Abstract
Gene silencing by methylation of promoter CpG islands is deeply involved in cancers, but its involvement in polyclonal disorders is still unclear. Here, we analyzed the presence of gene silencing in intestinal metaplasia (IM) of the stomach, a polyclonal disorder, in which multiple gastric glands aberrantly differentiate into those with intestinal characteristics. By a genome-wide screening, CpG islands in the putative promoter regions of four genes (ZIK1, ZNF141, KAL1, and FGF14) were found to be specifically methylated in glands with IM, and their expression was markedly decreased. When demethylation was induced in cell lines with their methylation by 5-aza-2′-deoxycytidine, expression of ZIK1, KAL1, and FGF14 was restored, supporting causal roles of methylation in their silencing. Analysis of ZIK1 methylation in a single gland showed that the vast majority of DNA molecules isolated from a gland with IM were methylated and that those from a gland without IM were not. ZIK1 methylation was present in glands isolated from physically distant positions within a stomach, showing that methylation occurred multifocally. These data indicate that methylation of multiple genes occurs independently in multiple glands, each of which has its own stem cell, demonstrating that involvement of aberrant gene silencing in noninherited polyclonal human disorders needs more attention.
Methylation of promoter CpG islands (CGIs) causes silencing of their downstream genes,1,2 and that of tumor-suppressor genes is involved in development and progression of various cancers.2–4 Methylation is induced by aging and chronic inflammation and is present not only in cancers, which are clonal disorders, but also in noncancerous tissues.5–9 However, it is still unclear whether methylation is induced as a rare and random event, like a mutation,10 or as an event that occurs in multiple cells and targets nonrandom genes. If methylation of promoter CGIs occurs as relatively frequent and nonrandom events, aberrant methylation could be involved not only in cancers but also in polyclonal disorders, such as aberrant differentiation and degenerative disorders.
Gastric mucosae provide an informative model to examine the origin of aberrant methylation. It is known that a single gastric gland is made of differentiated cells produced from multiple progenitor cells that originate from a single stem cell.11 If aberrant gene silencing occurs in a stem cell, all of the cells in the entire gland produced from it are expected to have silencing of the same gene. A single gastric gland can be collected using the gland isolation technique,12,13 and the methylation status of a gene can be analyzed using DNA from the single gland. By examining the presence of methylation of a specific gene in multiple glands, we can explore whether or not its silencing was induced independently in multiple glands, possibly in multiple stem cells.
As a polyclonal disorder of the stomach, intestinal metaplasia (IM) is highly prevalent and well defined.11 When IM develops in an individual, glands with intestinal characteristics, such as the presence of goblet cells and expression of intestinal enzymes, initially appear multifocally in the pyloric area and then gradually spread into the fundic area. The polyclonal origin of IM has been demonstrated by analysis of inactivation patterns of X chromosomes in females and by use of chimeric mice.11 If we can find genes whose promoter CGIs are consistently methylated in IM, such genes could be involved in development of IM by causing irreversible changes. Although LOX, HAND1, FLNc, THBD, and HRASLS, whose promoter CGIs were found to be methylated in noncancerous gastric mucosae, were candidates, the extent of their methylation did not correlate with the degree of IM.8
In this study, we performed a genome-wide screening for genes silenced in IM using methylation-sensitive-representational difference analysis (MS-RDA)4,14,15 and analyzed the presence of methylation of the identified genes in multiple gastric glands with and without IM isolated from distant positions within a stomach.12,13
Materials and Methods
Gland Isolation, Assessment of IM, and DNA/RNA Extraction
Fundic and pyloric glands in the anterior and posterior walls were freshly obtained from the stomachs of patients who underwent gastrectomy due to gastric cancers at Aichi Cancer Center Hospital with written informed consents. To isolate glands with and without IM, the submucosal tissue was gently split off from the mucosa after washing in 0.1 mol/L phosphate-buffered saline,13 and the mucosa was incubated at 37°C in calcium- and magnesium-free Hanks’ balanced salt solution with 30 mmol/L ethylenediamine tetraacetic acid for 30 minutes with gentle shaking, followed by vigorous agitation by vortex. Gastric glands were fixed in 70% ethanol and stained with Alcian blue. A gland with positive staining of goblet cells was considered as one with IM, and a gland without was considered as one without IM. Fourteen samples of gastric glands without IM were prepared from 14 gastric cancer cases. Sixteen samples with IM were prepared from 16 cases, 11 of which were the same with those used for samples without IM. For cases used for MS-RDA (cases 156 and 163), large quantities of glands with and without IM (>10,000 glands each) were collected. For the other cases, 100 to 500 glands were collected. DNA and RNA were extracted by the phenol and chloroform extraction method and ISOGEN (Nippon Gene, Tokyo, Japan), respectively.
