Abstract
In this study, the heteromeric N-methyl-d-aspartate (NMDA) receptor channels composed of NR1a and NR2A subunits were expressed, purified, reconstituted into liposomes, and characterized by using the patch clamp technique. The protein exhibited the expected electrophysiological profile of activation by glutamate and glycine and internal Mg2+ blockade. We demonstrated that the mechanical energy transmitted to membrane-bound NMDA receptor channels can be exerted directly by tension developed in the lipid bilayer. Membrane stretch and application of arachidonic acid potentiated currents through NMDA receptor channels in the presence of intracellular Mg2+. The correlation of membrane tension induced by either mechanical or chemical stimuli with the physiological Mg2+ block of the channel suggests that the synaptic transmission can be altered if NMDA receptor complexes experience local changes in bilayer thickness caused by dynamic targeting to lipid microdomains, electrocompression, or chemical modification of the cell membranes. The ability to study gating properties of NMDA receptor channels in artificial bilayers should prove useful in further study of structure–function relationships and facilitate discoveries of new therapeutic agents for treatment of glutamate-mediated excitotoxicity or analgesic therapies.
Keywords: arachidonic acid, mechanosensation, NMDA receptor, patch clamp, bilayer model
The ionotropic, glutamate-activated N-methyl-d-aspartate (NMDA) receptors are ligand-gated cation channels that play an important role in both physiological and pathological processes, including long-term potentiation and synaptic plasticity (1, 2), neuronal excitotoxicity, and cognitive deficits attributable to aging and pain (3–8). NMDA receptors are formed by heterooligomers of various NR1 and NR2 subunits (9). The secondary structure of the NMDA receptor monomer predicts four transmembrane segments (M1 to M4) with an extracellular N terminus and the C terminus located intracellularly. The M2 domain forms a cytoplasmic reentrant loop that lines the channel pore (9, 10). This region harbors a narrow constriction, forming the selectivity filter that controls voltage-dependent Mg2+ block (10–12). The C-terminal tail binds to cytoskeletal complexes, including kinases and structural proteins, which further modulate the function of the NMDA receptor channel (3, 13).
Mechanosensitivity is an important signal transduction mechanism that underlies a number of key biological processes ranging from cellular growth, cell volume and blood pressure regulation, touch, and pain sensation to cardiac arrhythmia, muscular dystrophy, and neuronal degeneration (14, 15). In prokaryotes, the mechanosensory transduction is carried out by mechanosensitive (MS) channels of small (MscS-like) and large (MscL-like) conductance (14, 16, 17). Many eukaryotic ion channels can be gated by mechanical forces. For example, the signaling properties of some membrane proteins, including ion channels, can change dramatically after undergoing a dynamic targeting to lipid microdomains within the cell membrane (18–20). Furthermore, the activity of several mammalian ion channels was found to be modulated by polyunsaturated fatty acids, including arachidonic acid and membrane phospholipids (21–25). The lipid bilayer and protein–lipid interaction are critical for MS gating and modulation of neuronal K2P channels such as TREK-1 and TRAAK (23, 24, 26). Several transient receptor potential channels can be activated by polymodal means such as membrane stretch, osmotic forces, heat, and exogenous lipids (27–30). The MEC superfamily of ion channels respond to body touch sensations in the worm Caenorhabditis elegans and, if mutated, induce death of touch cells (31). Stretch-activated calcium-dependent K+ channels are activated by both membrane stretch and amphipathic molecules that insert preferentially into one leaflet of the bilayer (32). Currently, there are two models that describe gating of MS channels by mechanical force: bilayer and tethered. Because prokaryotic cells lack a cytoskeleton, it has been shown that the lipid bilayer is the tension-bearing element transmitting the mechanical force to the MS channels (33–36). On the other hand, eukaryotic cells possess the excess membrane area supported by a contractile cortical cytoskeleton that locally regulates the mechanosensitivity of MS channels (37). However, recent evidence has demonstrated that eukaryotic MS channels also can be gated by the bilayer mechanism (27, 30, 38).
NMDA receptor channel activity is modulated by arachidonic acid (21, 39). Arachidonic acid cascade has been implicated in NMDA receptor-mediated synaptic plasticity, and arachidonic acid has been shown to enhance Ca2+ entry into hippocampal neurons after application of NMDA (40, 41). The enhancement of NMDA receptor responses by arachidonic acid, osmotic forces, membrane phospholipids, and membrane stretch led to a proposal that NMDA receptor channels are MS (22, 39). However, a direct effect of membrane tension on gating properties of the heteromeric channel complexes in cytoskeleton-free membrane preparations has not been investigated. The mechanism by which the mechanical stretch of neurons alters the voltage-dependent Mg2+ block and triggers Ca2+ influx through NMDA receptor channels after traumatic brain injury also remains poorly understood and requires further investigation (42–45). This study examines the mechanism by which recombinant NMDA receptor channel proteins reconstituted into cytoskeleton-free membrane preparations interact with the lipid environment. Our results demonstrate that the mechanical energy transmitted to membrane-bound NMDA receptor complexes is exerted via the lipid bilayer. In addition, we have found a correlation between membrane tension (induced by either mechanical or chemical stimuli) and internal Mg2+ block by using liposome patch clamp recording techniques.
Results
NMDA Receptor Protein Expression and Purification.
