Abstract
MicroRNAs are small noncoding 18- to 24-nt RNAs that are predicted to regulate expression of as many as 30% of protein-encoding genes. In prostate adenocarcinoma, 39 microRNAs are up-regulated, and six microRNAs are down-regulated. Production and function of microRNA requires coordinated processing by proteins of the microRNA machinery. Dicer, an RNase III endonuclease, is an essential component of the microRNA machinery. From a gene array analysis of 16 normal prostate tissue samples, 64 organ-confined, and four metastatic prostate adenocarcinomas, we identified an up-regulation of major components of the microRNA machinery, including Dicer, in metastatic prostate adenocarcinoma. Immunohistochemical studies on a tissue microarray consisting of 232 prostate specimens confirmed up-regulation of Dicer in prostatic intraepithelial neoplasia and in 81% of prostate adenocarcinoma. The increased Dicer level in prostate adenocarcinoma correlated with clinical stage, lymph node status, and Gleason score. Western blot analysis of benign and neoplastic prostate cell lines further confirmed Dicer up-regulation in prostate adenocarcinoma. Dicer up-regulation may explain an almost global increase of microRNA expression in prostate adenocarcinoma. The presence of up-regulated microRNA machinery may predict the susceptibility of prostate adenocarcinoma to RNA interference-based therapy.
MicroRNAs (miRNA or miR) are a class of small noncoding 18- to 24-nt RNAs that regulate diverse cellular and molecular processes, including cell death and proliferation.1 Recent data implicate differential expression of miR in the development of multiple types of human malignancies, including prostate adenocarcinoma (PCa): 39 miRs are up-regulated and six miRs are down-regulated in PCa.2–4 Targets of dysregulated miRs include major tumor suppressor genes (eg, retinoblastoma, RB1).4 The list of miR with known cancer gene targets continues to grow: let-7 negatively regulates Ras,5 miR-17-5p and miR-20a control E2F,6 and miR-16-1 and miR-15a repress Bcl-2.7
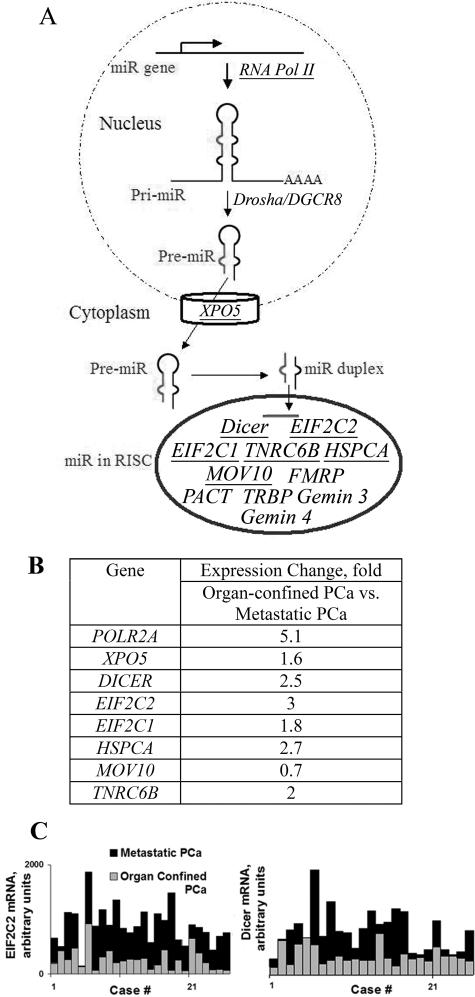
Production and function of miR requires a set of proteins collectively referred to as the miR machinery. MiRs encoded in the genome are transcribed by RNA polymerase II into primary transcripts, pri-miRNAs.8 The next step of miR maturation is the nuclear cleavage of pri-miRNA and release of a 60- to 70-nt stem-loop intermediate, an miRNA precursor (pre-miRNA). This step requires a ∼650-kd microprocessor complex that comprises Drosha, an RNase III endonuclease, and its cofactor, DGCR8.2 Pre-miRs are actively transported from the nucleus to the cytoplasm by the Exportin 5/RanGTP complex.9 In the cytoplasm, Dicer and the human immunodeficiency virus (HIV)-1 transactivating response RNA-binding protein cut both strands of the pre-miRNA duplex, generating a mature ∼21-nucleotide miR duplex.10,11 Chendrimada et al isolated a 500-kd protein complex containing transactivating response RNA-binding protein, Dicer, and EIF2C212. This complex also includes Gemin 3 and Gemin 4 and links miRNA processing with the assembly of the RNA-induced silencing complexes.12,13 Dicer has also been found to stably associate with HSPCA and EIF2C1.14,15 In addition, MOV10, TNRC6B, and protein arginine methyltransferase 5 specifically associate with EIF2C proteins.14 PACT (protein kinase, interferon-inducible double-stranded RNA-dependent activator, PRKRA) has been shown to interact with Dicer without facilitating its pre-miRNA cleavage activity.16 Finally, fragile X mental retardation protein interacts with the mammalian EIF2C2 and is associated with Dicer activity.17,18 Maturation of miR is summarized in Figure 1A.
Figure 1.
Up-regulation of the components of the miR machinery in prostate cancer. A: Schematic representation of miR maturation. Up-regulated genes are underlined. B: Up-regulation of genes encoding miR machinery components in metastatic prostate adenocarcinoma. The fold change over organ-confined prostate adenocarcinoma is shown. POLR2A, polymerase (RNA) II polypeptide A; XPO5, exportin 5; EIF2C2, eukaryotic translation initiation factor 2C (Ago2); HSPCA, heat shock 90-kd protein 1, alpha; TNRC6B, trinucleotide repeat containing 6B; MOV10, Moloney leukemia virus 10, homolog; EIF2C1, eukaryotic translation initiation factor 2C1 (Ago1). C: Dicer mRNA and EIF2C2 mRNA levels in organ-confined and metastatic PCa (P < 0.05) assessed by gene array. Representative cases are shown.
