Abstract
In this communication, we report an in-depth structure-based analysis of the human CYP1b1 protein carrying disease-causing mutations that are discovered in patients suffering from primary congenital glaucoma (PCG). The “wild-type” and the PCG mutant structures of the human CYP1b1 protein obtained from comparative modeling were subjected to long molecular dynamics simulations with an intention of studying the possible impact of these mutations on the protein structure and hence its function. Analysis of time evolution as well as time averaged values of various structural properties—especially of those of the functionally important regions: the heme binding region, substrate binding region, and substrate access channel—gave some insights into the possible structural characteristics of the disease mutant and the wild-type forms of the protein. In a nutshell, compared to the wild-type the core regions in the mutant structures are associated with subtle but significant changes, and the functionally important regions seem to adopt such structures that are not conducive for the wild-type-like functionality.
INTRODUCTION
Primary congenital glaucoma (PCG) is a severe eye disorder occurring at birth or early childhood (seen up to 3 years) and is a major cause of blindness in infancy. This disease is characterized by a developmental abnormality of the trabecular meshwork in the anterior chamber angle of the eye leading to increased intraocular pressure, which results in optic nerve damage and permanent loss of vision (1). PCG has been found to be mainly autosomal recessive in inheritance. The prevalence of the disease has been reported to vary geographically, with incidence ratios as low as 1:10,000 in the West to as high as 1:1,250 among the Slovak gypsies (2). High incidence ratios have usually been found in populations that practice inbreeding and consanguinity.
Genetic linkage studies have indicated that PCG is, genetically, a heterogeneous disease, mapping onto at least three different loci. The first locus is GLC3A, located in the region 2p21 of chromosome 2 (3); the second is GLC3B, located in the region 1p36 of chromosome 1 (4); and the third is GLC3C, located in the region 14q24.3 on chromosome 14 (5). In the majority of the PCG instances the candidate locus has been found to be GLC3A, which codes for a cytochrome p450 protein called CYP1b1.
The human CYP1b1 is a 543 amino acid long protein made up of three regions: the 53 residue long membrane-bound N-terminal region, a 10 residue long proline-rich region called “the hinge”, and the 480 residue long cytosolic globular domain. It is thought that CYP1b1 participates in the development of the trabecular meshwork of eye, which serves as a filter for drainage of anterior chamber fluid (6). To date, the mechanism or the pathway by which CYP1b1 functions in the development of the trabecular meshwork is not known. However, it is suspected that CYP1b1 may be involved in the elimination of a metabolite, the presence of which may have a toxic effect on eye development. It has also been thought that the protein may be involved in the generation of a regulatory molecule which controls the expression of genes involved in the development of the anterior chamber angle of the eye (7). It is certain that mutations having deleterious effects on protein function do hamper the normal development of the trabecular meshwork.
Sequence analyses have so far revealed several mutations in the CYP1b1 gene, of which some are found only in the PCG affected individuals (6,8–17). Recently, Panicker and co-workers (18,19) reported eight mutations in the Indian PCG patients. Our preliminary study had indicated that some of these nonsynonymous mutations occur in the conserved structural regions of the protein (18). It was therefore tempting to carry out detailed studies to explore the possible impact of these mutations on the protein structure and function. In this study, the wild-type (WT) and the mutant (MT) structures corresponding to the eight PCG mutations, A-115P, M-132R, Q-144P, P-193L, E-229K, S-239R, R-368H, and G-466D, have been obtained by comparative modeling approaches. The models were subjected to long molecular dynamics (MD) simulations with the intention of studying the time evolution as well as time averaged values of structural properties, especially of the functionally important regions (FIRs). Our investigations have revealed that the MT structures show differential structural properties during the simulation period compared to the WT. Very importantly, structural characteristics of FIRs are quite different in the MT structures than the WT, and these altered structural properties may not be conducive to the enzymatic function.
MATERIALS AND METHODS
Modeling of wild-type and mutant structures
The human CYP1b1 structure was modeled by means of comparative modeling procedures using the human CYP2c9 as the template. The sequence alignment of CYP1b1 and CYP2c9 was carried out using the CLUSTALX (http://www.ebi.ac.uk/clustalw) program and was further refined manually at some loop regions of the template. The academic version of MODELLER v.4.0 (20) (http://salilab.org/modeller) installed on the SGI-O2 system was used for model building. Of the 20 models generated by MODELLER, the one with the best G-score of PROCHECK (21) and with the best VERIFY-3D (22) profile was subjected to energy minimization (EM) followed by MD simulation for 300 ps (details of the protocol used are given in the following sections). All the instantaneous structures generated were assessed for their stereochemical quality using PROCHECK G-score, and the one with the best G-score was selected as the representative of WT structure. The WT structure was also further used as the starting structure to generate disease MT structures by replacing WT residues with disease associated residues. Overall, eight disease MT structures were generated corresponding to the eight point mutations A-115P, M-132R, Q-144P, P-193L, E-229K, S-239R, R-368H, and G-466D (18,23). The residue replacements were carried out using the MUTATE_MODEL command of MODELLER.
Identification of functionally important regions
CYPs are characterized by a highly conserved structural core divisible into α-rich and β-rich domains comprising ∼14 helices and four to six β-sheets. Functionally, CYP structure can also be characterized by means of five FIRs, viz., the heme binding region (HBR), the substrate binding region (SBR), the substrate access channel (SAC), the residues involved in charge relay, and the surface region that binds to CYP reductase protein while receiving electrons from it. For the purpose of this study some of these regions, viz., HBR, SBR, and SAC, were putatively identified as follows.
HBR
The HBR is defined by a set of residues that are in contact with the heme cofactor and such residues were identified as follows. The solvent accessible surface area (SASA) of all residues in the protein with and without the heme were calculated, and the residues that showed a decrease in SASA in the presence of heme were denoted as those comprising the HBR.
SBR
To identify the SBR, the crystal structures of various CYP protein-ligand complexes (Protein Data Bank (PDB) codes: 2cpp, 1pha, 6cp4, 1fag, 1e9x, 1egy, 1ea1, 1f4t) were used. The SBR residues in each structure were identified as those showing an increase in SASA upon the removal of the bound ligand in the CYP-substrate complexes. The putative SBR residues in CYP1b1 were then identified, as those found at the structurally equivalent positions in CYP1b1 in the multiple structural alignment of CYP1b1 with CYP-substrate complex structures.
SAC
The substrate access into the active site is thought to occur through an opening between the F/G loop and the B′-helix. The pathway (SAC) from outside to the active site of the protein is bounded by the F- and G-helices and the B/C loop. Ludemann et al. (24,25) have studied the substrate access pathway in P450cam and P450BM-3. They computationally demonstrated that these structures are very likely to be involved in substrate entry and exit. For our analysis we considered the SAC region as the residues comprising the SAC entrance, viz., the F/G loop and B′-helix.
Molecular dynamics simulations
The GROMACS (26) program package (http://www.gromacs.org) adopting the GROMOS96 force field parameters was used for EM and MD simulations. For the MD simulation studies, the structures were solvated (spc216 water representation) and Cl− counterions (3,3,4,3,3,5,4,2, and 2 for WT, A-115P, M-132R, Q-144P, P-193L, E-229K, S-239R, R-368H, and G-466D, respectively) were added as required to neutralize the total charge of the system by using the GENION option of GROMACS. The system consisted of 41820, 41821, 41823, 41824, 41825, 41828, 41836, 41820, and 41832 atoms, respectively, in WT, A-115P, M-132R, Q-144P, P-193L, E-229K, S-239R, R-368H, and G-466D. The solvated structures were energy minimized using the steepest descent method, terminating when maximum force is found smaller than 100 KJ/mol−1/nm−1. The energy-minimized structures were subjected to position restrained dynamics for 25 ps keeping the whole protein molecule fixed and allowing only the water molecules to move to equilibrate with respect to the protein structure. This followed dynamics simulations of the full system (protein and water) without any positional restraints.