Cell Lines and Treatment with 5-Aza-2′-Deoxycytidine (5-Aza-dC)
HSC39, HSC44, and HSC57 gastric cancer cell lines were kind gifts from Dr. K. Yanagihara, National Cancer Center Research Institute, Tokyo, Japan, and TMK1 was from Dr. W. Yasui, Hiroshima University, Hiroshima, Japan. KATOIII, MKN28, MKN74, and NUGC3 were obtained from the Japanese Collection of Research Bioresources (Osaka, Japan), and MKN45 was from RIKEN Bioresources Center (Tsukuba, Japan). A gastric cancer cell line (AGS), a cell line from the normal small intestine (FHs 74 Int), and a cell line from the normal colon (CCD 841 CoN) were purchased from the American Type Culture Collection (Manassas, VA). For treatment with 5-aza-dC, cells were seeded on day 0, media containing 5-aza-dC were freshly added on days 1 and 3, and cells were harvested on day 4.
MS-RDA
For MS-RDA,4,14,15 an R adaptor was ligated to 1 μg of genomic DNA digested with HpaII or SacII (New England Biolabs, Beverly, MA), and the ligation product was amplified by polymerase chain reaction (PCR) with R oligonucleotide in the presence of 1 mol/L betaine (Sigma, St. Louis, MO). By this procedure, DNA fragments derived from unmethylated CGIs were enriched in the PCR product (amplicon), but those from methylated CGIs were not. A J adaptor was ligated only to the tester amplicon, and 200 ng of it was mixed with 40 μg of the driver amplicon. The DNA mixture underwent heat denaturation and reannealing (competitive hybridization), and dsDNA with the J adaptor on both ends was selectively amplified using J oligonucleotide (selective amplification). After switching the J adaptor of the product to a new N adaptor, 40 ng of the product with the N adaptor was mixed with 40 μg of the driver amplicon, and the second competitive hybridization and selective amplification were performed. The product was cloned, and 96 clones were sequenced. Chromosomal positions and relative locations to CGIs16 were analyzed at the GenBank web site.
Bisulfite Modification, Methylation-Specific PCR (MSP), and Bisulfite Sequencing
For bisulfite modification, genomic DNA (500 ng) was restricted with BamHI (New England Biolabs) and denatured in 0.3 mol/L NaOH.17 In 3.1 mol/L NaHSO3 (pH 5.0) and 0.6 mmol/L hydroquinone, DNA underwent 15 cycles of denaturation at 95°C for 30 seconds and incubation at 50°C for 15 minutes. The product was desalted with the Wizard DNA cleanup system (Promega, Madison, WI) and desulfonated in 0.6 N NaOH. The sample was ethanol-precipitated and dissolved in 20 μl of Tris-EDTA buffer.
For MSP, 1 μl of the sodium-bisulfite-treated DNA was amplified with primers specific to methylated or unmethylated sequences (Table 1). DNA methylated in vitro with SssI methylase and DNA amplified by a GenomiPhi DNA amplification kit (Amersham Biosciences, Little Chalfont, UK) were used to determine annealing temperatures that specifically amplify methylated and unmethylated sequences as well as minimum PCR cycles to obtain visible bands with 100% methylated or unmethylated samples. For test samples, four cycles were added for their analysis.
Table 1.
List of Primers for MSP, RT-PCR, and Bisulfite Sequencing
| Gene symbol | Analysis | Forward | Reverse | Tempera-ture (°C) | Number of cycles |
|---|---|---|---|---|---|
| ZIK1 | MSP-M | 5′-GTTTGAGGTGACGTTGGGC-3′ | 5′-GACCCTTTTCTCAACGCGA-3′ | 64 | 33 |
| MSP-U | 5′-TTTGAGGTGATGTTGGGTG-3′ | 5′-AACAACCCTTTTCTCAACACA-3′ | 60 | 33 | |
| RT-PCR | 5′-AGAACTTTGCCCTTGTAGCC-3′ | 5′-GTGGATGAACCTGCCTTTG-3′ | 64 | n/a | |
| Sequencing (X) | 5′-AGGGTTTAGGTTAGAGTAGGAG-3′ | 5′-AAACTAAAACCAAATCCACAC-3′ | 52 | 30 | |
| Sequencing (Y) | 5′-AGTTTTTTGAGTTTGAATTAATG-3′ | 5′-ACAAAACCTTCTAAATTCTATAATC-3′ | 56 | 30 | |
| LOC-285300 | MSP-M | 5′-ATTTAAGCGGCGGTTCGT-3′ | 5′-CACGCGAAAAAACCAACG-3′ | 62 | 33 |