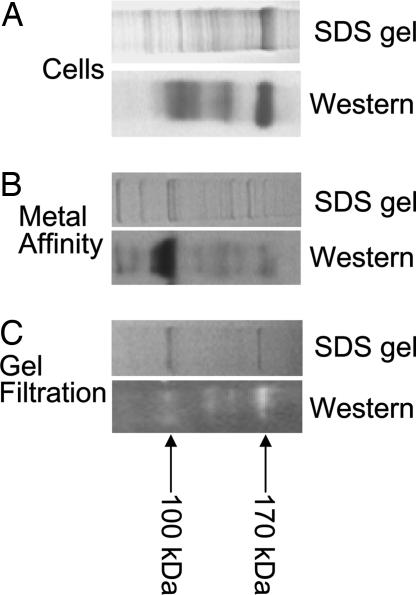
Samples of pelleted Spodoptera frugiperda (Sf9) insect cells coinfected with baculovirus expressing NR1a and NR2A subunits produced two bands of 170 and 100 kDa on an SDS/PAGE gel that immunoreacted with the 6xHis tag-specific antibodies (Fig. 1A). This band was not observed in uninfected cells (data not shown). Two bands of similar molecular mass were observed after purification of detergent-solubilized protein extract by metal chelate (Fig. 1B) followed by gel-filtration chromatography (Fig. 1C). This active fraction was used in experiments described below. The total yield from the Sf9 cells was ≈100 μg of pure receptor protein per liter of cell culture. Although metal-affinity chromatography resulted in several low-molecular-mass bands visible on SDS/PAGE gels and Western blots, the gel-filtration chromatography purification resulted in two main bands corresponding to 170 kDa for the NR2A subunit and 100 kDa for the NR1a subunit. A faint band of low-molecular mass still was visible on the Western blot after gel-filtration chromatography. It is most likely that the low-molecular-mass bands are breakdown products of NR1a or NR2A subunits. We have tested the gel-filtration chromatography fractions containing these low-molecular-mass protein bands and observed no channel activity after reconstitution into liposomes (n = 10, data not shown).
Fig. 1.
Purification of NR1a + NR2A NMDA receptor channel proteins. SDS/PAGE and corresponding 6xHis tag Western blot analysis of NMDA receptor protein samples isolated from Sf9 cells (A) after metal-affinity chromatography (B) and after gel-filtration chromatography (C). Arrows mark the positions of the NR1a and NR2A monomers.
Functional Properties of NMDA Receptors Reconstituted into Liposomes.
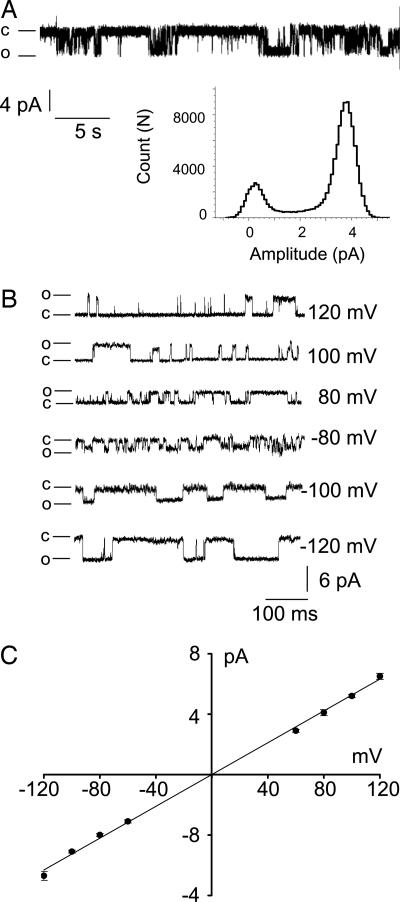
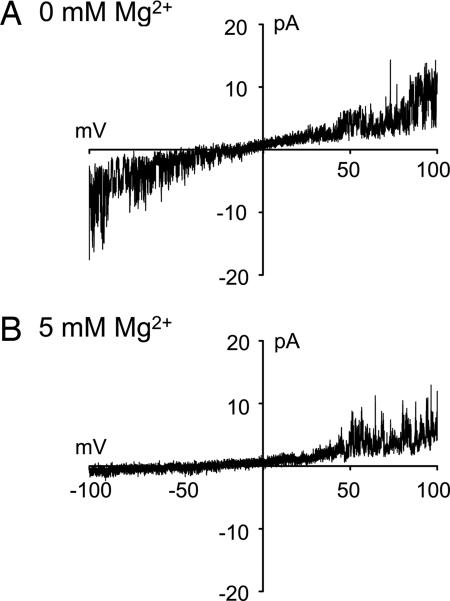
The activity of recombinant rat NMDA receptor channels composed of NR1a + NR2A subunits was examined after functional reconstitution of 20 μg of detergent-solubilized proteins, obtained after fractionation by using gel-filtration chromatography, into 5 mg of phosphatidylcholine (azolectin) lipids. This ratio of 1:250 was found to be optimal for channel activity. Reconstitution of both NR1a and NR2A subunits was necessary to obtain functional channels. No channel activity was observed from reconstituted fractions containing only one subunit (data not shown). Fig. 2 shows activity of recombinant NMDA receptor channels composed of NR1a and NR2A subunits in isolated azolectin liposome patches. Inclusion of receptor agonists (typically 10 μM glutamate and 100 μM glycine) in the pipette solution (facing the outside of the membrane) evoked single-channel currents in response to applied voltage (Fig. 2A). No unitary currents were recorded from control patches where either glutamate or glycine was not included in the pipette solution in the absence and presence of membrane stretch (n = 20 patches, data not shown). In symmetric recording solution lacking Mg2+, the current–voltage (I–V) relation of single-channel currents and I–V curves obtained by using voltage ramps were linear and reversed at 0 mV (Figs. 2B and 3A). The glutamate-activated NMDA receptor channel had a unitary cord conductance of ≈60 pS (Fig. 2) when calculated from the linear regression function or 58 ± 5.4 pS at +80 mV and 59 ± 4.1 pS at −80 mV (n = 5). However, the I–V curves produced by voltage ramps recorded in the presence of 5 mM internal Mg2+ (facing the inside of the patch membrane), and after inclusion of 10 μM glutamate and 100 μM glycine in the pipette solution, markedly rectified, indicating block of NMDA receptor channels by internal Mg2+ (n = 5 of 15 patches, Fig. 3B). Simultaneous addition of Mg2+ to the pipette and bath solutions abolished single-channel currents in all patches (n = 10). This observation is consistent with the hypothesis that intra- and extracellular Mg2+ binding sites are distinct and account for the two types of block (46).