All the simulations were performed in the NVT ensemble at constant temperature (300 K) and pressure (1 bar) with a time step of 2 fs. The nonbonded pair list was updated every five steps. The electrostatic interactions were calculated using the particle mesh Ewald summation method (27). During the simulation, constraints were applied on all bonds using the LINCS (28) algorithm, with parameters LINCS-order-4 and LINCS-warn-angle = 30. Coordinates were saved at regular time intervals of every 1 ps.
Analysis of MD trajectories
Most of the structural properties, such as root mean-square deviation (RMSD), root mean-square fluctuation (RMSF), SASA, secondary structure calculation, hydrogen bonds (Hbs), etc., were computed using the built-in functions of GROMACS. Individual Hb interactions were calculated using the HBPLUS (29) program. The volume of SBR was computed using the program POCKET (30) with a water probe radius of 1.4 Å. All the average properties, including Hb interactions, were computed after the time of equilibration of the systems. The SWISS-PDB VIEWER (31) and PSA (32) programs were used for superimpositions and solvent accessibility calculations, respectively.
RESULTS
Modeling CYP1b1 and its mutant forms
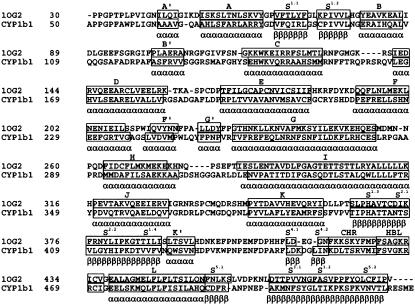
The experimentally (x-ray or NMR) determined structure of the human CYP1b1 is not yet available, and therefore we built a model by means of a comparative modeling procedure. A recently reported x-ray structure of CYP2c9 (33) (1OG2) was used as the structural template. The sequence alignment used for model building is shown in Fig. 1. The alignment is characterized by some insertions and deletions in the loop regions. Since the first 49 residues from the N-terminal end and the last 16 residues at the C-terminal (residues 528–543) in CYP1b1 did not have corresponding equivalent regions in CYP2c9, the modeling was carried out from the 50th to the 527th residue, followed by a rigorous refinement of the model by means of MD and EM (see the Materials and Methods section for the protocol used). The model (referred to as the wild-type and denoted by WT) has 92% of the residues in the most favored regions of the Ramachandran Map with a PROCHECK's (21) G-score value of −0.21 and a satisfactory VERIFY-3D (22) profile (Supplementary Material). The model was used to identify the structural parts which make up the HBR, the SBR, and the SAC, i.e., the three functionally FIRs (see the Materials and Methods section). The disease structure's MTs were modeled by replacing the side chains of the WT with the PCG mutations. In total, eight MTs were obtained for the eight PCG mutations.
FIGURE 1.
Sequence alignment of CYP1b1 (residues 50–527) with 1OG2 template (residues 30–490). The α-helices (boxed regions) are named in alphabetical order (A–L). The β-strands (boxed regions) are named using the sheet number (S1–S5) in which they occur followed by the strand number. The CHR and HBL regions are also represented by boxed regions. The nomenclature for secondary structures was adopted from the nomenclature generally followed in CYP1b1 literature especially from Akarsu et al. (4). Minor helices are indicated with a prime (A′, B′, F′, G′, and K′).
Mapping of PCG mutations on the CYP structure
We examined the mutation sites in the WT vis à vis the three FIRs, viz., HBR, SBR, and SAC. It is observed that some of the mutation sites are either among the residues comprising the FIRs or near to them, whereas the others are far from any of the FIRs (Supplementary Material). Mutations in spatially close sites can be expected to exert an influence on FIRs. Comparison of the WT and the MTs in the vicinity of the mutations gave rise to the following observations.
A-115P
This mutation occurs within the B/B′ loop at the C-terminal end of B-helix and is in close proximity to the HBR. A to P substitution not only restricts conformational freedom at the site but also precludes hydrogen-bonding interaction capability due to the absence of the amide hydrogen.
M-132R
This mutation site is in the HBR, in the loop connecting B′- and C-helices. In WT the side chain of M extends into the interior and packs well between I-helix (residues N-319–Y-349) and one of the heme propionate groups. The N of M-132 donates an Hb to D-326 of I-helix and accepts Hb from G-135, W-141, and R-468, thus interconnecting the I-helix, C-helix, and heme binding loop (HBL). Mutation to R leads to congestion in the packing in the immediate vicinity of the mutation site, thereby potentially harming the H-bond interactions.
Q-144P
This mutation occurs in the middle of C-helix and is also very close to the HBR. Mutation to P, a helix-breaking residue, can potentially break the continuity of the C-helix. The side chain of WT residue Q is in a polar environment surrounded by the side chains of residues S-131, H-140, R-314, and N-319 and makes Hbs with W-141 and A-147. Apart from this Q-144 interacts with R-314 in the N-terminal of I-helix, N-319, T-323 of I-helix, and S-131 of B′/C loop; thus this site is located at the crucial junction interconnecting helix I, helix C, and the B′/C-helix.
P-193L
This site is far away from any of the FIRs and is in the N-cap position of E-helix. Replacement of P, which is a better N-cap residue than L, can affect stability of the E-helix, thereby affecting the packing in this region.
E-229K
This mutation occurs in the C-terminal of the F-helix in the vicinity of the SBR. Substitution of E to K leads to a change from a negatively charged residue to a positively charged side chain and this in turn affects the local charge distribution. This mutation disturbs an important cluster of salt bridges. In WT, R-194∷:E-229, R-194∷:D-333, and D-333∷:K-512 form a triangle of ionic bond interactions, holding I-helix with F-helix and β-strand S3.2. Upon mutation, the R-194∷:E-229 interaction is lost and has the potential to destabilize the other ionic interactions.
S-239R
Position 239 is located in the F/G loop region and is an exposed site and thus may not have severe consequences. Nevertheless, since the region is close to the SAC, mutation of S to R could affect the structure and dynamics of this region. S makes main chain Hbs with T-234 and A-237 at the C-terminal of F-helix, whereas its side chain makes a Hb with T-256 and T-234.
R-368H
This is in between the J- and K-helices in an exposed loop, and the consequence of this change is not immediately apparent except that positively charged R is replaced by H, whose charge state depends on its protonation state. In WT R-368 interacts with G-365, D-367, V-363, Q-362, and D-374. Upon mutation the bonds to D-367 and D-374 are weakened. How this affects the conformation and functionality of the protein is not clear.
G-466D
This is located in the middle of the heme-binding loop (HBL) and is a site involved in the interaction with reductase protein. Presence of G is completely conserved in all CYP proteins at this position. From the conformational point of view, G is found in a turn and is accommodated well in the limited volume between HBL and B-helix. This site being a major docking area of the reductase protein, the mutation from G to an acidic residue D may be undesirable for the reductase binding since neutral/basic residues are preferred in this reductase binding region (RBR). The presence of G is completely conserved in all CYP proteins at this position.
MD simulation studies
Although the observations given above give some qualitative aspects of the possible deleterious nature of the mutations, an estimation of how these residue changes propagate into the protein structure leading to a functionally disruptive effect can be obtained by subjecting the MTs as well as the WT to MD simulations. Hence, the WT and the MT structures were subjected to MD simulations for 30 ns. The instantaneous structures (snapshots) saved at every successive 1 ps from the start of the simulation, totaling 30,000 for each structure, were used for analysis of time evolution of various structural properties analyzed in this study.