| MSP-U | 5′-TTAAGTGGTGGTTTGTGGGTAT-3′ | 5′-AAAAAACAAAACCACCCACAC-3′ | 65 | 33 | |
| RT-PCR | 5′-CCGTTGCCGTGATAGTC-3′ | 5′-ACAGGAATCCGTGGTCAG-3′ | 60 | n/a | |
| CD8A | MSP-M | 5′-GTTTGGTTAGGTTGTTTCGACG-3′ | 5′-ATCACCCGAAACCCCCG-3′ | 64 | 32 |
| MSP-U | 5′-AGGTTGTTTTGATGGTTTTATTTATG-3′ | 5′-ATCACCCAAAACCCCCAC-3′ | 62 | 33 | |
| RT-PCR | 5′-CCGAGAGAACGAGGGCTAC-3′ | 5′-TCGCTGGCAGGAAGACC-3′ | 64 | n/a | |
| ZNF141 | MSP-M | 5′-CGGTTGTTATTTTCGTTATATTTC-3′ | 5′-ACTCCAATACGCTAATTAAATACG-3′ | 60 | 32 |
| MSP-U | 5′-TACAACTCCAATCTCCCCA-3′ | 5′-TAGGTTAAGTATGAAGAGAAAGTTTT-3′ | 59 | 31 | |
| RT-PCR | 5′-CCTTTAGACGGTCCACAGAT-3′ | 5′-TATGTTGACTCAGGAGCGAA-3′ | 62 | n/a | |
| KAL1 | MSP-M | 5′-GTGCGAACGGGAGAGGC-3′ | 5′-GTCAACTACGAACCCGAACG-3′ | 64 | 33 |
| MSP-U | 5′-AAAACCCATAAACCAATCTCA-3′ | 5′-TGAATGGGAGAGGTGTTTGT-3′ | 58 | 33 | |
| RT-PCR | 5′-TTGACAATGAGTGCTCTGGG-3′ | 5′-AATCGTAACTCTTTTCTGGGCT-3′ | 62 | n/a | |
| FGF14 | MSP-M | 5′-TTGTTTAGCGCGATTCGGATCGAC-3′ | 5′-CGAAAAATAAAACCGACCGACG-3′ | 60 | 33 |
| MSP-U | 5′-TGTTTAGTGTGATTTGGATTGAT-3′ | 5′-ACAAAAAATAAAACCAACCAACA-3′ | 59 | 33 | |
| RT-PCR | 5′-GGCAACCTGGTGGATATCTT-3′ | 5′-CCTTGCCTGCAATATAACCT-3′ | 59 | n/a | |
| NKX2–8 | MSP-M | 5′-TTTTATTTTTTTCGGCGTTTTC-3′ | 5′-GAACGATAACCACCACCCG-3′ | 59 | 32 |
| MSP-U | 5′-GTTTTTATTTTTTTTGGTGTTTTT-3′ | 5′-AAACAATAACCACCACCCA-3′ | 59 | 32 | |
| RT-PCR | 5′-AGCAGGACGCGCAACAC-3′ | 5′-AGTCTGGCGGGCTGGTC-3′ | 62 | n/a | |
| NID2 | MSP-M | 5′-GTTTTTAGAGTTAGGAGCGTTC-3′ | 5′-CGAATACGACTACTAACCTACG-3′ | 64 | 33 |
| MSP-U | 5′-AGGTTAGTAGTTGTATTTGTTGTTGT-3′ | 5′-TCATCCAAACTACCCCACA-3′ | 58 | 33 | |
| RT-PCR | 5′-CTTACGAGGAGGTCAAACGC-3′ | 5′-CCGTTGGCAGGATAAAGAAA-3′ | 59 | n/a | |
| LOC-284801 | MSP-M | 5′-TGCGTTTACGTTCGTATTTGTC-3′ | 5′-GTAAAACCCGAAAAACCCG-3′ | 62 | 32 |
| MSP-U | 5′-GTTGTGTTTATGTTTGTATTTGTT-3′ | 5′-CACCCCTCATAAAACCCA-3′ | 58 | 33 | |
| RT-PCR | 5′-GGCAATAAGAATGAAGACCT-3′ | 5′-ATCAACAGCCCTTAAACATC-3′ | 60 | n/a | |
| RAX | MSP-M | 5′-AGTTGGTTTTTCGTTTTTAGCGTC-3′ | 5′-CTAAATCAAACTCCGCGAACG-3′ | 65 | 33 |
| MSP-U | 5′-TAGTTGGTTTTTTGTTTTTAGTGTT-3′ | 5′-CCTAAATCAAACTCCACAAACA-3′ | 58 | 32 | |
| RT-PCR | 5′-TGCGTCTGAAAGCCAAGG-3′ | 5′-AGAGGATGCGGGTACGGT-3′ | 62 | n/a | |
| GAPDH | RT-PCR | 5′-GCCAGTGGACTCCACGAC-3′ | 5′-CAACTACATGGTTTACATGTTC-3′ | 62 | n/a |
M, primers specific to methylated DNA; n/a, not available; U, primers specific to unmethylated DNA.
For bisulfite sequencing, 1 μl of the sodium-bisulfite-treated DNA was amplified with primers common to methylated and unmethylated DNA sequences. The PCR product was cloned into a pGEM-T Easy Vector (Promega), and 8 to 12 clones were sequenced using an ABI PRISM 310 sequencer (Applied Biosystems, Foster City, CA). Primer sequences and PCR conditions are shown in Table 1.
Quantitative Reverse Transcriptase (RT)-PCR
For quantitative RT-PCR, cDNA was synthesized from 1 μg of total RNA with oligo (dT)12–18 primer and Superscript II reverse transcriptase (Life Technologies, Gaithersburg, MD). Real-time PCR analysis was performed using an iCycler iQ detection system (Bio-Rad Laboratories, Hercules, CA) with SYBR Green PCR Core Reagents (Applied Biosystems) and 200 nmol/L of primers. The primer sequences and PCR conditions are shown in Table 1. Specific amplification of target sequences was confirmed by electrophoresis of the PCR products and by observation of their melting curves. To quantify the number of molecules of a specific gene, a standard curve was generated using templates that contained 10 to 106 copies of the gene. The number of cDNA molecules of a gene of interest was normalized to that of glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
Statistical Analysis
The differences of expression levels were analyzed by using the t-test, Welch method (both-sided).