Fig. 2.
Activity of recombinant NMDA receptor channels reconstituted into azolectin liposomes. (A) Current–amplitude frequency histogram constructed from 20 sec of single-channel data shown above recorded at +70 mV. The main open channel conductance level is ≈60 pS. (B) Representative single-channel currents recorded at different pipette potentials. The pipette solution contained 10 μM glutamate, 100 μM glycine, 200 mM KCl, and 5 mM Hepes (pH 7.3). The bath solution (i.e., facing the inside-out membrane) contained 200 mM KCl and 5 mM Hepes (pH 7.3). Closed and open channel states are marked c and o, respectively. (C) I–V relation of channels recorded from an isolated liposome patch containing reconstituted NR1a and NR2A receptor subunits. Single-channel conductance calculated from the slope of the fitted line was 60 pS (n = 5 patches).
Fig. 3.
Voltage-dependent block of liposome reconstituted recombinant NR1a + NR2A receptor proteins by internal Mg2+. Membrane currents were obtained in response to voltage ramps from −100 mV to +100 mV in the absence (A) and presence (B) of 5 mM Mg2+ in the bath solution. Membrane currents were recorded in the inside-out recording configuration. The external pipette solution contained 10 μM glutamate, 100 μM glycine, 200 mM KCl, and 5 mM Hepes (pH 7.3). The internal bath solution facing the inside-out membrane contained 200 mM KCl, 5 mM MgCl2, and 5 mM Hepes-KOH (pH 7.3) (n = 5 patches).
Mechanosensitivity of NMDA Receptor Channels in Liposome Patches.
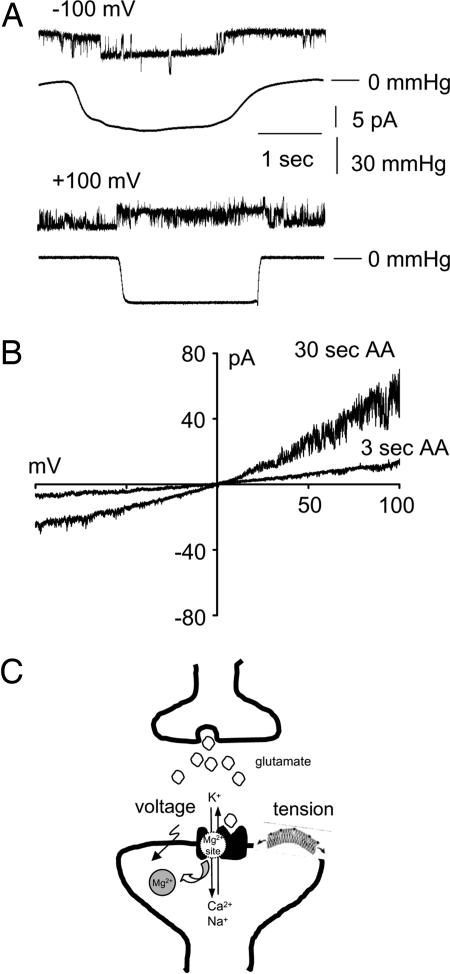
In liposome patches, the NMDA receptor channel activity was modified by negative pressure applied to the back of the patch pipette. In the presence of 5 mM Mg2+ in the bath solution, application of the negative pressure markedly reduced voltage-dependent internal Mg2+ block, resulting in opening of the channel to its full conductance at negative potentials (n = 5 of 16 patches, i.e., 11 patches have no channel activity). Similarly, at positive potentials the single-channel activity was enhanced when negative pressure was applied to the liposome membrane patch (Fig. 4A) (n = 5 of 15 patches, i.e., 10 patches had no channel activity). The pressure required for increased channel activity was between −50 and −80 mmHg (1mmHg = 133 Pa). We further examined the effect of the amphipathic molecule, arachidonic acid, coapplied with agonists (10 μM glutamate and 100 μM glycine) to the external face of the liposome membrane under the same experimental conditions (i.e., 5 mM Mg2+ in the bath solution). Arachidonic acid (5 μM) potentiated currents through open NMDA receptors (NR1a + NR2A) within ≈3 sec after seal formation by ≈2-fold compared with patches where arachidonic acid was not included (Fig. 4B) (n = 5 of 10 patches, i.e., 5 patches had no channel activity). There was a further 3-fold potentiation of NMDA receptor-mediated currents within 10–30 sec of arachidonic acid application. In the presence of Mg2+, the potentiating effect of arachidonic acid was more pronounced at positive pipette potentials compared with negative potentials. However, the rectification pattern of I–V curves, obtained by using voltage ramps, was less pronounced compared with patches lacking arachidonic acid, and there was a reduction in Mg2+ block at negative pipette potentials in the presence of arachidonic acid. Arachidonic acid had no significant effect on the reversal potential of the NMDA receptor-mediated current. Arachidonic acid alone did not activate detectable currents in control patches, that is, patches obtained from liposomes without reconstituted proteins (n = 20) or patches without agonists in the pipette solution (n = 20) (data not shown).