Overall structural changes
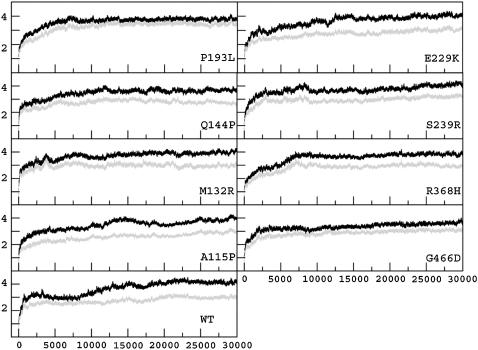
RMSD trajectories. The overall structural deviation of the WT and the MTs from their starting structures during the entire course of simulations is shown in Fig. 2. It should be noted that the starting structures of WT and MTs were identical except for the side chains of the residues at the mutation sites. The trajectories shown in Fig. 2 do not take precisely the same course; all the structures initially evolve rapidly during the first 1.5–2 ns and seem to stabilize at different times (Table 1). All further analyses were carried out using the data collected after RMSD stabilization. Whereas the all-Cα RMSD trajectories take long times for stabilization, the trajectories of the secondary structures (shown in gray in Fig. 2) quickly stabilize in all the structures. It may be noted that some of the RMSD trajectories seem to stabilize for short periods before drifting to stable structures. For example, in the case of A-115P and WT, both the structures after an initial increase of RMSDs seem to stabilize for some time (∼5 ns), followed by a gradual drift into another stabilized structure. An examination of the structures revealed that some loop regions undergo slow conformational transitions, during which time the RMSD trajectories show slow drift-like variation. Owing to the fact that structures simulated are from comparative modeling, we discarded the early parts of stabilization and used only those which seemed stabilized until the end of simulation. The average value of the overall RMSD after stabilization is ∼4.1 Å in WT, whereas in MTs it ranges from 3.4 Å to 3.9 Å (Table 1). The average values of RMSD after stabilization for the secondary structures are ∼2.9 Å in WT and between 2.8 Å to 3.4 Å in the MTs. Higher RMSDs of the secondary structures in some MTs than the WT reflect relative structural deviations occurring in the core of the MTs.
FIGURE 2.
Trajectories of the overall Cα RMSDs (in angstroms) (black line) of the WT and MT structures with respect to the starting structure over 30,000 ps MD simulation. The x axis represents the simulation time in picoseconds. The y axis represents RMSD in angstrom units. Gray lines indicate the Cα RMSD trajectories calculated for only the secondary structures. This figure was prepared using XMGRACE (50) (http://plasmagate.weizmann.ac.il/Grace).
TABLE 1.
Average properties of WT and MT structures computed after RMSD stabilizations (T-stab)
| RMSD (Å)
|
RMSF (Å)
|
||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All cα
|
Sec-str cα
|
heme
|
All cα
|
Sec-str cα
|
Cys-Fe (Å)
|
SBR (Å3)
|
SAC (Å)
|
Hbs
|
|||||||||||
| T-stab (ns) | avg | sd | avg | sd | avg | sd | ≤1 Å | >1 Å | ≤1 Å | >1 Å | avg | sd | avg | sd | avg | sd | avg | sd | |
| WT | 15 | 4.1 | 0.2 | 2.9 | 0.1 | 2.9 | 0.4 | 54 | 46 | 66 | 34 | 2.1 | 0.1 | 403.0 | 79.0 | 7.3 | 0.3 | 351.3 | 9.5 |
| A-115P | 15 | 3.7 | 0.2 | 2.8 | 0.2 | 4.5 | 0.3 | 60 | 40 | 73 | 27 | 2.9 | 0.1 | 392.2 | 110.7 | 8.3 | 0.3 | 342.2 | 9.5 |
| M-132R | 8 | 3.8 | 0.2 | 3.0 | 0.1 | 3.6 | 0.3 | 43 | 57 | 54 | 48 | 2.3 | 0.1 | 306.5 | 86.0 | 10.7 | 0.7 | 352.2 | 9.9 |
| Q-144P | 12 | 3.6 | 0.1 | 2.8 | 0.1 | 3.6 | 0.4 | 49 | 51 | 60 | 40 | 2.5 | 0.1 | 308.2 | 69.6 | 9.6 | 0.8 | 341.6 | 10.8 |
| P-193L | 7 | 3.8 | 0.1 | 3.4 | 0.1 | 2.3 | 0.3 | 41 | 59 | 53 | 47 | 3.3 | 0.4 | 610.0 | 172.6 | 7.9 | 1.2 | 347.0 | 10.0 |
| E-229K | 15 | 3.9 | 0.1 | 2.9 | 0.2 | 2.6 | 0.4 | 53 | 47 | 61 | 39 | 2.1 | 0.1 | 377.5 | 152.7 | 9.9 | 1.0 | 358.5 | 9.6 |
| S-239R | 8 | 3.8 | 0.2 | 3.0 | 0.2 | 2.6 | 0.4 | 35 | 65 | 43 | 57 | 1.9 | 0.1 | 355.3 | 106.4 | 7.7 | 0.4 | 355.1 | 9.7 |
| R-368H | 8 | 3.8 | 0.1 | 3.0 | 0.1 | 3.6 | 1.1 | 41 | 59 | 54 | 46 | 2.3 | 0.3 | 254.9 | 83.6 | 10.5 | 0.6 | 356.8 | 10.1 |
| G-466D | 5 | 3.4 | 0.2 | 2.9 | 0.1 | 2.3 | 0.3 | 51 | 49 | 61 | 39 | 2.2 | 0.1 | 747.1 | 295.3 | 9.2 | 0.4 | 342.3 | 9.5 |
“RMSD” is the root mean-square deviation from the starting structure; “RMSF” is the average residue fluctuations from their mean positions; “Cys-Fe” is the coordination distance between the C470:sulfur atom and the Fe of heme; “SBR” is the volume of the putative SBR; “SAC” is the size of the substrate access channel entrance formed by the structure's F/G loop and the B′-helix; and “Hbs” are the average number of hydrogen bonds present in the structure. For each property, the average (avg) and the standard deviations (sd) are given.
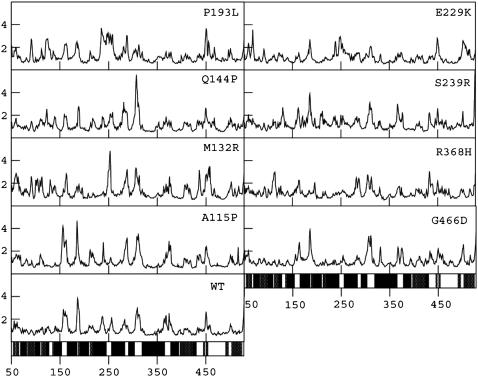
RMSF. The distribution of Cα RMSF values is shown in Fig. 3. In the WT 54% of the residues have RMSF values <1 Å (see Table 1). This could be considered the basal level fluctuation for the WT protein. All the MTs other than A-115P have larger percentages of residues having RMSF values >1 Å compared to the WT. Thus, on the whole, the MTs are characterized by increased protein flexibilities. Similarly, the percentage of residues having RMSF >1 Å in the protein core comprising secondary structural elements is greater in MTs (except A-115P) than WT, thus indicating that these MTs are associated with increases in flexibilities in some of the residues in the core region. The RMSF profiles in Fig. 3 further illustrate residue-wise differences between WT and MTs, which are conspicuously evident in terms of peak heights that represent magnitudes of regional flexibilities. Comparison of these regional flexibilities further revealed characteristic decrease and increase in flexibilities in each of the MTs in certain loops, helices, and β-sheets as given in Table 2.
FIGURE 3.
Residue-wise RMSF (in angstroms) profiles of Cα atoms of the WT and MT structures computed after stabilization of the RMSD trajectories. The x axis represents the residue number. The y axis represents RMSF in angstrom units. The bars at the bottom of the figure indicate the location of α-helices (black) and β-strands (gray). This figure was prepared using XMGRACE.
TABLE 2.