In an miR-guided fashion, the miR machinery regulates the expression of multiple tumor suppressor genes and oncogenes.19 Several components of miR machinery are implicated in carcinogenesis. PACT is up-regulated in bronchioloalveolar carcinoma, and a higher level of PACT in small-size peripheral adenocarcinomas of the lung was associated with shorter survival periods.20 Jaronczyk et al have observed an elevated expression of Argonaute mRNA, another component of the miR machinery, in multiple human tumor cell lines.21 It has been shown that the reduced expression of Dicer mRNA in non-small cell lung carcinomas is associated with shorter postoperative survival.22 In addition, a fourfold increased expression of Dicer mRNA was shown in the Burkitt’s lymphoma-derived cell line EB-3.23
In addition to being an important RNA interference (RNAi) effector, it has been shown that Dicer mutations are associated with defects independent of global errors induced by dysfunction of the miR machinery. In yeast, independently of the RNAi pathway, Dicer has been shown to regulate G1 arrest in response to nitrogen-limiting conditions and initiate the Cdc2-dependent DNA replication and DNA damage checkpoints.24,25 Dicer is crucial for proper structuring of centromeric heterochromatin, a process important for accurate sister chromosome segregation.26 In a chicken-human hybrid cell line, loss of Dicer leads to chromosome mis-segregation and to the accumulation of cells in the G2/M phase of the cell cycle.27 These findings point to the role of Dicer in controlling checkpoints in response to mutagenic stress. In conclusion, it is important to consider the aberrant expression of Dicer and other components of the miR machinery in human malignancies.
In an effort to elucidate the mechanism of miR up-regulation in PCa, we characterized the alterations in expression of genes encoding proteins of miR machinery. Our gene array analysis demonstrated an increased transcription of Dicer and most of its partners in metastatic PCa. The up-regulation of Dicer with PCa progression was confirmed by immunohistochemical analysis of clinical samples and immunocytochemical and Western blot analysis of prostate cell lines.
Materials and Methods
Clinical Profile of Cases and Paraffin Tissue Microarray
The tissue microarray set consists of 232 prostate tissue specimens from 166 different patients arrayed onto slides in quadruplicate. The demographic and clinicopathologic features of the patients in this study are listed in Table 1. Regional lymph nodes were not assessed in two cases (NX). Two specimens were obtained from patients of Asian race, 10 from African Americans, and 200 from Caucasians. The race of 20 patients was not available. Samples were procured as previously described.28
Table 1.
Summary of the Clinicopathologic and Demographic Features of the Patients
| Variable | Number |
|---|---|
| Age | 63.3 (46 to 81 years) |
| Normal prostate tissue | 11 |
| Benign prostatic hypertrophy | 15 |
| Normal adjacent to tumor | 42 |
| Prostatic intraepithelial neoplasia | 38 |
| Prostate adenocarcinoma | |
| Stage II | 41 |
| Stage III | 39 |
| Stage IV | 30 |
| Lymph node status, N0 | 83 |
| Lymph node status, N1 | 22 |
| T2a | 5 |
| T2b | 41 |
| T3a | 32 |
| T3b | 22 |
| T4 | 8 |
| Gleason score, <7 | 15 |
| Gleason score, 7 | 52 |
| Gleason score, >7 | 40 |
| Metastases | 13 |
Immunohistochemical Stains and Statistical Analysis
For immunostaining, 4-μm sections of tissue array were cut and mounted on glass slides. The sections were heated at 60°C for 12 hours and deparaffinized in xylene and ethanol. Antigen retrieval was performed using 25 mmol/L sodium citrate buffer (pH 6.0) at 90°C for 15 minutes, followed by treatment of 3% H2O2 to block endogenous peroxidase. The slides were incubated at 4°C overnight with anti-Dicer antibodies at 1:600 dilution. The sections were then incubated with biotinylated anti-mouse IgG (Vector Elite Kit; Vector Laboratories, Burlingame, CA) for 30 minutes. This was followed by incubating the section with Vectastain Elite ABC reagent (Vector Laboratories, Burlingame, CA) and 3,3′-diaminobenzidine solution (DAKO, Carpinteria, CA) to develop the stain. Hematoxylin was used for counterstaining. Only staining of the luminal epithelial cells was scored. The staining of the basal cells was used as a positive internal control. Strongly positive (“3”) staining of Dicer was defined as homogenous cytoplasm staining more intense than in basal cells. Moderately positive (“2”) score was assigned to a granular cytoplasm staining with apical cytoplasmic or perinuclear concentration. This level of intensity was equal to that normally seen in basal cells. Weakly positive (“1”) staining was less intense than in basal cells and limited to the basal cytoplasm. “0” reflected the lack of Dicer immunoreactivity and was the most common pattern seen in benign luminal cells. Three pathologists (R.D., T.M., and S.C.) scored stained slides. Scores for all cores from one case were averaged. Cases with all cores missing on all examined tissue microarray sections were excluded from the analysis. All statistical analysis of immunohistochemical studies of the tissue microarray set was performed with Sigma Stat (SYSTAT, Point Richmond, CA). Two sample comparisons were performed with the Mann-Whitney rank sum test. Multiple samples were compared using analysis of variance on ranks or one-way analysis of variance.