Fig. 4.
Modulation of Mg2+ block by stretch and arachidonic acid. (A) Stretch response of NMDA receptor channels. Representative currents were recorded from a liposome patch containing reconstituted recombinant NR1a and NR2A receptor subunits. The external pipette solution contained 10 μM glutamate, 100 μM glycine, 200 mM KCl, and 5 mM Hepes (pH 7.3). The internal bath solution facing the inside-out membrane contained 200 mM KCl, 5 mM MgCl2, and 5 mM Hepes (pH 7.3). The traces located below each current recording show the corresponding pressure recorded during applied suction (n = 5 patches). (B) Potentiation of NMDA receptor currents by arachidonic acid in isolated liposome membrane patches. Representative I–V curves for NR1a + NR2A subunit combinations obtained in the presence of 5 μM arachidonic acid evoked by voltage ramps from −100 mV to +100 mV. The recordings were acquired at 3 and 30 sec after the initial application of arachidonic acid to the liposome patch. The external pipette solution contained 10 μM glutamate, 100 μM glycine, 5 μM arachidonic acid, 200 mM KCl, and 5 mM Hepes (pH 7.3). The internal bath solution facing the inside-out membrane contained 200 mM KCl, 5 mM MgCl2, and 5 mM Hepes (pH 7.3) (n = 5 patches). (C) A schematic diagram of polymodal modulation of NMDA receptor currents. At the glutamatergic synapse, coincidental release of glutamate and membrane depolarization and/or membrane stretch releases the resting Mg2+ block, leading to ion influx.
Discussion
The study of recombinant ion channel proteins reconstituted into lipid bilayers is challenging because of the difficulty of expressing native protein complexes in the required amounts, homogeneity, and preserved functionality. In this study, we report the expression and purification of functional heteromeric NMDA receptor channel proteins composed of NR1a and NR2A subunits (Fig. 1). This work has permitted us to reconstitute the NMDA receptor complexes into azolectin liposomes, similar to the reconstitution of bacterial and archaeal MS channels (47–49), and reexamine the MS properties of the channel (22, 39). Recombinant NMDA NR1a + NR2A receptor proteins reconstituted into liposomes yield functional ion channels that exhibited gating properties similar to those observed in neuronal cells or Xenopus oocytes (21, 39, 46). A single open conductance level was observed in standard recording solution in the presence of glutamate and glycine facing the outside of the membrane. In contrast, previous studies of reconstituted glutamate receptor complexes isolated from brain membranes have reported multiconductance state behavior (50, 51). This discrepancy is most likely a result of procedures used to isolate and purify the receptor proteins. The glutamate receptor channel complexes isolated from brain membranes did not contain detectable amounts of NR1 subunit. This subunit is essential for NMDA receptor channel activity and plays an important role in Mg2+ block (12). Most likely the proteins reconstituted into bilayers were not NMDA receptor channels composed of NR1 and NR2 subunits as reported in our study but functional aggregates of multiple glutamate-activated NMDA and non-NMDA receptor channels that are present in rat and Xenopus brain preparations (50, 51). In addition to different subunit stoichiometry of channels, other features of protein reconstitution such as a source of proteins, lipid environment, or ionic conditions also may have affected the conductance state behavior of the NMDA receptor channels.
Stretching of the liposomal membrane resulted in increased NMDA receptor channel activity (Fig. 4). Previous studies have shown that both hypoosmotic solution and positive hydrostatic pressure potentiated NMDA receptor currents, whereas hyperosmotic solution and negative pressure had the opposite effect, indicating involvement of NMDA receptor channels in mechanotransduction (22, 39). However, patch clamp experiments investigating MS properties of NMDA receptors were performed in native membrane patches with an intact cytoskeleton. The cellular cytoskeleton network is a complex structure associated with many signaling molecules. NMDA receptors can interact with scaffolding proteins and second messenger system molecules (e.g., kinases), and their function can be affected by the integrity of actin (52–55). Therefore, it remained unclear whether interaction with other proteins within the lipid domain of the plasma membrane or tension transmitted via the lipid bilayer modulates the gating properties of NMDA receptor channels. In the present study, we demonstrate that mechanical energy transmitted to NMDA receptors exerted via the lipid bilayer alone is sufficient to modulate the channel activity without interaction with the cytoskeletal proteins. This occurs because liposome preparations are free from the cytoskeletal network. Furthermore, detergent-solubilized and homogeneity-purified NMDA receptor subunits have to undergo reassembly into the lipid bilayer during the reconstitution process to form functional channels that lack associating complexes. Thus, in an artificial lipid bilayer system such as liposomes, the effect of complex signaling pathways is eliminated when testing the effect of membrane deformation on the biophysical properties of ion channels.