Regions (α-helices, β-sheets, and loops) showing an average increase or decrease of RMSF in the MTs compared to the WT; RMSF of a particular structure is taken to be increased or decreased if there is an average change in RMSF of >0.3 Å in at least 50% of its residues
| Increase | Decrease | |
|---|---|---|
| A-115P | D; S4.2, S3.2; C/D, B/B′, E/F, G/H, H/I, J/K, K′/L′ | B′, F; D/E, K/K′ |
| M-132R | B, H; S1.1, S1.2, S2.1, S2.2, S4.2; B/B′, B′/C, G/H, H/I, K′/L′, CHR, HBL, F/G | F; C/D, D/E, E/F, J/K, K/K′ |
| Q-144P | B′, G, H; S4.1; B/B′, G/H, H/I, HBL | C/D, D/E, E/F, J/K |
| P-193L | B′, D, F, G; S1.1, S1.2, S4.1, S4.2; B/B′, B′/C, F/G, G/H, L′/L, CHR, HBL | C/D, D/E, H/I, J/K, K/K′ |
| E-229K | G; S4.1, S3.1, S3.2; F/G | J; C/D, D/E, E/F, H/I, J/K |
| S-239R | D, E; S5.1; B′/C, E/F, J/K, K′/L′, HBL | C/D, D/E |
| R-368H | B, B′, E, K′; S1.2; B/B′, G/H, K′/L′, HBL | F; C/D, D/E, E/F, J/K, L′/L |
| G-466D | D, I, K′; H/I, K′/L′ | B′, F; C/D, D/E, E/F, L′/L |
See Fig. 1 for the definitions of α-helices and β-sheets. Loops are denoted by their flanking helices. HBL denotes heme binding loop; CHR denotes charge relay region.
Structural changes at the mutation sites. As mentioned earlier substitutions of residues at the mutation sites are expected to bring out certain structural changes at the mutation sites. To know the changes brought out by the mutations, we computed the average values of the φ, ψ dihedral angles and number of times a given Hb interaction is observed during simulations (referred to as the occupancy of Hb interaction) at the mutation site in WT and MTs, and the results are tabulated in Table 3.
TABLE 3.
Average φ, ψ dihedral angle conformations and the occupancies of the Hb interactions involving the mutation positions in the WT and the MTs
| Wild-type
|
MUTANT
|
|||||||
|---|---|---|---|---|---|---|---|---|
| MT site | φ | ψ | Hb | Occ | φ | ψ | Hb | Occ |
| A-115P | −117.1 | −23.9 | [HBL] 468ARG(NH1)∷:115ALA(O)[B/B′] | 91 | −81.2 | −30.9 | ||
| [B/B′] 115ALA(N)∷:404THR(OG1)[S1.3] | 75 | |||||||
| M-132R | −106.8 | −71.8 | [B′/C] 135GLY(N)∷:132MET(O) [B′/C] | 57 | −71.5 | −50.9 | [HBL] 468ARG(NH1)∷:132ARG(O) [B′/C] | 74 |
| [HBL] 468ARG(NH2)∷:132MET(O) [B′/C] | 31 | [B′/C] 132ARG(N)∷:326ASP(OD1) [I] | 34 | |||||
| [B′/C] 132MET(N)∷:326ASP(OD1)[I] | 15 | [HBL] 468ARG(NH2)∷:132ARG(O) [B′/C] | 31 | |||||
| [B′/C] 132MET(N)∷:326ASP(OD2)[I] | 10 | [B′/C] 132ARG(N)∷:326ASP(OD2) [I] | 22 | |||||
| [B′/C] 132ARG(NE)∷:326ASP(OD1) [I] | 16 | |||||||
| [B′/C] 132ARG(NH2)∷:144GLN(OE1) [C] | 16 | |||||||
| [B′/C] 132ARG(NH2)∷:323THR(OG1) [I] | 13 | |||||||
| [B′/C] 132ARG(NE)∷:322ALA(O) [I] | 12 | |||||||
| [B′/C] 132ARG(NH1)∷:322ALA(O) [I] | 11 | |||||||
| [B′/C] 132ARG(NE)∷:322ASP(OD2) [I] | 10 | |||||||
| [B′/C] 132ARG(NH1)∷:323THR(OG1) [I] | 10 | |||||||
| Q-144P | −58.6 | −46.7 | [C]148ALA(N)∷:144GLN(O) [B′/C] | 98 | −54.8 | −46.5 | [E]148ALA(N)∷:144PRO(O)[C] | 99 |
| [C]144GLN(N)∷:140HIS(O)[C] | 92 | [E]147ALA(N)∷:144PRO(O)[C] | 10 | |||||
| [I]323THR(OG1)∷:144GLN(OE1)[C] | 26 | |||||||
| [C]144GLN(NE2)∷:131SER(OG) [B′/C] | 15 | |||||||
| [I]319ASN(ND2)∷:144GLN(OE1)[C] | 15 | |||||||
| [C]144GLN(NE2)∷:140HIS(O)[C] | 14 | |||||||
| P-193L | −58.2 | −37.2 | [E]197THR(N)∷:193PRO(O)[E] | 97 | −57.7 | −39.7 | [E]197THR(N)∷:193LEU(O)[E] | 91 |
| [E]196LEU(N)∷:193PRO(O)[E] | 40 | [E]193LEU(N)∷:516PHE(O)[S5.2] | 72 | |||||
| [E]196LEU(N)∷:193LEU(O)[E] | 32 | |||||||
| [E]197THR(OG1)∷:193LEU(O)[E] | 12 | |||||||
| [E]193LEU(N)∷:515SER(OG) [S3.2] | 10 | |||||||
| E-229K | −57.7 | −49.7 | [F]229GLU(N)∷:225LEU(O)[F] | 94 | −58.8 | −47.8 | [F]229LYS(N)∷:225LEU(O)[F] | 95 |
| [F]233ARG(N)∷:229GLU(O)[F] | 60 | [F]233ARG(N)∷:229LYS(O)[F] | 77 | |||||
| [F]234THR(N)∷:229GLU(O)[F] | 16 | [F]229LYS(N)∷:226SER(O)[F] | 14 | |||||
| [F]229GLU(N)∷:226SER(O)[F] | 15 | |||||||
| [F]232GLY(N)∷:229GLU(O)[F] | 10 | |||||||
| S-239R | −64.1 | −40.9 | [F]243VAL(N)∷:239SER(O)[F/G] | 79 | −43 | −46.2 | [F]243VAL(N)∷:239ARG(O)[F/G] | 94 |
| [F/G]239SER(N)∷:234THR(O)[F] | 23 | [F/G]239ARG(NH2)∷:234THR(O)[F] | 40 | |||||
| [F/G]239SER(N)∷:235VAL(O)[F] | 20 | [F/G]239ARG(NH2)∷:233ARG(O)[F] | 26 | |||||
| [F/G]242ASP(N)∷:239SER(O)[F/G] | 14 | [F/G]239ARG(NH2)∷:256THR(O)[G] | 21 | |||||
| [F/G]239ARG(NH1)∷:233ARG(O)[F] | 17 | |||||||
| [F/G]239ARG(NE)∷:255ARG(O)[F/G] | 15 | |||||||
| [F/G]242ASP(N)∷:239ARG(O)[F/G] | 12 | |||||||
| [F/G]239ARG(NE)∷:234THR(O)[F] | 11 | |||||||
| [F/G]239ARG(N)∷:236GLY(O)[F] | 10 | |||||||
| [F/G]239ARG(NH1)∷:255ARG(O)[F/G] | 10 | |||||||
| R-368H | −78.9 | 96.3 | [J/K]365GLY(N)∷:368ARG(O)[J/K] | 83 | −99.5 | 116.7 | [J/K]368HIS(N)∷:365GLY(O)[J/K] | 43 |
| [J/K]368ARG(NH1)∷:362GLN(O)[J] | 14 | [L]489HIS(NE2)∷:368HIS(O)[J/K] | 40 | |||||
| [J/K]365GLY(N)∷:368HIS(O)[J/K] | 32 | |||||||
| [J/K]368HIS(NE2)∷:374ASP(OD1)[J/K] | 10 | |||||||
| [J/K]368HIS(NE2)∷:374ASP(OD2)[J/K] | 10 | |||||||
| G-466D | −101.8 | −106 | [HBL]469ARG(NE)∷:466GLY(O)[HBL] | 30 | −53.7 | 104.1 | [HBL]469ARG(NH1)∷:466ASP(OD2) [HBL] | 10 |
| [HBL]469ARG(NH1)∷:466GLY(O)[HBL] | 17 | |||||||
| [HBL]469ARG(NH2)∷:466GLY(O)[HBL] | 15 | |||||||
| [HBL]466GLY(N)∷:528HEM(O1A)[HEME] | 10 | |||||||
“Occ” denotes the Hb occupancy percentage. Only Hbs with occupancies ≥10 are shown. For each Hb interaction, the secondary structures/loop regions involved in the interaction are indicated within square brackets.