Results
Isolation of 10 Putative Promoter CGIs Aberrantly Methylated in IM
MS-RDA was performed using an equal mixture of two samples without IM (cases 156 and 163) as the tester and an equal mixture of two samples with IM from the same cases as the driver. By this way of subtraction, DNA fragments methylated in gastric glands with IM commonly in the two cases were expected to be isolated. Two series of MS-RDA were performed using two methylation-sensitive restriction enzymes, HpaII and SacII.
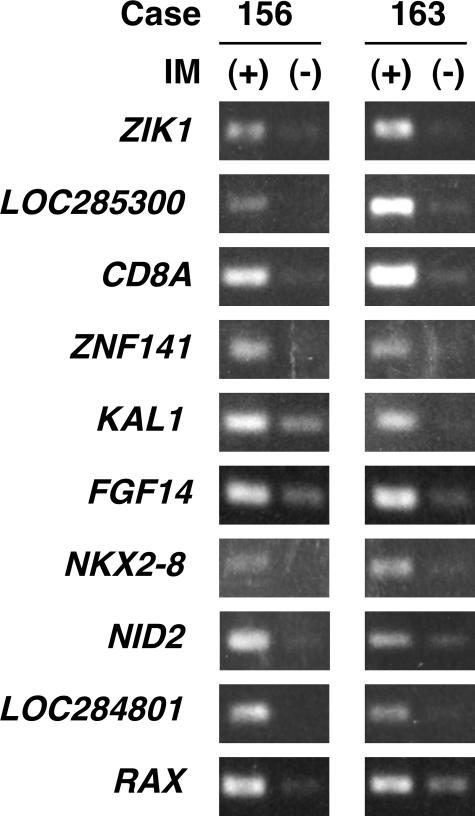
We sequenced 192 DNA fragments obtained by the two series of MS-RDA and found 65 nonredundant fragments. Genomic origins of these 65 fragments were searched for in the GenBank database, and 30 fragments were found to be derived from CGIs in the putative promoter regions of known genes. Methylation statuses of these 30 CGIs were analyzed by MSP in the samples used for MS-RDA, and 10 CGIs (Table 2) were found to be methylated in samples with IM (Figure 1).
Table 2.
List of 10 CGIs Differentially Methylated in the Initial Samples of IM
| DNA fragments isolated by MS-RDA
|
Genes in the downstream
|
|||||
|---|---|---|---|---|---|---|
| Length (bp) | Accession no. | From | To | Chromosome | Gene symbol | Full names or alternative names |
| MS-RDA with HpaII | ||||||
| 227 | AC003682 | 72722 | 72845 | 19q13 | ZIK1 | Zinc finger protein-interacting with K protein 1 |
| 254 | AC108724 | 85358 | 85612 | 3p14 | LOC285300 | |
| 204 | AC064848 | 16801 | 17005 | 2p12 | CD8A | CD8 antigen, a polypeptide (p32) |
| MS-RDA with SacII | ||||||
| 405 | AC079140 | 120795 | 121200 | 4p16 | ZNF141 | Zinc finger protein 141 (clone pHZ-44) |
| 394 | AC005184 | 22438 | 22832 | Xp22 | KAL1 | Kallmann syndrome 1 |
| 374 | AE014303 | 198013 | 198387 | 13q34 | FGF14 | Fibroblast growth factor 14 |
| 382 | AL079303 | 21708 | 22090 | 14q13 | NKX2-8 | NK2 transcription factor-related, locus 8 |
| 384 | AL118557 | 77025 | 77409 | 14q21 | NID2 | Nidogen 2 (osteonidogen) |
| 346 | AL121904 | 65293 | 65639 | 20p11 | LOC284801 | |
| 190 | AC067859 | 139250 | 139440 | 2q31 | RAX | Retina and anterior neural-fold homeobox |
After analysis of 190 clones isolated by MS-RDA, 30 fragments were found to be derived from putative promoter CGIs. MSP analysis showed that these 10 CGIs were methylated in the samples with IM used for MS-RDA.
Figure 1.
Specific methylation of the 10 putative promoter CGIs identified by MS-RDA. These CGIs were identified by MS-RDA using pooled samples of gastric glands with and without IM. Their methylation statuses were analyzed by MSP of the samples used for MS-RDA.