In the model of stretch-induced neuronal cell injury, the Mg2+ block of the NMDA receptor channel was significantly reduced (42). PKC only partially restored the block by Mg2+, and the exact mechanism by which mechanical injury alters the activity of NMDA receptor channels remains unknown. We observed a similar reduction in internal Mg2+ block upon application of pressure to the liposome patches containing reconstituted NMDA receptor channel. The effect of mechanical deformation of the lipid membrane on NMDA receptor response in the presence of internal Mg2+ could be mimicked by arachidonic acid applied externally. Arachidonic acid significantly reduced Mg2+ block and potentiated currents through NMDA receptor channels in liposome patches. Potentiation of NMDA receptor currents by arachidonic acid has been reported previously in native patches from mammalian central neurons (21, 22). This effect could be explained in terms of the tension induced by differential incorporation of asymmetrical, cone-shaped lipophilic compounds into the lipid bilayer, creating mechanical stress on the channels in a manner similar to their effect on bacterial MS channels (34–36, 56), which further suggests that the membrane tension and/or the curvature of the lipid bilayer are important modulators of the NMDA receptor function. Mg2+ block confers on the NMDA receptor the capacity to act as a molecular coincidence detector and is an important factor in synaptic transmission. Because the physiological intracellular Mg2+ concentration is similar to the extracellular concentration, it is likely that the NMDA receptor acts as a bidirectional rectifier during synaptic transmission (46, 57). Therefore, a physiological consequence of membrane deformation and its interference with Mg2+ block could be reduced inhibition of the channel activity during synaptic transmission, which can result in intracellular Ca2+ transients.
Changes in intracellular [Ca2+] can play a detrimental role in traumatic and ischemic injury in the central nervous system (58). Stretch-induced deformation of neuronal cell membrane evoked intracellular Ca2+ transients by activation of synaptic NMDA receptor channels (44, 45). Ca2+ signaling via NMDA receptors also is an important modulator of long-term potentiation and synaptic plasticity (1, 2, 4). At CA1 synapses of hippocampus, where long-term potentiation is mediated via NMDA receptor channel, the long-term increase in synaptic transmission could be prevented by inhibition of phospholipase A2, which prevents release of arachidonic acid (59–61). Furthermore, arachidonic acid enhanced NMDA receptor-mediated Ca2+ entry into hippocampal neurons (40).
Our study demonstrates that, in the liposome system, arachidonic acid and membrane stretch both alleviate Mg2+ block of NMDA receptor channels. Similarly, the signaling properties of native NMDA receptor channels could be modulated directly by bilayer tension induced locally as a result of membrane deformation during dynamic targeting of proteins to lipid microdomains, electrocompression, or chemical modification of the bilayer (such as the arachidonic acid cascade system). Deformation of the cell membrane occurs in many physiological and pathological conditions such as during the motility of growth cones, elongation of neurites, and tissue trauma. Indeed, experimental evidence suggests that NMDA receptors may act as mechanomodulators during formation of dendritic spine synapses (62).
In conclusion, our results demonstrate that the activity of NMDA receptor channels can be enhanced by mechanical energy developed in the lipid bilayer alone. We propose a model where the gating properties of NMDA receptors are modulated by polymodal stimuli. In addition to a membrane depolarization, locally introduced membrane tension may lead to unblocking of NMDA receptor channels, allowing Ca2+ influx into the postsynaptic cell (Fig. 4C). The ability to express, purify, and reconstitute specific NMDA receptor subunits into liposomes may prove a useful method in further studies of the structure–function relationship and investigation of the role of the lipid environment in electrical signaling with further application to a variety of voltage- and ligand-gated ion channels. Knowledge of MS regulation of NMDA receptors may be instrumental in providing improved methods for treatment of glutamate-mediated neuronal degeneration, analgesic therapies, and development of in vitro models of cellular injury (i.e., brain trauma). Further study of MS channels reconstituted into lipid bilayers could be crucial for the discovery of new therapeutic agents for treatment of disorders that involve MS proteins.
Materials and Methods
Recombinant Baculoviruses Construction.
cDNA clones of the rat NR1a and NR2A NMDA receptors were a gift of L. Wollmuth (State University of New York, Stony Brook, NY) and J. Boulter (University of California, Los Angeles, CA). The NR1a and NR2A subunits were cloned separately into pENTR/D-TOPO (Invitrogen, Carlsbad, CA) gateway vector, and a TEV protease cleavage site was introduced on the 3′ end of the gene to allow the removal of tag if required. Forward and reverse primers used to amplify and clone NR1a subunit were 5′-CACCATGAGCACCATGCACCTGCTGAC-3′ and 5′-TCAATGGTGATGGTGATGATGGCTCTCCCTATGACGGGAACAC-3′, respectively. The NR2A subunit was amplified and cloned by using forward primer 5′-CACCATGGGCAGATTGGGCTACTGG-3′ and reverse primer 5′-TCAATGGTGATGGTGATGATGAACATCAGATTCGATACTAGG-3′. The sequences of both clones were verified by sequencing. To create recombinant baculovirus expressing each gene, the sequenced pENTR clones were recombined with BaculoDirect C-term Linear DNA (Invitrogen) and transfected into Sf9 insect cells according to the manufacturer's protocol. At 7 days postinfection, the budded virus of each construct was harvested and used for subsequent budded virus amplification.
Protein Expression in Insect Cells.
Sf9 cells were grown in Sf-900II (Invitrogen) serum-free media with shaking at 120 rpm at 27°C. For coexpression of NR1a and NR2A subunits, the cells were coinfected at the same multiplicity of infection (moi) with both NR1a and NR2A recombinant baculoviruses at 3 × 106 cells per ml by using a moi of 2–5 pfu per cell. The optimal time of harvest for the coexpression of NR1a and NR2A subunits was 72 h postinfection. The culture was centrifuged at 8,000 × g for 15 min at 4°C, and the cell pellet was kept at −80°C until required.