Immunoblot Detection of Dicer
The cells were washed with phosphate-buffered saline and lysed using a cell disruption buffer (mirVANA PARIS; Ambion, Austin, TX). The proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (10% polyacrylamide gel) and blotted onto a nitrocellulose membrane. The membrane was blocked with 5% powdered nonfat milk in Tris-Tween 20 buffer, pH 7.4, overnight at 4°C, followed by 2 hours of incubation with a 1:600 dilution of anti-Dicer antibody. The membrane was then washed three times with Tris-Tween 20 buffer and incubated with anti-mouse IgG for 1 hour at room temperature. Dicer expression was detected with the Super SignalWest Femto Maximum Sensitivity substrate (Pierce, Rockford, IL) according to the manufacturer’s protocol.
Gene Array: cRNA Preparation, Affymetrix Chip Hybridization, Data Preprocessing, and Statistical Analysis
cRNA was prepared and hybridized to Affymetrix HGU95Av2, B, and C chips as previously described.29 The raw scanned array images were first processed through the Affymetrix Microarray Analysis Suite 5.0 (MAS; Affymetrix Corporation, Santa Clara, CA) to generate probe cel intensity (*.cel) files. The *.cel files were then analyzed using MAS 5.0 to generate gene expression signal values for each probe set. Data normalization to remove variation in overall chip intensities was performed by global scaling to a chip mean target intensity of 200 (MAS 5.0). We analyzed 16 normal prostate samples, organ-confined prostate cancer tissue samples from 64 prostate cancer patients, and 24 metastatic tissues from four patients. To identify differentially regulated genes, Affymetrix U95Av2, B, and C chip data were combined, and organ-confined PCa were compared with donors and to metastatic samples by significance analysis of microarrays.30 Before analysis, genes that show low variation across all samples were removed by using the filtering option in the Avadis (Strand Genomics, Bangalore, India) data analysis tool.
Antibodies, Cell Lines, and Cell Culture
Dicer antibody was from Clonegene (Hartford, CT) and was used at a 1:600 dilution. Tubulin antibody was from Oncogene Research Products (San Diego, CA). DU145 and RWPE-1 cell lines were from American Type Culture Collection (Manassas, VA). RWPE-1 was maintained in complete keratinocyte serum-free medium containing 50 μg/ml bovine pituitary extract, 5 ng/ml epidermal growth factor, and 1% antibiotic/antimycotic mixture. Medium was changed every 48 hours. Cells were passaged on confluence and seeded at 1 to 2 × 106 cells/T-25 flask. DU145 were maintained in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (Gibco, Sydney, Australia). The primary prostate cancer cells, passage 0, were derived from prostatectomy specimen.
Results
Gene Array Analysis of miR Machinery in Prostate Adenocarcinoma
Using Affymetrix HGU95 chips we analyzed gene expression in 64 patients with organ-confined PCa and 24 metastatic PCa samples from four patients. The gene expression analysis of the organ-confined PCa and normal prostate samples was previously described by our group.28 All of the genes encoding miR machinery components showed a similar level of expression in normal prostate and in the organ-confined PCa. However, expression of several genes was up-regulated in metastatic PCa (summarized in Figure 1). POLR2A, polymerase RNA II, was up-regulated 5.1-fold in metastatic PCa. Drosha (RNASE3L) and DGCR8 did not show differential expression in normal prostate versus organ-confined PCa versus metastatic PCa.
Dicer and its closest partner, EIF2C2, were up-regulated 2.5- and threefold, respectively, in metastatic PCa compared with organ-confined PCa. Several major functional partners of Dicer were up-regulated in metastatic PCa as well. In our analysis, XPO5 was up-regulated 1.6-fold. A significantly increased Exportin 5 level in metastatic PCa has been previously shown.31 EIF2C1 and HSPCA were up-regulated 1.8- and 2.7-fold, respectively, in metastatic samples compared with organ-confined PCa.
Immunohistochemical Analysis of Dicer Expression in Benign Human Prostate Tissue
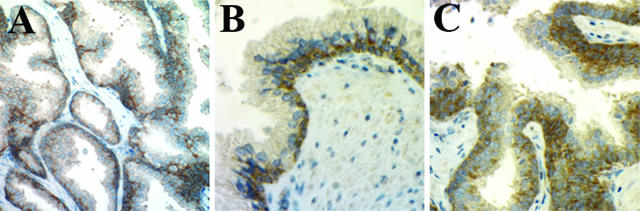
The normal human prostatic epithelium consists of basal, intermediate, secretory luminal, and neuroendocrine cells.32 Within the layer of the basal cells there are stem cells.32 The expression of Dicer was examined in normal prostate tissue, normal tissue adjacent to the PCa (normal adjacent to tumor), and benign prostatic hypertrophy specimens. In normal prostate tissue, Dicer immunoreactivity was limited to the cytoplasm of the basal cells (Figure 2, A and B). No nuclear staining was identified. Weak Dicer immunoreactivity was rarely detected in fibromuscular stroma. Dicer was negative in the luminal cells of the glands involved with basal cell hyperplasia, whereas the foci of basal cell hyperplasia were distinctly positive (Figure 2C).
Figure 2.
Expression of Dicer in benign prostate epithelium. A, B: Basal cells express a high level of Dicer, whereas luminal cells are negative, as determined by immunohistochemistry; original magnification: ×200 (A) and ×400 (B). C: Dicer highlights basal cell hyperplasia, as determined by immunohistochemistry; original magnification, ×400.
Of the benign prostate specimens, 84% (54 of 64) showed immunoreactivity of ≤0.5, and the mean Dicer immunoreactivity in all benign samples (n = 64) was 0.2. Although luminal cells in the normal adjacent to tumor samples tended to show higher Dicer immunoreactivity, no statistically significant difference was observed between normal prostates, normal adjacent to tumor, and benign prostatic hypertrophy specimens. Of note, urethral urothelium and peripheral ganglion cells within the prostate demonstrated strong cytoplasmic Dicer immunoreactivity. The results of the immunohistochemical analysis of Dicer expression in normal prostate tissue are summarized in Table 2.