Decreased Expression of the Four Genes in Gastric Glands with IM
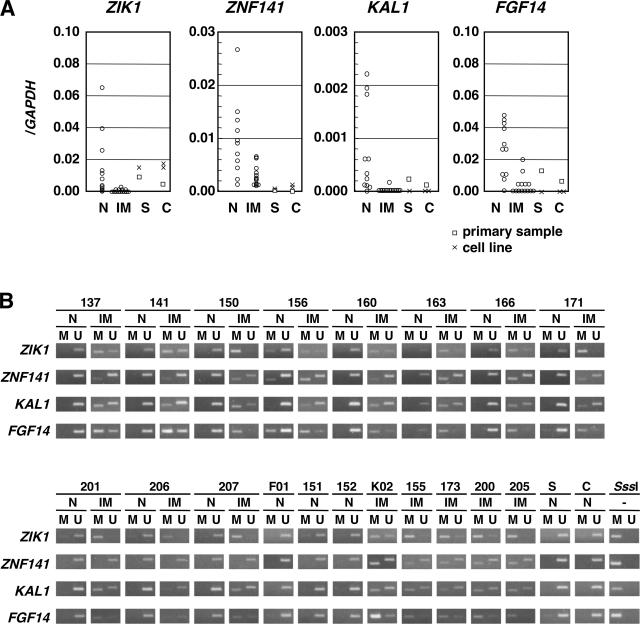
To examine the effect of methylation of the putative promoter regions on mRNA expression, we analyzed the expression levels of the 10 genes in 11 samples of gastric glands without IM (including cases 156 and 163), 14 samples with IM (including cases 156 and 163), two samples of the normal small intestine (including one cell line, FHs 74 Int), and three samples of the normal colon (including one cell line, CCD 841 CoN). mRNA expression of ZIK1, ZNF141, KAL1, and FGF14 was markedly decreased in gastric gland samples with IM (Figure 2A). The mRNA expressions of ZIK1, KAL1, and FGF14 especially were lost in most samples with IM. In the normal small intestine and colon, ZNF141, KAL1, and FGF14 were not, or were barely, expressed, whereas ZIK1 was expressed at low levels. As for the remaining six genes, five genes (LOC285300, NKX2-8, NID2, LOC284801, and RAX) were not expressed even in gastric gland samples without IM. CD8A did not show decreased expression in samples with IM (data not shown), which was considered to be due to erroneous positioning of the putative promoter region.
Figure 2.
mRNA expression and methylation of the four genes in additional primary samples. A: mRNA expression levels. The mRNA expression levels of the 10 genes listed in Table 2 were quantitatively analyzed in 11 samples of gastric glands without IM (N), 14 samples with IM (IM), two samples of the normal small intestine (one primary sample and one cell line; S), and three samples of the normal colon (one primary sample and two cell lines; C). The expression levels of the four genes were markedly decreased in gastric gland samples with IM and were also very low in the normal small intestine and colon. There is a possibility that their silencing in gastric epithelial cells produced a cellular environment similar to that of the small intestine and colon. B: Methylation statuses of the four genes in 16 samples of gastric glands with IM and 14 samples without (11 paired glands; N), one sample of the normal small intestine (S), and one sample of the normal colon (C). All four genes were specifically methylated in the samples of gastric glands with IM although not methylated in the samples of gastric glands without the normal small intestine and the normal colon.
Methylation statuses of these four genes were also analyzed by MSP in the expanded panel of samples (Figure 2B). ZIK1 was methylated in all of the 16 samples with IM, although slightly methylated only in one (case 156) of the 14 samples without IM. ZNF141 was methylated in 12 of the 16 samples with IM, although slightly methylated only in one (case 156) of the 14 samples without IM. KAL1 was methylated in 14 of the 16 samples with IM while in none of the samples without IM. FGF14 was methylated in 15 of the 16 samples with IM, although only in one (case 156) of the 14 samples without IM. These data confirmed that all these four genes were specifically and consistently methylated in gastric gland samples with IM, and this led to their decreased expression in glands with IM. Contamination of glands with IM into a sample that was supposed not to contain such glands was suggested for case 156.
Role of the Methylation of the Putative Promoter Regions in the Decreased Expression
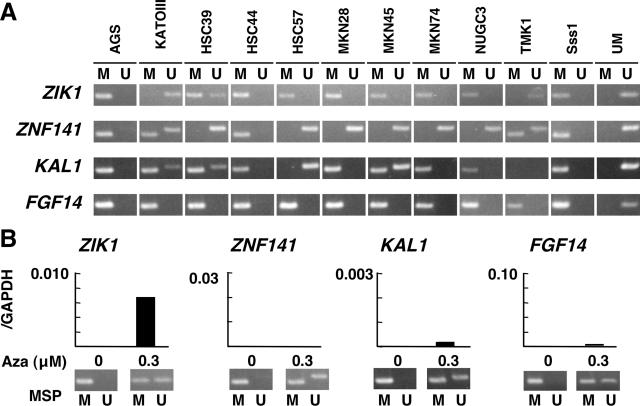
To analyze further the role of methylation of the putative promoter CGIs of the four genes in their decreased expression, we analyzed gene re-expression after treating cell lines with a demethylating agent, 5-aza-dC. To find cell lines suitable for this purpose, 10 gastric cancer cell lines (AGS, KATOIII, HSC39, HSC44, HSC57, MKN28, MKN45, MKN74, NUGC3, and TMK1) were screened. ZIK1, ZNF141, KAL1, and FGF14 were methylated in eight, four, eight, and all 10 of them, respectively, and AGS and HSC44 cell lines had methylation of all of the four genes (Figure 3A).
Figure 3.