Protein Purification.
For lysis, the cell pellet from a 1-liter expression was resuspended in 100 ml of buffer A (20 mM sodium phosphate, 500 mM NaCl, 10 mM imidazole, and 1.5% octyl glucoside, pH 7.8) with EDTA-free protease inhibitor tablets (Roche, Basel, Switzerland). Sample was placed on a rotating wheel for 20 min, followed by sonication three times with a probe at 75 W for 15 sec with 10- to 20-sec cooling periods on ice. The octyl glucoside concentration was adjusted to 3% after sonication and placed on a rotating wheel for another 20 min. The lysed sample was clarified by centrifugation at 18,000 × g for 15 min at 4°C. The clarified lysate was applied to TALON resin (Clontech, Mountain View, CA) for binding. Then, 12.5 ml of lysate was applied to 1.25 ml of TALON and placed on a rotating wheel for 20–30 min. Unbound proteins were removed by centrifugation at 700 × g for 2 min at 4°C. After binding, each 1.25 ml of TALON was washed for 10 min with 50 ml of buffer A. Elution of the protein was performed by transferring 2.5 ml of TALON to a 10-ml disposable column (Bio-Rad, Hercules, CA), and protein was eluted with 1 ml of buffer A containing 150 mM imidazole. For gel filtration, 500 μl of TALON-purified protein sample was loaded onto a Superdex 75 10/300 GL column (GE Healthcare, Rydalmere, NSW, Australia) at 0.25 ml/min with 20 mM sodium phosphate, 300 mM NaCl buffer at pH 7.5. Fractions (0.5 ml) were collected for analysis by Western blotting and reconstitution into proteoliposomes.
Preparation of Proteoliposomes.
Detergent-solubilized protein fractions were incubated with phosphatidylcholine (azolectin, P3644; Sigma, Castle Hill, NSW, Australia) liposomes at various protein ratios for 30 min at room temperature. Bio-Beads (Bio-Rad) were added, and the suspension was rocked for 4 h to remove detergent. The supernatant then was centrifuged for 30 min at 90,000 rpm (TL-100; Beckman, Gladesville, NSW, Australia). The pelleted proteoliposomes were resuspended in 50 μl of 10 mM Hepes-KOH (pH 7.2), and aliquots of liposomes were spotted onto glass slides followed by dehydration at 4°C inside a desiccator. A small aliquot of rehydrated liposomes was placed in a patch clamp chamber filled with the recording solution of 200 mM KCl and 5 mM Hepes (pH 7.3) in the absence or presence of 5 mM MgCl2. Presence of Mg2+ in the bath solution was a necessary requirement to increase the patch stability during stretching of the liposome membrane. Pipettes were back-filled with a solution containing 10 μM glutamate, 100 μM glycine, 200 mM KCl, and 5 mM Hepes (pH 7.3). Gigaohm seals were formed either on contact between the pipette and a blister or by a brief application of suction.
Electrophysiological Recordings.
Membrane currents were recorded from the isolated liposome patches by using the improved patch clamp techniques (63). Pipettes were made from borosilicate glass (Sigma) by using a micropipette puller (P-87; Sutter Instrument Co., Novato, CA) and had an average resistance of 5–6 MΩ in a recording solution containing 200 mM KCl and 5 mM Hepes-KOH. To stretch the membrane patch, suction was applied to the pipette holder with a syringe, and the applied pressure was monitored by piezoelectric transducer (Omega Engineering, Manchester, U.K.). Glutamate-activated single-channel currents were filtered at 2 kHz, digitized at 5 kHz, and analyzed by using pCLAMP 9 data acquisition and analysis software (Axon Instruments, Union City, CA). Current recordings were viewed with the Clampfit for Windows program (Axon Instruments), and current amplitudes were determined by measuring the difference between the cursor aligned at the peak and baseline currents. Voltage ramps were applied from −100 to +100 mV over 2.64 sec.
Not all proteoliposome patches contained active channels, and in general the success rate of obtaining quality recordings in the absence of Mg2+ was ≈20%. This low success rate was mainly because of the poor quality of blisters formed in the absence of Mg2+ as well as liposome membrane patch instability. The addition of 5 mM Mg2+ to the bath solution improved patch stability and resulted in a higher success rate.
Abbreviations
- MS
mechanosensitive
- I–V
current–voltage
- NMDA
N-methyl-d-aspartate.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS direct submission.