Table 2.
Expression of Dicer in Benign Prostate Tissue and Prostatic Intraepithelial Neoplasia
| Histology | No. of cases | Mean ± SEM | Immunoreactivity score
|
|||
|---|---|---|---|---|---|---|
| 0 | 0.1 to 0.5 | 0.6 to 1 | 1.1 to 2 | |||
| Normal prostate | 11 | 0.3 ± 0.1 | 55% (6) | 18% (2) | 27% (3) | 0 |
| BPH | 15 | 0.1 ± 0.2 | 60% (9) | 40% (6) | 0 | 0 |
| NAT | 38 | 0.3 ± 0.1 | 58% (22) | 24% (9) | 13% (5) | 5% (2) |
| All benign | 64 | 0.2 ± 0.1 | 37 (58%) | 17 (25%) | 8 (12%) | 2 (3%) |
| PIN | 36 | 0.6 ± 0.1 | 11 (31%) | 8 (22%) | 10 (28%) | 7 (19%) |
No samples of benign prostate tissue or PIN showed Dicer immunoreactivity of >2. NAT, normal adjacent to the tumor; BPH, benign prostatic hypertrophy.
Dicer Expression in Human Prostate Adenocarcinoma Samples Correlates with Clinical Features
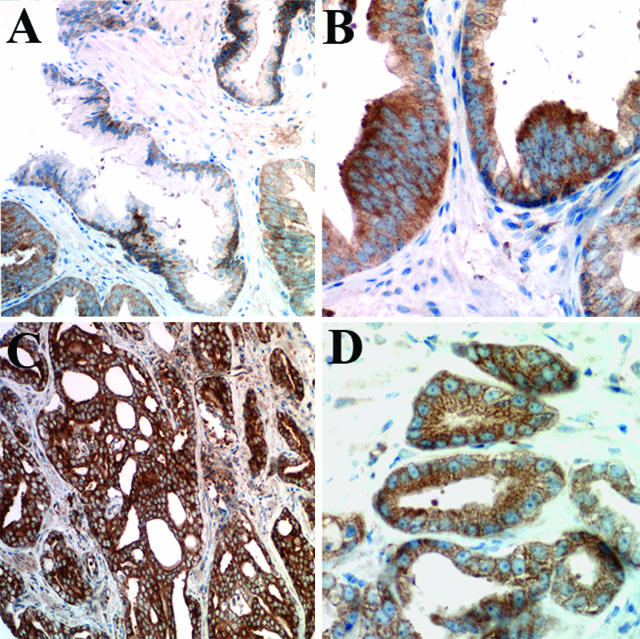
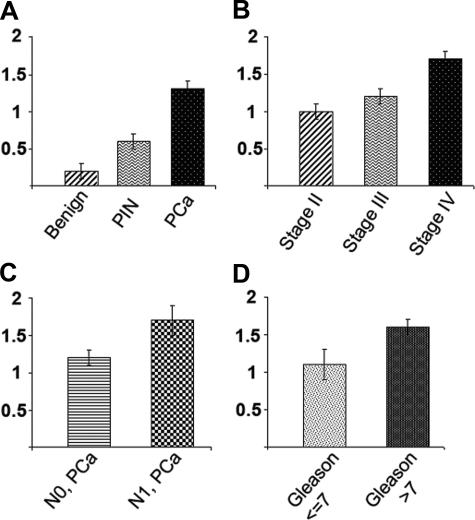
In prostatic intraepithelial neoplasia (PIN) and PCa, Dicer was observed not only in basal cells but also in the cytoplasm of the luminal cells (Figure 3). In PIN specimens, the mean Dicer immunoreactivity was 0.6, a twofold increase over normal prostate tissue. Of the PIN specimens, 53% (19 of 36) showed immunoreactivity of ≥0.5. The majority of prostate cancers showed overexpression of Dicer: 81% (89 of 107) of PCa cases demonstrated Dicer intensity of ≥0.5 with mean immunoreactivity of 1.2 for N0 PCa (no positive regional lymph nodes, n = 83) and mean of 1.7 for N1 PCa (metastases in regional nodes, n = 22) (P = 0.006) (Table 3). The difference in mean Dicer immunoreactivity in benign prostate samples, PIN, and PCa was statistically significant (P < 0.001). Dicer expression increased with the clinical stage of the PCa (Table 4) (P = 0.001) and pathological T stage (P < 0.009). Higher Dicer levels were seen in PCa with Gleason score >7. There was a significant difference in mean immunoreactivity between tumors with Gleason score >7 and those with Gleason score ≤7 (P < 0.025) (Table 5). Mean Dicer immunoreactivity in benign and malignant prostate tissue is summarized in Figure 4.
Figure 3.
Expression of Dicer in neoplastic prostate tissue. A: The largest benign gland in the center of the field shows Dicer immunoreactivity limited to the basal cells; glands in the right and left lower corners are affected by PIN, characterized by multilayered high-columnar cells with enlarged nuclei, prominent nucleoli, and higher cytoplasmic levels of Dicer, shown by immunohistochemistry; original magnification, ×200. B: Prostatic intraepithelial neoplasia examined by Dicer immunohistochemistry; original magnification, ×400. C: Prostate adenocarcinoma with smaller, haphazardly arranged glands of varying size, with high Dicer immunoreactivity, shown by immunohistochemistry; original magnification, ×200. D: Strong Dicer immunoreactivity in neoplastic cells of prostate adenocarcinoma, immunohistochemistry; original magnification, ×400.
Table 3.