Silencing of the four genes in gastric cancer cell lines. A: Methylation statuses of the four genes were analyzed in 10 gastric cancer cell lines. ZIK1, ZNF141, KAL1, and FGF14 were found to be methylated in eight, four, eight, and all, respectively, of the 10 cell lines. B: AGS gastric cancer cells were treated with 5-aza-dC, and demethylation and re-expression were analyzed. Re-expression in association with demethylation was clearly observed for ZIK1 and weakly for KAL1 and FGF14 but not for ZNF141. The scales for the y axis were unified to those of Figure 2A for comparison. Similar results were obtained using HSC44 gastric cancer cell line.
After treatment of AGS and HSC44 cells with 5-aza-dC, abundant transcription in association with demethylation of the putative promoter CGI was observed for ZIK1, and weak transcription was observed for KAL1 and FGF14 (Figure 3B, note that the scales in the y axis were unified to those in Figure 2A). Re-expression of ZNF141 was not observed. Similar results were obtained for both AGS and HSC44 cell lines, and it was indicated that methylation of the CGIs in the putative promoter regions caused transcriptional repression of ZIK1, KAL1, and FGF14. There remains a possibility that transcription factors for ZNF141 had been lost in the two cell lines and that this was responsible for the lack of its re-expression.
Occurrence of Methylation in Multiple Single Glands
The above analyses were performed using samples prepared by pooling 100 to 10,000 gastric glands. Some samples with IM had both methylated and unmethylated DNA molecules (for example, cases 141 and K02 for ZIK1 in Figure 2B), whereas the other samples with IM had only methylated DNA molecules (cases 171, 201, and 207). This could be due to coexistence of cells with and without methylation in a single gland or coexistence of glands with and without methylation.
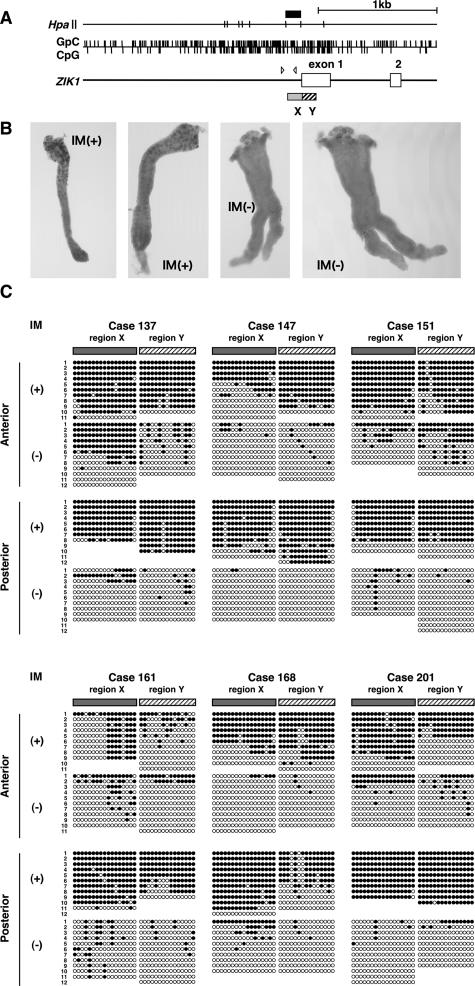
Having the four genes that are specifically methylated in gastric glands with IM, we proceeded to methylation analysis in a single gastric gland. Due to the limitation of DNA material obtained from a single gland, we selected ZIK1, whose methylation clearly caused gene silencing, for this analysis. The CGI in its putative promoter region was analyzed by bisulfite sequencing of its two regions, X and Y (Figure 4A). Single gastric glands with and without IM were obtained from anterior and posterior walls of the stomach in six cases to make sure that they were from different origins (representative results in Figure 4B). For each gland, eight to 12 DNA molecules were analyzed (Figure 4C).
Figure 4.
Methylation analysis in single glands with and without IM. A: Genomic structure of the putative promoter region of ZIK1. The region was divided into X and Y regions for bisulfite sequencing. Closed box, DNA fragment isolated by MS-RDA; arrowheads, locations of MSP primers. B: Representative photos of single glands with and without IM isolated by single gland isolation technique. C: Results of bisulfite sequencing of single glands in six cases. Gastric glands were isolated from the anterior and posterior walls of the six cases, and the presence or absence of IM was determined by Alcian blue staining. DNA was extracted from a single gland, and bisulfite sequencing was performed. The vast majority of DNA molecules obtained from a single gland with IM were methylated, whereas those from a single gland without IM were unmethylated. Closed circle, methylated CpG site; open circle, unmethylated CpG site.
First, it was noted that almost all DNA molecules in a single gland with IM were methylated, whereas most DNA molecules in a single gland without IM were unmethylated. This finding experimentally confirmed that a gastric gland is a unit of occurrence of methylation of ZIK1 and possibly for the other three genes. It was also noted that methylation patterns were common to DNA molecules in a single gland [for example, region X of an IM(−) gland in the posterior wall of case 151, and region X in an IM(+) gland in the anterior wall of case 161] but were entirely different among different glands. This supported that methylation patterns in the cells in a gastric gland had a clonal origin and that methylation of specific genes took place in glands with IM multifocally.