References
- 1.Bliss TV, Collingridge GL. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 2.Castellano C, Cestari V, Ciamei A. Curr Drug Targets. 2001;2:273–283. doi: 10.2174/1389450013348515. [DOI] [PubMed] [Google Scholar]
- 3.Waxman EA, Lynch DR. Neuroscientist. 2005;11:37–49. doi: 10.1177/1073858404269012. [DOI] [PubMed] [Google Scholar]
- 4.Cull-Candy S, Brickley S, Farrant M. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- 5.Rosi S, Ramirez-Amaya V, Hauss-Wegrzyniak B, Wenk GL. J Neuroinflam. 2004;1:12. doi: 10.1186/1742-2094-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Salter MW. Curr Top Med Chem. 2005;5:557–567. doi: 10.2174/1568026054367638. [DOI] [PubMed] [Google Scholar]
- 7.Brown DG, Krupp JJ. Curr Top Med Chem. 2006;6:749–770. doi: 10.2174/156802606777057571. [DOI] [PubMed] [Google Scholar]
- 8.Chizh BA, Headley PM. Curr Pharm Des. 2005;11:2977–2994. doi: 10.2174/1381612054865082. [DOI] [PubMed] [Google Scholar]
- 9.Dingledine R, Borges K, Bowie D, Traynelis SF. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- 10.Kuner T, Wollmuth LP, Karlin A, Seeburg PH, Sakmann B. Neuron. 1996;17:343–352. doi: 10.1016/s0896-6273(00)80165-8. [DOI] [PubMed] [Google Scholar]
- 11.Wollmuth LP, Kuner T, Seeburg PH, Sakman B. J Physiol. 1996;491:779–797. doi: 10.1113/jphysiol.1996.sp021257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kupper J, Ascher P, Neyton J. J Physiol. 1998;507:1–12. doi: 10.1111/j.1469-7793.1998.001bu.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cull-Candy SG, Leszkiewicz DN. Sci STKE. 2004;255:1–9. doi: 10.1126/stke.2552004re16. [DOI] [PubMed] [Google Scholar]
- 14.Martinac B. J Cell Sci. 2004;117:2449–2460. doi: 10.1242/jcs.01232. [DOI] [PubMed] [Google Scholar]
- 15.Hamill OP. Pflügers Arch. 2006;453:333–351. doi: 10.1007/s00424-006-0131-0. [DOI] [PubMed] [Google Scholar]
- 16.Martinac B, Kloda A. Prog Biophys Mol Biol. 2003;82:11–24. doi: 10.1016/s0079-6107(03)00002-6. [DOI] [PubMed] [Google Scholar]
- 17.Kung C, Blount P. Mol Microbiol. 2004;53:373–380. doi: 10.1111/j.1365-2958.2004.04180.x. [DOI] [PubMed] [Google Scholar]
- 18.Martens JR, Navarro-Polanco R, Coppock EA, Nishiyama A, Parshley L, Grobaski TD, Tamkun MM. J Biol Chem. 2000;275:7443–7446. doi: 10.1074/jbc.275.11.7443. [DOI] [PubMed] [Google Scholar]
- 19.Lockwitch TP, Liu X, Singh BB, Jadlowiec J, Weiland S, Ambudkar IS. J Biol Chem. 2000;275:11934–11942. doi: 10.1074/jbc.275.16.11934. [DOI] [PubMed] [Google Scholar]
- 20.Bravo-Zehnder M, Ori P, Norambuena A, Wallner M, Meera P, Toro L, Lattore R, Gonzales A. Proc Natl Acad Sci USA. 2000;97:13114–13119. doi: 10.1073/pnas.240455697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller B, Sarantis M, Traynelis SF, Attwell D. Nature. 1992;355:722–725. doi: 10.1038/355722a0. [DOI] [PubMed] [Google Scholar]
- 22.Casado M, Ascher P. J Physiol. 1998;513:317–330. doi: 10.1111/j.1469-7793.1998.317bb.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel AJ, Honoré E, Maingret F, Lesage F, Fink M, Duprat F, Lazdunski M. EMBO J. 1998;17:4283–4290. doi: 10.1093/emboj/17.15.4283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maingret F, Fosset M, Lesage F, Lazdunski M, Honoré E. J Biol Chem. 1999;274:1381–1387. doi: 10.1074/jbc.274.3.1381. [DOI] [PubMed] [Google Scholar]
- 25.Chemin J, Patel AJ, Duprat F, Lauritzen I, Lazdunski M, Honoré E. EMBO J. 2005;24:44–53. doi: 10.1038/sj.emboj.7600494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honoré E, Patel AJ, Chemin J, Suchyna T, Sachs F. Proc Natl Acad Sci USA. 2006;103:6859–6864. doi: 10.1073/pnas.0600463103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maroto R, Raso A, Wood TG, Kurosky A, Martinac B, Hamill OP. Nat Cell Biol. 2005;7:179–185. doi: 10.1038/ncb1218. [DOI] [PubMed] [Google Scholar]
- 28.Minke B, Cook B. Physiol Rev. 2002;82:429–472. doi: 10.1152/physrev.00001.2002. [DOI] [PubMed] [Google Scholar]
- 29.Clapham DE. Nature. 2003;426:517–522. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- 30.Ramsey IS, Delling M, Clapham DE. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 31.Tavernarakis N, Driscoll M. Annu Rev Physiol. 1997;59:659–689. doi: 10.1146/annurev.physiol.59.1.659. [DOI] [PubMed] [Google Scholar]
- 32.Qi Z, Chi S, Su S, Naruse K, Sokabe M. Mol Membr Biol. 2005;22:519–527. doi: 10.1080/09687860500370703. [DOI] [PubMed] [Google Scholar]