Higher Dicer Immunoreactivity in N1 PCa
| N stage | No. of cases | Mean ± SEM | Immunoreactivity score
|
||||
|---|---|---|---|---|---|---|---|
| 0 | 0.1 to 0.5 | 0.6 to 1 | 1.1 to 2 | 2.1 to 3 | |||
| N0 | 83 | 1.2 ± 0.1 | 12% (10) | 13% (11) | 27% (22) | 35% (29) | 13% (11) |
| N1 | 22 | 1.7 ± 0.2 | 5% (1) | 5% (1) | 18% (4) | 36% (8) | 36% (8) |
The difference in the mean immunoreactivity among PCa with N0 and N1 is statistically significant (P = 0.006).
Table 4.
Increase of Dicer in Neoplastic Prostate Tissue and PCa Stage
| Stage | No. of cases | Mean ± SEM | Immunoreactivity score
|
||||
|---|---|---|---|---|---|---|---|
| 0 | 0.1 to 0.5 | 0.6 to 1 | 1.1 to 2 | 2.1 to 3 | |||
| II | 39 | 1 ± 0.1 | 23% (9) | 15% (6) | 21% (8) | 33% (13) | 8% (3) |
| III | 38 | 1.2 ± 0.1 | 5% (2) | 13% (5) | 34% (13) | 29% (11) | 18% (7) |
| IV | 30 | 1.7 ± 0.1 | 7% (2) | 3% (1) | 17% (5) | 43% (13) | 30% (9) |
| Total | 107 | 13 | 12 | 26 | 37 | 19 | |
The difference in the mean values among the stages II, III, and IV is statistically significant (P = 0.001).
Table 5.
Expression of Dicer in PCa Correlates with Gleason Score
| Gleason score | No. of cases | Mean ± SEM | Immunoreactivity score
|
||||
|---|---|---|---|---|---|---|---|
| 0 | 0.1 to 0.5 | 0.6 to 1 | 1.1 to 2 | 2.1 to 3 | |||
| ≤7 | 67 | 1.1 ± 0.2 | 18% (12) | 13% (9) | 25% (17) | 31% (21) | 12% (8) |
| >7 | 40 | 1.6 ± 0.1 | 3% (1) | 8% (3) | 23% (9) | 40% (16) | 28% (11) |
| Total | 107 | 13 | 12 | 26 | 37 | 19 | |
The difference in the mean immunoreactivity among PCa with Gleason score ≤7 and >7 are statistically significant (P = 0.025).
Figure 4.
Schematic representation of mean Dicer immunoreactivity in benign and malignant prostate tissue. A: Mean Dicer immunoreactivity in benign prostate, PIN, and PCa (P = 0.001). B: Mean Dicer immunoreactivity distinguishes PCa by stage (P = 0.001). C: Mean Dicer immunoreactivity is higher in N1 PCa (N0, no positive regional lymph nodes; N1, metastases in regional nodes) (P = 0.006). D: Mean Dicer immunoreactivity is higher in PCa with Gleason score >7 (P = 0.025).
These results suggest that the increase in Dicer expression parallels the progression of PIN to organ-confined prostate cancer and, furthermore, from locally aggressive to metastatic PCa. When 17 cases of PCa with PSA relapse were analyzed, no significant correlation between Dicer immunoreactivity and PSA relapse was found. The extranodal metastasis status of the majority of PCa cases in our series was unknown (89 of 105 cases), making the statistical analysis of Dicer expression in PCa metastatic to visceral organs unreliable.
Western Blot and Immunocytochemical Analysis of Dicer Expression in Prostate Cell Lines
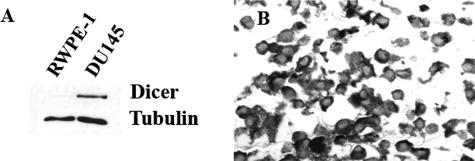
Our findings in clinical PCa samples raised the question as to whether Dicer is up-regulated in neoplastic prostate cell lines. We compared Dicer levels in RWPE-1 and DU145 prostate cell lines. Of note, RWPE-1 is not tumorigenic.33 The DU145 cell line was derived from a human prostate adenocarcinoma metastatic to the brain.34 Western blot analysis of these human prostate cell lines incubated under routine cell culture conditions showed dramatically higher Dicer expression in DU145 compared with RWPE-1 (Figure 5A). Likewise, Dicer was detected at a higher level in primary prostate cancer cell lines when compared with primary benign prostate cells (data not shown).
Figure 5.
Western blot and immunocytochemical analysis of Dicer expression in DU145 and RWPE-1 prostate cancer cell lines. A: Expression of Dicer in RWPE-1 and DU145 cell lines. DU145 cell line demonstrates higher level of Dicer by Western blot. When 70 μg of total protein extract were loaded, Dicer was detected in RWPE-1 cells but at a level dramatically lower than in DU145. B: Immunocytochemistry of Dicer in DU145 cell line; original magnification, ×400.
In addition, high Dicer levels were shown by immunocytochemistry in the cytoplasm of DU145 (Figure 5B) and three other human PCa cell lines: PPC-1, LNCAP, and PC3 (data not shown). Increased level of Dicer detected in the androgen-receptor-negative cell line DU145 further suggested that it might be a useful marker of aggressive androgen-independent tumors.
Discussion
In this study, we evaluated the expression and clinical relevance of Dicer in human prostate cancer. We present definitive evidence that in a significant fraction of PCa the expression of Dicer is up-regulated and is associated with aggressive cancer features. Furthermore, other components of the miR machinery (XPO5, EIF2C2, EIF2C1, HSPCA, MOV10, and TNRC6B) are up-regulated. To the best of our knowledge, this is the first report demonstrating alterations in expression of Dicer and other miR machinery components in human cancer.