Discussion
It was shown here that ZIK1, KAL1, and FGF14, and potentially ZNF141, were silenced by methylation of their promoter CGIs in gastric glands with IM. Analysis of single glands showed that a gastric gland, known to be produced from a single stem cell, was a unit of methylation induction and that methylation of the same genes could occur in multiple glands within a stomach in association with development of IM. This study showed that silencing of specific genes could occur in a sizable population of cells in a tissue and suggested that such consistent gene silencing could potentially impair cellular functions in multiple cells within the tissue. This emphasizes that epigenetic abnormalities could be involved in polyclonal (nontumorous) acquired disorders and marks the necessity of more intense analysis of epigenetic abnormalities in these disorders.
All four genes were expressed in gastric glands without IM but were barely expressed in the normal small intestine and colon, without methylation of their promoter CGIs. This suggested that, in the small intestine and colon, physiological lack of their expression is associated with intestinal phenotypes and that, in the gastric epithelium, their aberrant silencing could lead to intestinal phenotypes. It is still unclear what proportion of various cellular phenotypes in IM can be explained by silencing of these four genes. However, the clear and consistent association between the presence of their silencing and development of IM indicates that their silencing is at least partly involved in the irreversible changes of cellular phenotypes.
ZIK1 (19q13.43), previously known as LOC284307 and cDNA clone IMAGE:4442601, was recently identified as a “zinc finger protein-interacting with K protein 1.”18 Except that it is a zinc finger protein and a potential transcription factor, little is known about its function. ZNF141 (4p16.3) was originally identified as a candidate gene responsible for Wolf-Hirschhorn syndrome,19 which is characterized by multiple developmental defects associated with chromosomal aneusomy. ZNF141 is also a zinc finger protein and is almost ubiquitously expressed at low levels. KAL-1 (Xp22.32) is a gene responsible for Kallmann syndrome, which consists of idiopathic hypogonadotropic hypogonadism and anosmia.20 Its product, anosmin-1, plays a key role in axonal branching and guidance21 but, at the same time, is considered to be important in the genesis of various organs.22 FGF14 (13q34), also known as FHF4, belongs to a subclass of fibroblast growth factors23 and is a gene responsible for cerebellar ataxia.24 Interestingly, all four genes are deeply involved in normal development and tissue differentiation. Because critical roles of aberrant expression of intestine-specific homeobox genes, CDX1 and CDX2, in the development of IM have been reported,25–27 the effect of the silencing of the four or three genes on the CDX1 and CDX2 expression or vice versa is of interest.
It is necessary to clarify how silencing of the four genes was induced. We recently found that Helicobacter pylori infection potently induced methylation of CGIs and that some CGIs were methylated at higher levels than others.8 Methylation of the four genes identified here was almost absent in gastric mucosae without IM even in the presence of H. pylori infection (data not shown). There is a possibility that methylation of the four genes is also induced by H. pylori but that the methylation cannot be detected in samples without IM because it is closely associated with IM. As a mechanism for induction of methylation in preferential genes, decreased transcription is recognized as a trigger of promoter methylation.9,28,29 Decreased transcription of specific genes in response to H. pylori infection could underlie the induction of aberrant methylation of preferential genes in multiple cells.
IM is well known as a lesion often associated with intestinal-type gastric cancers.11 Some researchers believe that IM is a precancerous lesion.27,30 In this case, intestinal-type gastric cancers originated from IM must always have epigenetic alterations present in IM. Other researchers believe that common factors, such as H. pylori infection, induce both IM and intestinal-type gastric cancers.31 In this case, even if intestinal-type gastric cancers are closely associated with IM, some of them are expected to lack epigenetic alterations consistently present in IM. We analyzed methylation of the four genes in 15 intestinal-type gastric cancers, 35 diffuse-type gastric cancers, and seven gastrointestinal stromal tumors. ZIK1 was methylated in 11 of 15 intestinal-type gastric cancers, 28 of 35 diffuse-type gastric cancers, and none of the gastrointestinal stromal tumors. ZNF141 was methylated in 2/15, 14/35, and 0/7; KAL1 was in 4/15, 18/35 and 0/7; and FGF14 was in 15/15, 31/35, and 0/7, respectively. Although an essential role of any of these four genes in IM has not been established, it was more likely that IM and intestinal-type gastric cancers simply shared common causes.
In the colon, the presence of aberrant methylation of SFRP1 in very early neoplastic lesions, aberrant crypt foci, has been reported.32,33 The presence of methylation of ESR and other genes in the non-neoplastic colonic mucosae is also reported.7 Taken together with our findings in a polyclonal disorder, IM, aberrant methylation seems to be present in very early stages of some neoplastic diseases and polyclonal disorders.
In summary, we identified three, and possibly one additional, genes that were silenced in a polyclonal disorder, IM, of the stomach. The presence of aberrant gene silencing in a sizable population of cells in a tissue emphasized the potential role of epigenetic abnormalities in polyclonal disorders.