- 33.Martinac B, Delcour AH, Buechner M, Adler J, Kung C. In: Comparative Aspects of Mechanoreceptor Systems. Ito F, editor. Berlin: Springer; 1992. pp. 3–18. [Google Scholar]
- 34.Hamill OP, Martinac B. Physiol Rev. 2001;81:685–740. doi: 10.1152/physrev.2001.81.2.685. [DOI] [PubMed] [Google Scholar]
- 35.Perozo E, Kloda A, Cortes DM, Martinac B. Nat Struct Biol. 2002;9:696–703. doi: 10.1038/nsb827. [DOI] [PubMed] [Google Scholar]
- 36.Perozo E, Cortes DM, Sompornpisut P, Kloda A, Martinac B. Nature. 2002;418:942–948. doi: 10.1038/nature00992. [DOI] [PubMed] [Google Scholar]
- 37.Hamill OP, McBride DW. Trends Neurosci. 1994;17:439–443. doi: 10.1016/0166-2236(94)90125-2. [DOI] [PubMed] [Google Scholar]
- 38.Zhang Y, Gao F, Popov VL, Wen JW, Hamill OP. J Physiol. 2000;523:117–130. doi: 10.1111/j.1469-7793.2000.t01-1-00117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paoletti P, Ascher P. Neuron. 1994;13:645–655. doi: 10.1016/0896-6273(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 40.Richards DA, Bliss TVP, Richards CD. Eur J Neurosci. 2003;17:2323–2328. doi: 10.1046/j.1460-9568.2003.02671.x. [DOI] [PubMed] [Google Scholar]
- 41.Dumuis A, Sebben M, Haynes L, Pin JP, Bockaert J. Nature. 1988;336:68–70. doi: 10.1038/336068a0. [DOI] [PubMed] [Google Scholar]
- 42.Zhang L, Rzigalinski BA, Ellis EF, Satin LS. Science. 1996;274:1921–1923. doi: 10.1126/science.274.5294.1921. [DOI] [PubMed] [Google Scholar]
- 43.Regan RF, Choi DW. Brain Res. 1994;633:236–242. doi: 10.1016/0006-8993(94)91544-x. [DOI] [PubMed] [Google Scholar]
- 44.LaPlaca MC, Thibault LE. J Neurosci Res. 1998;52:220–229. doi: 10.1002/(SICI)1097-4547(19980415)52:2<220::AID-JNR10>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 45.Geddes-Klein DM, Serbest G, Mesfin MN, Cohen AS, Meaney DF. J Neurosci. 2006;97:462–474. doi: 10.1111/j.1471-4159.2006.03761.x. [DOI] [PubMed] [Google Scholar]
- 46.Johnson JW, Ascher P. Biophys J. 1990;57:1085–1090. doi: 10.1016/S0006-3495(90)82626-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Delcour AH, Martinac B, Adler J, Kung C. Biophys J. 1989;56:631–635. doi: 10.1016/S0006-3495(89)82710-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Le Dain AC, Saint N, Kloda A, Ghazi A, Martinac B. J Biol Chem. 1998;273:12116–12119. doi: 10.1074/jbc.273.20.12116. [DOI] [PubMed] [Google Scholar]
- 49.Kloda A, Martinac B. Biophys J. 2001;80:229–240. doi: 10.1016/S0006-3495(01)76009-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Aistrup GL, Szentirmay M, Kumar KN, Babcock KK, Schowen RL, Michaelis EK. FEBS Lett. 1996;394:141–148. doi: 10.1016/0014-5793(96)00938-6. [DOI] [PubMed] [Google Scholar]
- 51.Kerry CJ, Sudan HL, Abutidze K, Mellor IR, Barnard EA, Usherwood PNR. Mol Pharmacol. 1992;44:142–152. [PubMed] [Google Scholar]
- 52.Rosenmund C, Westbrook GL. Neuron. 1993;10:805–814. doi: 10.1016/0896-6273(93)90197-y. [DOI] [PubMed] [Google Scholar]
- 53.Kornau HC, Seeburg PH, Kennedy MB. Curr Opin Neurobiol. 1997;7:368–373. doi: 10.1016/s0959-4388(97)80064-5. [DOI] [PubMed] [Google Scholar]
- 54.Wyszynski M, Lin J, Rao A, Nigh E, Beggs AH, Craig AM, Shen M. Nature. 1997;385:439–442. doi: 10.1038/385439a0. [DOI] [PubMed] [Google Scholar]
- 55.Chen L, Huang LYM. Nature. 1992;356:521–523. doi: 10.1038/356521a0. [DOI] [PubMed] [Google Scholar]
- 56.Martinac B, Adler J, Kung C. Nature. 1990;348:261–263. doi: 10.1038/348261a0. [DOI] [PubMed] [Google Scholar]
- 57.Alvarez-Leefmans FJ, Gamino SM, Giraldez F, Gonzales-Serratos H. J Physiol. 1986;378:461–483. doi: 10.1113/jphysiol.1986.sp016230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tymianski M, Tator CH. Neurosurgery. 1996;38:1176–1195. doi: 10.1097/00006123-199606000-00028. [DOI] [PubMed] [Google Scholar]
- 59.Okada D, Yamagishi S, Sugiyama H. Neurosci Lett. 1989;100:141–146. doi: 10.1016/0304-3940(89)90674-5. [DOI] [PubMed] [Google Scholar]
- 60.Williams JH, Bliss TVP. Neurosci Lett. 1988;88:81–85. doi: 10.1016/0304-3940(88)90319-9. [DOI] [PubMed] [Google Scholar]
- 61.Linden DJ, Sheu FS, Murakami K, Routtenberg A. J Neurosci. 1987;7:3783–3792. doi: 10.1523/JNEUROSCI.07-11-03783.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korkotian E, Segal M. Neuron. 2001;30:751–758. doi: 10.1016/s0896-6273(01)00314-2. [DOI] [PubMed] [Google Scholar]
- 63.Hamill OP, Marty A, Neher E, Sakmann B, Sigworth FJ. Pflügers Arch. 1981;391:85–100. doi: 10.1007/BF00656997. [DOI] [PubMed] [Google Scholar]