From the table it can be seen that the G-466D mutation is characterized by a major conformational change at the mutation site compared to WT. The mutation site is marked by a large change in the ψ angle from −106° (WT) to +104° (MT), characterizing the flipping of the peptide unit between the residues 466 and 477. Concomitantly the Hb interactions involving the C O group at the mutation site have also been severed. Compared to G-446D, the other mutations have not led to such major conformational changes at their respective sites. With regard to Hb interaction, absence or presence of Hbs in the MTs has been determined by the absence or presence of functional groups capable of forming Hb interaction. For example, in A-115P, A in WT is involved in two Hbs, of which one is formed by the amide hydrogen. P in the MT was not involved in any such Hbs. The C
O group at the mutation site have also been severed. Compared to G-446D, the other mutations have not led to such major conformational changes at their respective sites. With regard to Hb interaction, absence or presence of Hbs in the MTs has been determined by the absence or presence of functional groups capable of forming Hb interaction. For example, in A-115P, A in WT is involved in two Hbs, of which one is formed by the amide hydrogen. P in the MT was not involved in any such Hbs. The C O of P instead has formed Hb with a water molecule as it is on the surface. In some cases, MTs are characterized by the formation of more numbers of Hb interactions, albeit with very low occupancies.
O of P instead has formed Hb with a water molecule as it is on the surface. In some cases, MTs are characterized by the formation of more numbers of Hb interactions, albeit with very low occupancies.
Structural properties of the functionally important regions.
a. HBR. The heme moiety is held in place by hydrophobic and Hb interactions, formed by the HBL, C-helix, β-strand S1.3, and B/B′ and B′/C loops (see Fig. 1 for the definitions of secondary structures). The list of Hb interactions and their occupancies in the WT and MTs during simulations is given in Table 4. WT forms Hbs with the surrounding structures with ∼50% or more occupancy. Comparatively all the MTs show a reduction or complete absence of Hb interactions from one or many of these structures, implying altered binding of the heme in all the MTs than WT. It may be noted that decreased Hb interactions in the case of MTs with the four structures surrounding the propionates correlate with their increased RMSF fluctuations in those structures (Table 2). The positional alteration of the heme in MTs is also indicated by the Fe-Cys coordination. Only in E-229K is the coordination bond length similar to that found in WT; in all other MTs the coordination bond length is either shorter or longer than that of the WT, with P-193L showing the highest deviation (Table 1). We also computed the RMSD for the heme with respect to its starting position. The RMSD values (Table 1) indicate that the position of the heme undergoes more deviation in all the MTs, except P-193L and G-466D, compared to the WT.
b. SBR. We calculated the volume of the continuous void over the heme moiety bounded by helices B′, I, F, and G, β-strand S1.3, and B/C loop to represent the volume of the SBR. Although the volume measured this way may be more than the actual volume occupied by any hypothetical substrate, it nevertheless gives a convenient representation of SBR for comparison purposes. In WT, on average, the volume is ∼403 Å3 (Table 1) and the value hardly fluctuates. The MTs R-368H, M-132R, Q-144P, S-239R, E-229K, and A-115P show a reduction in the size of their SBRs, whose volumes are, respectively, 63%, 76%, 76%, 88%, 94%, and 97% of that of WT. In the case of P-193L and G-466D the volumes of SBR have increased to 151% and 185%, respectively, of that of the WT. This result indicates that the MTs are characterized by a distorted substrate-binding pocket, either smaller or larger than that found in the WT.
c. SAC. The catalytic site of CYP1b1 or the SBR is not directly accessible from the surface of the protein. Thermal motion pathway analysis (34) in p450cam had revealed the existence of three potential substrate entry/exit pathways (denoted by pw1, pw2, and pw3). Random expulsion MD (REMD) ligand egress (24,25) trajectories further revealed three subpathways (a, b, and c) in the pathway pw2. Pathway pw2a, comprising the F/G loop and the B′-helix, has been found to be a common pathway among the p450s (24,25,35–40), and hence we monitored the size of the opening between the F/G loop and the B′-helix during the entire period of simulation (Supplementary Material). In the WT the average distance is ∼7 Å and seems to be stabilized by the Hb R-124∷L-247 formed between the F/G loop and the B′-helix. An earlier study involving “REMD” simulations (24) had revealed the breakage and formation of similar Hb interactions between the F/G loop and the B′-helix. The MTs A-115P and S-239R also show hydrogen-bonding patterns similar to WT. In correlation, the SAC size of A-115P and S-239R is similar to WT. Whereas the MTs M-132R, E-229K, R-368H, and G-466D show a complete absence of Hbs between the F/G loop and the B′-helix, the MTs Q-144P and P-193L have irregular patterns of Hbs. Accordingly, in these MTs the conformation of SAC is wider and associated with noticeably large fluctuations.
TABLE 4.
Occupancies (in %) of Hbs between “heme” and protein residues from structures surrounding heme in the WT and MTs; only occupancies ≥5% are shown
| B/B′ loop
|
B′/C loop
|
C-helix
|
β-strand S1.3
|
HBL
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 117 | 135 | 137 | 141 | 145 | 398 | 399 | 401 | 464 | 465 | 466 | 467 | 468 | 469 | 470 | 471 | |
| WT | 48 | – | – | 84 | 73 | 31 | 48 | – | 41 | – | 17 | 10 | 94 | 22 | – | – |
| A-115P | 24 | – | – | – | 25 | – | – | – | – | – | 97 | – | – | 46 | – | – |
| M-132R | – | – | – | 72 | 18 | – | – | 55 | – | – | 48 | 10 | 67 | 46 | – | – |
| Q-144P | – | – | 54 | – | 32 | – | – | – | 61 | 36 | 61 | 37 | 74 | 74 | – | – |
| P-193L | 27 | – | – | – | 88 | – | – | – | 89 | – | – | – | 71 | – | – | 18 |
| E-229K | 99 | – | – | 79 | 16 | – | – | 96 | 61 | – | 96 | 83 | 99 | 20 | – | – |
| S-239R | 82 | 13 | – | – | 19 | – | – | 28 | 94 | – | 37 | – | 93 | 71 | – | – |
| R-368H | 47 | – | 87 | – | – | – | – | – | – | 27 | 70 | 53 | 79 | 30 | – | – |
| G-466D | 98 | – | 10 | – | 77 | – | – | 66 | 91 | – | – | – | – | 59 | – | – |
DISCUSSION
The CYP1b1 model
Modeling studies reported earlier (11,16) have used the bacterial CYPs (CYP102, CYP108, and CYPc17) as structural templates. Of these templates, CYP102 (p450BM-3) was considered a good template for modeling eukaryotic CYPs because it shares more features in common with the eukaryotic CYPs than the other bacterial CYPs. After elucidation of the first mammalian CYP structure, viz., CYP2c5 (33), Shimada and co-workers (41) used it as a template to model CYP1b1 for their study on the binding mode of selected substrates with regard to the effects of allelic variants of CYP1b1 on their metabolism. Recently the structure of human CYP2c9 (PDB-CODE: 1OG2) (42) was reported, and it has a sequence identity of 31.5% with CYP1b1. Given the fact that CYPs within a species are expected to have more structural similarity, CYP2c9 has been used as the template in this study. The model of CYP1b1 reported in this study can be considered better than the earlier reported models because of the following reasons. The loop region between the F- and G-helices, a FIR, has been modeled better due to good correspondence of alignment in the template. Templates used for modeling in earlier modeling studies lacked proper structural information for this region because of its highly variable nature and because some templates lacked coordinate data. Importantly, the region in CYP2c9 has short helices F′ and G′, a human-specific CYP structural feature not observed in bacterial and other mammalian CYP structures (42). Furthermore, the three-dimensional (3D) model obtained from MODELLER was further subjected to rigorous refinement by a combination of EM and short MD to relieve stereochemical strains.