Human Dicer is mapped to the subtelomeric region of chromosome 14 (14q32.13). An extensive search of publicly available databases for neoplasms associated with genetic abnormalities at 14q32.13 locus yielded a number of human malignancies.35–37 As for the underlying mechanisms of increased Dicer expression in PCa, it is possible that Dicer up-regulation is induced by a genomic instability at 14q32 (eg, amplification). The up-regulation of other miR machinery components might be secondary to the increased Dicer level. In lung adenocarcinoma, methylation of the Dicer promoter region does not appear to control the level of Dicer.22
A Global Up-Regulation of miRs and Up-Regulated miR Machinery in Prostate Cancer
miR arrays of several solid cancers revealed an almost global up-regulation of miRs as a common feature of oncogenesis in many tissue types.4 Specifically in prostate adenocarcinoma, 39 of 45 differentially expressed miRs are up-regulated.4 Because Dicer is essential for the generation of mature miR, it is likely that higher Dicer level may increase expression of multiple miR species in prostate cancer cells. The opposite effect of reduced Dicer on miR was reported in lung adenocarcinoma, where decreased Dicer levels corresponded to a decrease in miR let-7.38
Up-regulation of the central components of the miR machinery may account for the general up-regulation of miRs in prostate cancer. This hypothesis relies on the assumption that the up-regulated miR machinery is functional. The functionality of the miR machinery is supported by fact that both Dicer protein and its mRNA are up-regulated. Furthermore, many research groups demonstrated a functional RNAi pathway in most prostate adenocarcinoma cell lines by using small interfering RNA (siRNA)/short hairpin RNA constructs targeting mRNAs.39,40
Immunohistochemical Data Explain Gene Array Findings
Our gene array analysis detected no difference in Dicer expression between normal prostate tissue and organ-confined PCa. The immunohistochemical analysis of PCa samples offered an explanation for this finding. In normal prostate tissue, Dicer immunoreactivity was detected in basal cells only. Neoplastic epithelial cells exhibited increased Dicer expression. The hallmark of the invasive PCa is the disappearance of basal cells. Therefore, even though the absolute level of Dicer is probably unchanged when normal prostate tissue is compared with organ-confined PCa, the redistribution of Dicer from vanishing basal cells to proliferating neoplastic cells appears to be biologically significant. As PCa progresses and metastasizes to lymph nodes and visceral organs, the Dicer level continued to rise. Hence, Dicer was up-regulated 2.5-fold in metastatic PCa compared with the organ-confined PCa, as determined by gene array transcriptional profiling.
In cancer, the increasing number of proliferating neoplastic glands and the decrease of intervening stroma may account for a seemingly higher expression of some proteins. Proteins predominantly localized to epithelial glandular compartment of tissues are particularly susceptible to this bias, especially when quantified in protein extracts (by Western blot). In contrast, immunohistochemical analysis allows localization of the antigen of interest in tissue. It allowed us to evaluate Dicer expression only in the luminal cells of the individual glands, benign or malignant. The level of Dicer demonstrated by the most luminal cells within the glands in a particular tissue core was recorded. Our measurements (scoring scheme) were independent of the number of glands in a given microscopic field.
The Pattern of Dicer Expression
Luminal cells of benign prostatic glands do not express Dicer at detectable levels. In contrast, basal cells were most commonly strongly positive. The functional significance of this expression pattern may be linked to the antiapoptotic function of Dicer (most likely via miR-mediated action).41 Stem cells, their immediate progeny, and intermediate cells are known to express bcl-2, which prevents apoptosis, and are crucial for self-renewal of the normal prostatic luminal epithelium.32 Other proteins whose expression in benign prostate is limited to the basal cells and whose expression in luminal cells has also been associated with prostate cancer are ezrin,42 c-met,43 and prostate stem cell antigen PSCA.44 The expression pattern of these proteins supports the concept that prostate cancers arise from the transformation of basal/intermediate cells.45 The presence of Dicer in normal basal cells of prostate and its up-regulation in neoplastic luminal cells suggests that Dicer may play a role in the early steps of prostate cancer development, probably by potentiating an almost global miR up-regulation.
Dicer and miR/siRNA-Mediated Therapy
Hundreds of human miR are predicted to regulate 10 to 30% of protein-encoding genes by interactions with their 3′ untranslated regions.46,47 Perfectly complementary interactions between small RNA (such as siRNA or miRNA) and target mRNA result in mRNA degradation. In contrast, mismatched bp interactions between either siRNA or miRNA and the target mRNA can result in translational repression.48,49
Because mammalian Dicer functions in both siRNA and miR pathways,50,51 prostate cancer may become a useful model to study the competition between small RNA pathways: an endogenous miR pathway and experimental, or exogenous, RNAi pathway.50 With the development of RNAi-based treatments, it might be crucial to learn what branch of small RNA pathway is potentiated by increased levels of the protein components of the miR machinery.
When confirmed by a further validation study in an independent and larger cohort, increased expression of Dicer may be clinically useful for the prognosis of prostate adenocarcinoma patients. The significant up-regulation of Dicer in prostate cancers, together with its relative lack of expression in benign luminal cells, suggests that Dicer may become an attractive therapeutic target. Finally, understanding Dicer expression patterns will be required to predict the susceptibility to potential miRNA- and siRNA-based therapy.52
Acknowledgments
We thank Jennifer Hunt, M.D., for early helpful discussions and support; Geoffrey Murdoch, M.D., Christine McMahon, M.D., Suzanne Bakdash, M.D., and George Michalopolous, M.D., Ph.D., for review of the manuscript and helpful critiques; and William LaFramboise, Ph.D., and Patricia Petrosko, Ph.D., for the primary prostate cell lines. S.C. would like to thank Jeffrey Nickerson, Ph.D., and Anthony Imbalzano, Ph.D., who taught him the principles of research.
Footnotes
Address reprint requests to Simion Chiosea, M.D., Department of Pathology, University of Pittsburgh, University of Pittsburgh Medical Center Presbyterian, C920.1, 200 Lothrop St., Pittsburgh, PA 15213. E-mail: chioseasi@upmc.edu.
