. 1997;71:1265–1271. doi: 10.1128/jvi.71.2.1265-1271.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoffman M A, Banerjee A K. J Virol. 1997;71:4272–4277. doi: 10.1128/jvi.71.6.4272-4277.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He B, Paterson R G, Ward C D, Lamb R A. Virology. 1997;237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- 12.Bridgen A, Elliott R M. Proc Natl Acad Sci USA. 1996;93:15400–15404. doi: 10.1073/pnas.93.26.15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enami M, Luytjes W, Krystal M, Palese P. Proc Natl Acad Sci USA. 1990;87:3802–3805. doi: 10.1073/pnas.87.10.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neumann G, Zobel A, Hobom G. Virology. 1994;202:477–479. doi: 10.1006/viro.1994.1365. [DOI] [PubMed] [Google Scholar]
- 15.DuBridge R B, Tang P, Hsia H C, Leong P M, Miller J H, Calos M P. Mol Cell Biol. 1987;7:379–387. doi: 10.1128/mcb.7.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castrucci M R, Bilsel P, Kawaoka Y. J Virol. 1992;66:4647–4653. doi: 10.1128/jvi.66.8.4647-4653.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez D R, Donis R O. Virology. 1998;249:52–61. doi: 10.1006/viro.1998.9318. [DOI] [PubMed] [Google Scholar]
- 18.Huddleston J A, Brownlee G G. Nucleic Acids Res. 1982;10:1029–1037. doi: 10.1093/nar/10.3.1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niwa H, Yamamura K, Miyazaki J. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 20.Neumann G, Castrucci M R, Kawaoka Y. J Virol. 1997;71:9690–9700. doi: 10.1128/jvi.71.12.9690-9700.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mena I, Vivo A, Pérez E, Portela A. J Virol. 1996;70:5016–5024. doi: 10.1128/jvi.70.8.5016-5024.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luytjes W, Krystal M, Enami M, Parvin J D, Palese P. Cell. 1989;59:1107–1113. doi: 10.1016/0092-8674(89)90766-6. [DOI] [PubMed] [Google Scholar]
- 23.Goto H, Bethell R C, Kawaoka Y. Virology. 1997;238:265–272. doi: 10.1006/viro.1997.8810. [DOI] [PubMed] [Google Scholar]
- 24.Pleschka S, Jaskunas S R, Engelhardt O G, Zürcher T, Palese P, García-Sastre A. J Virol. 1996;70:4188–4192. doi: 10.1128/jvi.70.6.4188-4192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subbarao E K, Kawaoka Y, Murphy B R. J Virol. 1993;67:7223–7228. doi: 10.1128/jvi.67.12.7223-7228.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Enami M, Palese P. J Virol. 1991;65:2711–2713. doi: 10.1128/jvi.65.5.2711-2713.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horimoto T, Kawaoka Y. J Virol. 1994;68:3120–3128. doi: 10.1128/jvi.68.5.3120-3128.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li S, Xu M, Coelingh K. Virus Res. 1995;37:153–161. doi: 10.1016/0168-1702(95)00031-k. [DOI] [PubMed] [Google Scholar]
- 29.Castrucci M R, Kawaoka Y. J Virol. 1995;69:2725–2728. doi: 10.1128/jvi.69.5.2725-2728.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yasuda J, Bucher D J, Ishihama A. J Virol. 1994;68:8141–8146. doi: 10.1128/jvi.68.12.8141-8146.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enami M, Sharma G, Benham C, Palese P. Virology. 1991;185:291–298. doi: 10.1016/0042-6822(91)90776-8. [DOI] [PubMed] [Google Scholar]
- 32.Keitel W A, Piedra P A. In: Textbook of Influenza. Nickolson K G, Webster R G, Hay A, editors. Oxford: Blackwell; 1998. pp. 373–390. [Google Scholar]
- 33.Kovesdi I, Brough D E, Bruder J T, Wickham T J. Curr Opin Biotechnol. 1997;8:583–589. doi: 10.1016/s0958-1669(97)80033-x. [DOI] [PubMed] [Google Scholar]
- 34.Hoffmann E. Ph.D. thesis. Giessen, Germany: Justus-Liebig-University; 1997. [Google Scholar]