Deleterious nature of the mutations
The detailed structural analysis by means of MD simulations in this study confirms some of our earlier speculations about the effects of some of these mutations (19). The average structural properties, though, indicate similarity of the overall fold of MTs and WT. The precise differences in the RMSD trajectories, which indicate the differences in the path of transition of structures from the starting conformation to their final states despite the initial structures being identical (except at the mutation sites), clearly speaks of the influence of residue substitutions on the dynamics of the protein. The RMSF data indicate that mutations are characterized by a subtle but significant increase in the flexibility of the molecule. Further, the individual MTs have specific regions of increased or decreased fluctuations compared to WT, some of these located at spatially distant sites and forming part of FIRs.
It is also observed that mutations have an effect on the integrity of the active site pocket. From a functional point of view the active site regions of enzymes are fairly rigid structures, allowing for specific interactions of functional groups of the enzyme and substrate. Any change in the structure of the active site region can affect the substrate binding and functional interactions between the substrate and the enzyme. The volume of the SBR gives a measure of the disruptive effect due to mutations. As revealed in this study, the volume of the substrate-binding pocket is different in all MTs except A-115P compared to WT. In P-193L and G-466D the volumes of SBRs are larger than the WT, whereas in the other five MTs (M-132R, Q-144P, E-229K, S-239R, and R-368H) the SBRs are smaller in volume than the WT. A-115P, which shows a severe effect on HBR, seems to have no effect on SBR. In terms of the most affecting mutation on SBR, G-466D stands out distinctly.
The MTs also reveal changes in the conformation and dynamics of the SAC region wherein in all the MTs the size of the channel opening is greater than that of WT. Moreover, in some of the MTs (M-132R, Q-144P, P-193L, and E-229K) the SAC flutters irregularly, most severely in P-193L (Supplementary Material), during the course of the entire simulation, probably indicating disruption of substrate recognition and also its accessibility.
We also examined the correlation between the MTs and the WT for residue-wise Hb occupancies and the RMSFs, both of which give a measure of the nature of the 3D fold of the protein. It was found that MTs Q-144P, S-239R, R-368H, E-229K, and A-115P show poor correlation (Cij ≤ 0.7) for Hbs, whereas M-132R, E-229K, P-193L, R-368H, Q-144P, and G-466D show poor correlation for RMSFs. (Table 5). Furthermore, the extent of overall deviations of all the average properties of the MTs compared to the WT place the MTs Q-144P, G-466D, R-368H, A-115P, P-193L, M-132R, E-229K, and S-239R in descending order of deleteriousness (Table 1). But the deleteriousness observed in a specific FIR could be more severe in some MTs than others. For example, the Cys-heme coordination bond distance can be used to quantify the extent of structural deviation occurring in the HBR in MTs than the WT. In P-193L, this distance is ∼3.28 Å, which is more than the usual coordination bonding distance in CYPs (∼2 Å). In MTs A-115P, Q-144P, M-132R, R-368H, and G-466D the distance is a little over the WT ranging between 2.2 Å and 2.9 Å. E-229K exhibits the same distance as that of WT (2.1 Å), whereas S-239R shows 1.85 Å. Thus HBR, from the Cys-heme coordination bond point of view, is affected severely by P-193L mutation and less severely by A-115P than the other mutations. Other MTs having coordination distance between 1.85 Å and 2.55 Å may have proper heme binding. The Cys-heme distance roughly correlates with the RMSD of the heme cofactor from initial conformation. The other indicator of deleteriousness at the HBR, viz., RMSD of heme from its initial conformation, indicates A-115P as the most deleterious mutation.
TABLE 5.
Correlation coefficients (Cij) between WT and MTs, for the Hb occupancies and residue-wise Cα RMSF profiles
| WT versus MT | Hbs (Cij) | RMSF (Cij) |
|---|---|---|
| A-115P | 0.66 | 0.76 |
| M-132R | 0.71 | 0.42 |
| Q-144P | 0.55 | 0.66 |
| P-193L | 0.73 | 0.56 |
| E-229K | 0.61 | 0.50 |
| S-239R | 0.57 | 0.72 |
| R-368H | 0.58 | 0.50 |
| G-466D | 0.76 | 0.68 |
Generally, loss of protein function can happen when key residue substitutions occur in catalytic/substrate binding sites. However, several disease-causing mutations have been reported which do not occur in catalytic/substrate binding sites. It is argued that such mutations manifest their deleterious effects by bringing in changes in structural characteristics such as a change in surface charge distribution, disruption of packing in protein core regions, etc. In general, any amino acid substitution, whether pathogenic or nonpathogenic, has some impact on the protein structure. The extent of impact is dependent on the position and the nature of the amino acid that is replaced, as well as on the newly introduced amino acid. Some replacements are well accommodated in the protein structure either with or without any structural changes in the protein such as rearrangements in packing of secondary structural elements in the core (43–45). In the case of the pathogenic mutations investigated in this study, it has been found that they can be accommodated into the protein structure but are associated with some structural changes that are functionally significant.
Sunyaev et al. (46) as well as Wang and Moult (47) have proposed a few criteria to qualitatively judge whether an amino acid mutation can be deleterious or not. Accordingly, mutations M-132R, Q-144P, E-229K, S-239R, R-368H, and G-466D lead to a change in electrostatics and hydrophobicity; mutation Q-144P introduces the helix-breaking residue P in the middle of C-helix. A multiple sequence alignment of p450 sequences was taken from the PFAM database to study the profile of amino acids at the mutation sites. Many of the mutation sites have the WT allele occurring at a greater frequency than the MT residue. Positions 466 and 368 seem especially to be highly conserved among all the p450s. Other mutation positions, though not very much conserved among all p450s, are more specific to CYP1b1s. Mutation site 193 has a higher proportion of mutated residue L in general but the proportion of P increases as we consider specifically CYP1b1s. This might mean that CYP1b1 has adapted more specifically to P rather than L, and P might be important for its structural integrity. Positions 132 and 144 are not completely conserved among the three CYP1b1 members. But it is clear that none of the MT residues occur among the three mammalian CYP1b1 members, as well as other p450 proteins except for mutations P-193L and E-229K and S-239R, indicating the incompatibility of the MT residues for these positions in all CYPs in general. For P-193L, E-229K, and S-239R, though some proteins do have the MT residues, when it comes to the case of CYP1b1s the MT residues are no longer represented, indicating the structural specificity of CYP1b1 for the WT residues.
Severity of disease manifestation versus extent of structural disruption of MTs
Panicker et al. (48) have performed genotype-phenotype correlation studies and have indicated severity of disease manifestation for some of the PCG mutations found in the Indian population. They have reported, based on the clinically observed phenotype, the percentages of severe phenotypes in the Indian population associated with various mutations in at least one eye. They are P-193L (62.5%), E-229K (80%), and R-368H (72%). These percentages represent the cases in which severe disease phenotypes were observed, but they do not really compare the extent of disease severity associated with each mutation. Further, incomplete penetrance of the PCG mutations (49) makes the assessment of the deleteriousness of each mutation difficult. The MD simulation data presented here on the disruptive effect on specific FIRs could be validated with experimental evidence and then effectively used for disease prognosis.
The role of molecular dynamics in revealing the deleterious nature of disease mutations
A previous homology modeling study on CYP1b1 (11) suggested that some of the mutations (W-57C, G-61E, G-365W, P-379L, R-390H, E-387K, P-437L, and R-469W) might disrupt either the hinge region or the conserved core of the protein. In another study, homology modeling of MT forms of CYP1b1 indicated that one (R-444Q) out of the four (D-192V, A-330F, V-364M, and R-444Q) missense mutations in the protein caused significant structural changes whereas the remaining were structurally neutral (16).