Supported by grants from the Postdoctoral Pathology Research Training Program (PPRTP, RT-00147, CC-99FUND), the Department of Pathology, the University of Pittsburgh Medical Center (S.C.), and a Research Scholar grant (RSG-05-246-01-GMC) from the American Cancer Society (R.W.S.).
References
- Hwang HW, Mendell JT. MicroRNAs in cell proliferation, cell death, and tumorigenesis. Br J Cancer. 2006;94:776–780. doi: 10.1038/sj.bjc.6603023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, Prueitt RL, Yanaihara N, Lanza G, Scarpa A, Vecchione A, Negrini M, Harris CC, Croce CM. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D, Slack FJ. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, Wojcik SE, Aqeilan RI, Zupo S, Dono M, Rassenti L, Alder H, Volinia S, Liu CG, Kipps TJ, Negrini M, Croce CM. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli AE, Conte D, Li N, Parrish S, Ha I, Baillie DL, Fire A, Ruvkun G, Mello CC. Genes and mechanisms related to RNA interference regulate expression of the small temporal RNAs that control C. elegans developmental timing. Cell. 2001;106:23–34. doi: 10.1016/s0092-8674(01)00431-7. [DOI] [PubMed] [Google Scholar]
- Hutvagner G, McLachlan J, Pasquinelli AE, Balint E, Tuschl T, Zamore PD. A cellular function for the RNA-interference enzyme Dicer in the maturation of the let-7 small temporal RNA. Science. 2001;293:834–838. doi: 10.1126/science.1062961. [DOI] [PubMed] [Google Scholar]
- Chendrimada TP, Gregory RI, Kumaraswamy E, Norman J, Cooch N, Nishikura K, Shiekhattar R. TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature. 2005;436:740–744. doi: 10.1038/nature03868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056–2060. doi: 10.1126/science.1073827. [DOI] [PubMed] [Google Scholar]
- Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- Maniataki E, Mourelatos Z. A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes Dev. 2005;19:2979–2990. doi: 10.1101/gad.1384005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Hur I, Park SY, Kim YK, Suh MR, Kim VN. The role of PACT in the RNA silencing pathway. EMBO J. 2006;25:522–532. doi: 10.1038/sj.emboj.7600942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin P, Zarnescu DC, Ceman S, Nakamoto M, Mowrey J, Jongens TA, Nelson DL, Moses K, Warren ST. Biochemical and genetic interaction between the fragile X mental retardation protein and the microRNA pathway. Nat Neurosci. 2004;7:113–117. doi: 10.1038/nn1174. [DOI] [PubMed] [Google Scholar]
- Jin P, Alisch RS, Warren ST. RNA and microRNAs in fragile X mental retardation. Nat Cell Biol. 2004;6:1048–1053. doi: 10.1038/ncb1104-1048. [DOI] [PubMed] [Google Scholar]
- Hammond SM. MicroRNAs as oncogenes. Curr Opin Genet Dev. 2006;16:4–9. doi: 10.1016/j.gde.2005.12.005. [DOI] [PubMed] [Google Scholar]
- Roh MS, Kwak JY, Kim SJ, Lee HW, Kwon HC, Hwang TH, Choi PJ, Hong YS. Expression of double-stranded RNA-activated protein kinase in small-size peripheral adenocarcinoma of the lung. Pathol Int. 2005;55:688–693. doi: 10.1111/j.1440-1827.2005.01892.x. [DOI] [PubMed] [Google Scholar]
- Jaronczyk K, Carmichael JB, Hobman TC. Exploring the functions of RNA interference pathway proteins: some functions are more RISCy than others? Biochem J. 2005;387:561–571. doi: 10.1042/BJ20041822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karube Y, Tanaka H, Osada H, Tomida S, Tatematsu Y, Yanagisawa K, Yatabe Y, Takamizawa J, Miyoshi S, Mitsudomi T, Takahashi T. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaul D, Sikand K. Defective RNA-mediated c-myc gene silencing pathway in Burkitt’s lymphoma. Biochem Biophys Res Commun. 2004;313:552–554. doi: 10.1016/j.bbrc.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Carmichael JB, Provost P, Ekwall K, Hobman TC. ago1 and dcr1, two core components of the RNA interference pathway, functionally diverge from rdp1 in regulating cell cycle events in Schizosaccharomyces pombe. Mol Biol Cell. 2004;15:1425–1435. doi: 10.1091/mbc.E03-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Provost P, Silverstein RA, Dishart D, Walfridsson J, Djupedal I, Kniola B, Wright A, Samuelsson B, Radmark O, Ekwall K. Dicer is required for chromosome segregation and gene silencing in fission yeast cells. Proc Natl Acad Sci USA. 2002;99:16648–16653. doi: 10.1073/pnas.212633199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volpe TA, Kidner C, Hall IM, Teng G, Grewal SI, Martienssen RA. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002;297:1833–1837. doi: 10.1126/science.1074973. [DOI] [PubMed] [Google Scholar]
- Fukagawa T, Nogami M, Yoshikawa M, Ikeno M, Okazaki T, Takami Y, Nakayama T, Oshimura M. Dicer is essential for formation of the heterochromatin structure in vertebrate cells. Nat Cell Biol. 2004;6:784–791. doi: 10.1038/ncb1155. [DOI] [PubMed] [Google Scholar]