Although qualitative analysis and modeling studies give clues about the possible effects of disease-causing mutations in proteins, it remains still intriguing how precisely residue changes in sites other than FIRs can bring about changes in protein structure, which in turn have deleterious effects on protein function. In this context, our study becomes important. Further it also shows that MD simulation has vast potential to elucidate the structure-function relationships in greater detail to precisely understand the function impeding mechanisms. The current MD simulation study on CYP1b1 gives some clues about the possible underlying structural disruptions leading to loss of function due to disease mutations. It can be postulated in the context here that the PCG MT proteins carrying the pathogenic mutations fold into a WT-like structure, but with changed structural properties, which are detrimental to the WT-like function. Changes in WT-like structural properties may lead to impairment of one or more of the following: substrate access and binding abilities, interaction with the reductase, electron transfer ability, and efficient binding of heme. Any such functional impairment can potentially cause pathogenicity. In vitro testing of these MT forms for their catalytic activity on common CYP1b1 substrates is currently being carried out which can validate the inferences made from this study.
SUPPLEMENTARY MATERIAL
An online supplement to this article can be found by visiting BJ Online at http://www.biophysj.org.
Acknowledgments
The authors acknowledge the contribution of Mr. V. B. Sreenu during the initial phases of the project work. The authors gratefully acknowledge the two anonymous referees and the editor for their critical and helpful comments.
M.S.A. gratefully acknowledges University Grants Commission, government of India, for the Senior Research Fellowship. H.A.N. gratefully acknowledges the Core-grant from the Centre for DNA Fingerprinting and Diagnostics, government of India.
References
- 1.deLuise, V. P., and D. R. Anderson. 1983. Primary infantile glaucoma (congenital glaucoma). Surv. Ophthalmol. 28:1–19. [DOI] [PubMed] [Google Scholar]
- 2.Gencik, A., A. Gencikova, and V. Ferak. 1982. Population genetical aspects of primary congenital glaucoma. I. Incidence, prevalence, gene frequency, and age of onset. Hum. Genet. 61:193–197. [DOI] [PubMed] [Google Scholar]
- 3.Sarfarazi, M., A. N. Akarsu, A. Hossain, M. E. Turacli, S. G. Aktan, M. Barsoum-Homsy, L. Chevrette, and B. S. Sayli. 1995. Assignment of a locus (GLC3A) for primary congenital glaucoma (Buphthalmos) to 2p21 and evidence for genetic heterogeneity. Genomics. 30:171–177. [DOI] [PubMed] [Google Scholar]
- 4.Akarsu, A. N., M. E. Turacli, S. G. Aktan, M. Barsoum-Homsy, L. Chevrette, B. S. Sayli, and M. Sarfarazi. 1996. A second locus (GLC3B) for primary congenital glaucoma (Buphthalmos) maps to the 1p36 region. Hum. Mol. Genet. 5:1199–1203. [DOI] [PubMed] [Google Scholar]
- 5.Stoilov, I. R. S. M. 2002. The third genetic locus (GLC3C) for primary congenital glaucoma (PCG) maps to Chromosome 14q24.3. Invest. Ophthalmol. Vis. Sci. 43:3015. (Abstr.) [PubMed] [Google Scholar]
- 6.Stoilov, I. 2001. Cytochrome P450s: coupling development and environment. Trends Genet. 17:629–632. [DOI] [PubMed] [Google Scholar]
- 7.Stoilov, I., I. Jansson, M. Sarfarazi, and J. B. Schenkman. 2001. Roles of cytochrome p450 in development. Drug Metabol. Drug Interact. 18:33–55. [DOI] [PubMed] [Google Scholar]
- 8.Bejjani, B. A., R. A. Lewis, K. F. Tomey, K. L. Anderson, D. K. Dueker, M. Jabak, W. F. Astle, B. Otterud, M. Leppert, and J. R. Lupski. 1998. Mutations in CYP1B1, the gene for cytochrome P4501B1, are the predominant cause of primary congenital glaucoma in Saudi Arabia. Am. J. Hum. Genet. 62:325–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plasilova, M., E. Ferakova, L. Kadasi, H. Polakova, A. Gerinec, J. Ott, and V. Ferak. 1998. Linkage of autosomal recessive primary congenital glaucoma to the GLC3A locus in Roms (Gypsies) from Slovakia. Hum. Hered. 48:30–33. [DOI] [PubMed] [Google Scholar]
- 10.Plasilova, M., A. Gerinec, and V. Ferak. 1998. Molecular diagnosis of mutations responsible for recurrent and severe forms of primary congenital glaucoma. Cesk. Slov. Oftalmol. 54:281–288. [PubMed] [Google Scholar]
- 11.Stoilov, I., A. N. Akarsu, I. Alozie, A. Child, M. Barsoum-Homsy, M. E. Turacli, M. Or, R. A. Lewis, N. Ozdemir, G. Brice, S. G. Aktan, L. Chevrette, M. Coca-Prados, and M. Sarfarazi. 1998. Sequence analysis and homology modeling suggest that primary congenital glaucoma on 2p21 results from mutations disrupting either the hinge region or the conserved core structures of cytochrome P4501B1. Am. J. Hum. Genet. 62:573–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakiuchi, T., Y. Isashiki, K. Nakao, S. Sonoda, K. Kimura, and N. Ohba. 1999. A novel truncating mutation of cytochrome P4501B1 (CYP1B1) gene in primary infantile glaucoma. Am. J. Ophthalmol. 128:370–372. [DOI] [PubMed] [Google Scholar]
- 13.Bejjani, B. A., D. W. Stockton, R. A. Lewis, K. F. Tomey, D. K. Dueker, M. Jabak, W. F. Astle, and J. R. Lupski. 2000. Multiple CYP1B1 mutations and incomplete penetrance in an inbred population segregating primary congenital glaucoma suggest frequent de novo events and a dominant modifier locus. Hum. Mol. Genet. 9:367–374. [DOI] [PubMed] [Google Scholar]
- 14.Martin, S. N., J. Sutherland, A. V. Levin, R. Klose, M. Priston, and E. Heon. 2000. Molecular characterisation of congenital glaucoma in a consanguineous Canadian community: a step towards preventing glaucoma related blindness. J. Med. Genet. 37:422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohtake, Y., R. Kubota, T. Tanino, H. Miyata, and Y. Mashima. 2000. Novel compound heterozygous mutations in the cytochrome P4501B1 gene (CYP1B1) in a Japanese patient with primary congenital glaucoma. Ophthalmic Genet. 21:191–193. [PubMed] [Google Scholar]
- 16.Mashima, Y., Y. Suzuki, Y. Sergeev, Y. Ohtake, T. Tanino, I. Kimura, H. Miyata, M. Aihara, H. Tanihara, M. Inatani, N. Azuma, T. Iwata, and M. Araie. 2001. Novel cytochrome P4501B1 (CYP1B1) gene mutations in Japanese patients with primary congenital glaucoma. Invest. Ophthalmol. Vis. Sci. 42:2211–2216. [PubMed] [Google Scholar]
- 17.Stoilov, I. R., V. P. Costa, J. P. Vasconcellos, M. B. Melo, A. J. Betinjane, J. C. Carani, E. V. Oltrogge, and M. Sarfarazi. 2002. Molecular genetics of primary congenital glaucoma in Brazil. Invest. Ophthalmol. Vis. Sci. 43:1820–1827. [PubMed] [Google Scholar]
- 18.Panicker, S. G., A. B. Reddy, A. K. Mandal, N. Ahmed, H. A. Nagarajaram, S. E. Hasnain, and D. Balasubramanian. 2002. Identification of novel mutations causing familial primary congenital glaucoma in Indian pedigrees. Invest. Ophthalmol. Vis. Sci. 43:1358–1366. [PubMed] [Google Scholar]