- Chandran UR, Dhir R, Ma C, Michalopoulos G, Becich M, Gilbertson J. Differences in gene expression in prostate cancer, normal appearing prostate tissue adjacent to cancer and prostate tissue from cancer free organ donors. BMC Cancer. 2005;5:45. doi: 10.1186/1471-2407-5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, Michalopoulos G, Becich M, Luo JH. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J Clin Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, Shah RB, Chandran U, Monzon FA, Becich MJ, Wei JT, Pienta KJ, Ghosh D, Rubin MA, Chinnaiyan AM. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer Cell. 2005;8:393–406. doi: 10.1016/j.ccr.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Tokar EJ, Ancrile BB, Cunha GR, Webber MM. Stem/progenitor and intermediate cell types and the origin of human prostate cancer. Differentiation. 2005;73:463–473. doi: 10.1111/j.1432-0436.2005.00047.x. [DOI] [PubMed] [Google Scholar]
- Bello D, Webber MM, Kleinman HK, Wartinger DD, Rhim JS. Androgen responsive adult human prostatic epithelial cell lines immortalized by human papillomavirus 18. Carcinogenesis. 1997;18:1215–1223. doi: 10.1093/carcin/18.6.1215. [DOI] [PubMed] [Google Scholar]
- Stone KR, Mickey DD, Wunderli H, Mickey GH, Paulson DF. Isolation of a human prostate carcinoma cell line (DU 145). Int J Cancer. 1978;21:274–281. doi: 10.1002/ijc.2910210305. [DOI] [PubMed] [Google Scholar]
- Ihara Y, Kato Y, Bando T, Yamagishi F, Minamimura T, Sakamoto T, Tsukada K, Isobe M. Allelic imbalance of 14q32 in esophageal carcinoma. Cancer Genet Cytogenet. 2002;135:177–181. doi: 10.1016/s0165-4608(01)00654-9. [DOI] [PubMed] [Google Scholar]
- Shao J, Li Y, Wu Q, Liang X, Yu X, Huang L, Hou J, Huang X, Ernberg I, Hu LF, Zeng Y. High frequency loss of heterozygosity on the long arms of chromosomes 13 and 14 in nasopharyngeal carcinoma in Southern China. Chin Med J (Engl) 2002;115:571–575. [PubMed] [Google Scholar]
- Tzai TS, Chen HH, Chan SH, Ho CL, Tsai YS, Cheng HL, Dai YC, Lin JS, Yang WH, Chow NH. Clinical significance of allelotype profiling for urothelial carcinoma. Urology. 2003;62:378–384. doi: 10.1016/s0090-4295(03)00344-3. [DOI] [PubMed] [Google Scholar]
- Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y, Mitsudomi T, Takahashi T. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- Paduano F, Villa R, Pennati M, Folini M, Binda M, Daidone MG, Zaffaroni N. Silencing of survivin gene by small interfering RNAs produces supra-additive growth suppression in combination with 17-allylamino-17-demethoxygeldanamycin in human prostate cancer cells. Mol Cancer Ther. 2006;5:179–186. doi: 10.1158/1535-7163.MCT-05-0132. [DOI] [PubMed] [Google Scholar]
- Wannenes F, Ciafre SA, Niola F, Frajese G, Farace MG. Vector-based RNA interference against vascular endothelial growth factor-A significantly limits vascularization and growth of prostate cancer in vivo. Cancer Gene Ther. 2005;12:926–934. doi: 10.1038/sj.cgt.7700862. [DOI] [PubMed] [Google Scholar]
- Muljo SA, Ansel KM, Kanellopoulou C, Livingston DM, Rao A, Rajewsky K. Aberrant T cell differentiation in the absence of Dicer. J Exp Med. 2005;202:261–269. doi: 10.1084/jem.20050678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdman A, Fang X, Pang ST, Nilsson B, Ekman P, Egevad L. Ezrin expression in prostate cancer and benign prostatic tissue. Eur Urol. 2005;48:852–857. doi: 10.1016/j.eururo.2005.03.013. [DOI] [PubMed] [Google Scholar]
- van Leenders G, van Balken B, Aalders T, Hulsbergen-van de Kaa C, Ruiter D, Schalken J. Intermediate cells in normal and malignant prostate epithelium express c-MET: implications for prostate cancer invasion. Prostate. 2002;51:98–107. doi: 10.1002/pros.10073. [DOI] [PubMed] [Google Scholar]
- Reiter RE, Gu Z, Watabe T, Thomas G, Szigeti K, Davis E, Wahl M, Nisitani S, Yamashiro J, Le Beau MM, Loda M, Witte ON. Prostate stem cell antigen: a cell surface marker overexpressed in prostate cancer. Proc Natl Acad Sci USA. 1998;95:1735–1740. doi: 10.1073/pnas.95.4.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkhoff H. Role of the basal cells in premalignant changes of the human prostate: a stem cell concept for the development of prostate cancer. Eur Urol. 1996;30:201–205. doi: 10.1159/000474170. [DOI] [PubMed] [Google Scholar]
- Berezikov E, Guryev V, van de Belt J, Wienholds E, Plasterk RH, Cuppen E. Phylogenetic shadowing and computational identification of human microRNA genes. Cell. 2005;120:21–24. doi: 10.1016/j.cell.2004.12.031. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438–442. doi: 10.1101/gad.1064703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y, Yi R, Cullen BR. MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA. 2003;100:9779–9784. doi: 10.1073/pnas.1630797100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ketting RF. Partners in dicing. Genome Biol. 2006;7:210. doi: 10.1186/gb-2006-7-3-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Jaskiewicz L, Kolb FA, Pillai RS. Post-transcriptional gene silencing by siRNAs and miRNAs. Curr Opin Struct Biol. 2005;15:331–341. doi: 10.1016/j.sbi.2005.05.006. [DOI] [PubMed] [Google Scholar]
- Izquierdo M. Short interfering RNAs as a tool for cancer gene therapy. Cancer Gene Ther. 2005;12:217–227. doi: 10.1038/sj.cgt.7700791. [DOI] [PubMed] [Google Scholar]