- 19.Reddy, A. B., K. Kaur, A. K. Mandal, S. G. Panicker, R. Thomas, S. E. Hasnain, D. Balasubramanian, and S. Chakrabarti. 2004. Mutation spectrum of the CYP1B1 gene in Indian primary congenital glaucoma patients. Mol. Vis. 10:696–702. [PubMed] [Google Scholar]
- 20.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779–815. [DOI] [PubMed] [Google Scholar]
- 21.Laskowski, R. A., D. S. Moss, and J. M. Thornton. 1993. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 231:1049–1067. [DOI] [PubMed] [Google Scholar]
- 22.Luthy, R., J. U. Bowie, and D. Eisenberg. 1992. Assessment of protein models with three-dimensional profiles. Nature. 356:83–85. [DOI] [PubMed] [Google Scholar]
- 23.Reddy, A. B., S. G. Panicker, A. K. Mandal, S. E. Hasnain, and D. Balasubramanian. 2003. Identification of R368H as a predominant CYP1B1 allele causing primary congenital glaucoma in Indian patients. Invest. Ophthalmol. Vis. Sci. 44:4200–4203. [DOI] [PubMed] [Google Scholar]
- 24.Ludemann, S. K., V. Lounnas, and R. C. Wade. 2000. How do substrates enter and products exit the buried active site of cytochrome P450cam? 1. Random expulsion molecular dynamics investigation of ligand access channels and mechanisms. J. Mol. Biol. 303:797–811. [DOI] [PubMed] [Google Scholar]
- 25.Ludemann, S. K., V. Lounnas, and R. C. Wade. 2000. How do substrates enter and products exit the buried active site of cytochrome P450cam? 2. Steered molecular dynamics and adiabatic mapping of substrate pathways. J. Mol. Biol. 303:813–830. [DOI] [PubMed] [Google Scholar]
- 26.Lindahl, E., B. Hess, and D. van der Spoel. 2001. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J. Mol. Model. 7:306–317. [Google Scholar]
- 27.Darden, T., D. York, and L. Pedersen. 1993. Particle mesh Ewald: an N-log(N) method for Ewald sums in large systems. J. Chem. Phys. 98:10089–10092. [Google Scholar]
- 28.Hess, B., H. Bekker, H. J. C. Berendsen, and J. G. E. M. Fraaije. 1997. LINCS: a linear constraint solver for molecular simulations. J Comput. Chem. 18:1463–1472. [Google Scholar]
- 29.McDonald, I. K., and J. M. Thornton. 1994. Satisfying hydrogen bonding potential in proteins. J. Mol. Biol. 238:777–793. [DOI] [PubMed] [Google Scholar]
- 30.Levitt, D. G., and L. J. Banaszak. 1992. POCKET: a computer graphics method for identifying and displaying protein cavities and their surrounding amino acids. J. Mol. Graph. 10:229–234. [DOI] [PubMed] [Google Scholar]
- 31.Guex, N., and M. C. Peitsch. 1997. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 18:2714–2723. [DOI] [PubMed] [Google Scholar]
- 32.Mizuguchi, K., C. M. Deane, T. L. Blundell, M. S. Johnson, and J. P. Overington. 1998. JOY: protein sequence-structure representation and analysis. Bioinformatics. 14:617–623. [DOI] [PubMed] [Google Scholar]
- 33.Williams, P. A., J. Cosme, V. Sridhar, E. F. Johnson, and D. E. McRee. 2000. Mammalian microsomal cytochrome P450 monooxygenase: structural adaptations for membrane binding and functional diversity. Mol. Cell. 5:121–131. [DOI] [PubMed] [Google Scholar]
- 34.Lüdemann, S. K., O. Carugo, and R. C. Wade. 1997. Substrate access to cytochrome P450cam: a comparison of a thermal motion pathway analysis with molecular dynamics simulation data. J. Mol. Model. (Online). 3:369–374. [Google Scholar]
- 35.Oprea, T. I., G. Hummer, and A. E. Garcia. 1997. Identification of a functional water channel in cytochrome P450 enzymes. Proc. Natl. Acad. Sci. USA. 94:2133–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Winn, P. J., S. K. Ludemann, R. Gauges, V. Lounnas, and R. C. Wade. 2002. Comparison of the dynamics of substrate access channels in three cytochrome P450s reveals different opening mechanisms and a novel functional role for a buried arginine. Proc. Natl. Acad. Sci. USA. 99:5361–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hung, R. J., P. Boffetta, P. Brennan, C. Malaveille, A. Hautefeuille, F. Donato, U. Gelatti, M. Spaliviero, D. Placidi, A. Carta, A. Scotto di Carlo, and S. Porru. 2004. GST, NAT, SULT1A1, CYP1B1 genetic polymorphisms, interactions with environmental exposures and bladder cancer risk in a high-risk population. Int. J. Cancer. 110:598–604. [DOI] [PubMed] [Google Scholar]
- 38.Li, H., and T. L. Poulos. 2004. Crystallization of cytochromes P450 and substrate-enzyme interactions. Curr. Top. Med. Chem. 4:1789–1802. [DOI] [PubMed] [Google Scholar]
- 39.Poulos, T. L. 2003. Cytochrome P450 flexibility. Proc. Natl. Acad. Sci. USA. 100:13121–13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scott, E. E., Y. A. He, M. R. Wester, M. A. White, C. C. Chin, J. R. Halpert, E. F. Johnson, and C. D. Stout. 2003. An open conformation of mammalian cytochrome P450 2B4 at 1.6-Ǻ resolution. Proc. Natl. Acad. Sci. USA. 100:13196–13201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis, D. F., E. M. Gillam, S. A. Everett, and T. Shimada. 2003. Molecular modelling of human CYP1B1 substrate interactions and investigation of allelic variant effects on metabolism. Chem. Biol. Interact. 145:281–295. [DOI] [PubMed] [Google Scholar]
- 42.Williams, P. A., J. Cosme, A. Ward, H. C. Angove, D. Matak Vinkovic, and H. Jhoti. 2003. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 424:464–468. [DOI] [PubMed] [Google Scholar]
- 43.Nagarajaram, H. A., B. V. Reddy, and T. L. Blundell. 1999. Analysis and prediction of inter-strand packing distances between beta-sheets of globular proteins. Protein Eng. 12:1055–1062. [DOI] [PubMed] [Google Scholar]
- 44.Reddy, B. V., and T. L. Blundell. 1993. Packing of secondary structural elements in proteins. Analysis and prediction of inter-helix distances. J. Mol. Biol. 233:464–479. [DOI] [PubMed] [Google Scholar]
- 45.Reddy, B. V., H. A. Nagarajaram, and T. L. Blundell. 1999. Analysis of interactive packing of secondary structural elements in alpha/beta units in proteins. Protein Sci. 8:573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sunyaev, S., V. Ramensky, I. Koch, W. Lathe 3rd, A. S. Kondrashov, and P. Bork. 2001. Prediction of deleterious human alleles. Hum. Mol. Genet. 10:591–597. [DOI] [PubMed] [Google Scholar]
- 47.Wang, Z., and J. Moult. 2001. SNPs, protein structure, and disease. Hum. Mutat. 17:263–270. [DOI] [PubMed] [Google Scholar]
- 48.Panicker, S. G., A. K. Mandal, A. B. Reddy, V. K. Gothwal, and S. E. Hasnain. 2004. Correlation of genotype with phenotype in Indian patients with primary congenital glaucoma. Invest. Ophthalmol. Vis. Sci. 45:1149–1156. [DOI] [PubMed] [Google Scholar]
- 49.Bejjani, B. A., D. W. Stockton, and R. A. Lewis. 2000. Multiple CYP1B1 mutations and incomplete penetrance in an inbred population segregating primary congenital glaucoma suggest frequent de novo events and a dominant modifier locus. Hum. Mol. Genet. 9:367–374. [DOI] [PubMed] [Google Scholar]
- 50.Turner, P. J. 2005. XMGRACE, Version 5.1.19. Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology, Beaverton, OR.