Acknowledgments
The authors are grateful to Dr. E. Okochi-Takada for her critical reading of the manuscript. This study was supported by a grant-in-aid for Human Genome and Tissue Regeneration from the Ministry of Health, Labor and Welfare; and the Special Coordination Funds for Promoting Science and Technology from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
Footnotes
Address reprint requests to Toshikazu Ushijima, Carcinogenesis Division, National Cancer Center Research Institute, Tsukiji 5-chome, Chuo-ku, Tokyo 104, Japan. E-mail: tushijim@ncc.go.jp.
References
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. 2003;349:2042–2054. doi: 10.1056/NEJMra023075. [DOI] [PubMed] [Google Scholar]
- Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005;5:223–231. doi: 10.1038/nrc1571. [DOI] [PubMed] [Google Scholar]
- Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet. 1994;7:536–540. doi: 10.1038/ng0894-536. [DOI] [PubMed] [Google Scholar]
- Hsieh CJ, Klump B, Holzmann K, Borchard F, Gregor M, Porschen R. Hypermethylation of the p16INK4a promoter in colectomy specimens of patients with long-standing and extensive ulcerative colitis. Cancer Res. 1998;58:3942–3945. [PubMed] [Google Scholar]
- Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61:3573–3577. [PubMed] [Google Scholar]
- Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M, Tamura G, Saito D, Sugimura T, Ichinose M, Ushijima T. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12:989–995. doi: 10.1158/1078-0432.CCR-05-2096. [DOI] [PubMed] [Google Scholar]
- Ushijima T, Okochi-Takada E. Aberrant methylations in cancer cells: where do they come from? Cancer Sci. 2005;96:206–211. doi: 10.1111/j.1349-7006.2005.00035.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Loeb LA. The contribution of endogenous sources of DNA damage to the multiple mutations in cancer. Mutat Res. 2001;477:7–21. doi: 10.1016/s0027-5107(01)00091-4. [DOI] [PubMed] [Google Scholar]
- Tatematsu M, Tsukamoto T, Inada K. Stem cells and gastric cancer: role of gastric and intestinal mixed intestinal metaplasia. Cancer Sci. 2003;94:135–141. doi: 10.1111/j.1349-7006.2003.tb01409.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Bjerknes M, Amar J. Methods for the determination of epithelial cell kinetic parameters of human colonic epithelium isolated from surgical and biopsy specimens. Gastroenterology. 1984;86:78–85. [PubMed] [Google Scholar]
- Tsukamoto T, Inada K, Tanaka H, Mizoshita T, Mihara M, Ushijima T, Yamamura Y, Nakamura S, Tatematsu M. Down-regulation of a gastric transcription factor, Sox2, and ectopic expression of intestinal homeobox genes, Cdx1 and Cdx2: inverse correlation during progression from gastric/intestinal-mixed to complete intestinal metaplasia. J Cancer Res Clin Oncol. 2004;130:135–145. doi: 10.1007/s00432-003-0519-6. [DOI] [PubMed] [Google Scholar]
- Kaneda A, Takai D, Kaminishi M, Okochi E, Ushijima T. Methylation-sensitive representational difference analysis and its application to cancer research. Ann NY Acad Sci. 2003;983:131–141. doi: 10.1111/j.1749-6632.2003.tb05968.x. [DOI] [PubMed] [Google Scholar]
- Ushijima T, Morimura K, Hosoya Y, Okonogi H, Tatematsu M, Sugimura T, Nagao M. Establishment of methylation-sensitive-representational difference analysis and isolation of hypo- and hypermethylated genomic fragments in mouse liver tumors. Proc Natl Acad Sci USA. 1997;94:2284–2289. doi: 10.1073/pnas.94.6.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneda A, Kaminishi M, Sugimura T, Ushijima T. Decreased expression of the seven ARP2/3 complex genes in human gastric cancers. Cancer Lett. 2004;212:203–210. doi: 10.1016/j.canlet.2004.03.020. [DOI] [PubMed] [Google Scholar]
- Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, Kimura K, Makita H, Sekine M, Obayashi M, Nishi T, Shibahara T, Tanaka T, Ishii S, Yamamoto J, Saito K, Kawai Y, Isono Y, Nakamura Y, Nagahari K, Murakami K, Yasuda T, Iwayanagi T, Wagatsuma M, Shiratori A, Sudo H, Hosoiri T, Kaku Y, Kodaira H, Kondo H, Sugawara M, Takahashi M, Kanda K, Yokoi T, Furuya T, Kikkawa E, Omura Y, Abe K, Kamihara K, Katsuta N, Sato K, Tanikawa M, Yamazaki M, Ninomiya K, Ishibashi T, Yamashita H, Murakawa K, Fujimori K, Tanai H, Kimata M, Watanabe M, Hiraoka S, Chiba Y, Ishida S, Ono Y, Takiguchi S, Watanabe S, Yosida M, Hotuta T, Kusano J, Kanehori K, Takahashi-Fujii A, Hara H, Tanase TO, Nomura Y, Togiya S, Komai F, Hara R, Takeuchi K, Arita M, Imose N, Mutation K, Yuuki H, Oshima A, Sasaki N, Aotsuka S, Yoshikawa Y, Matsunawa H, Ichihara T, Shiohata N, Sano S, Moriya S, Momiyama H, Satoh N, Takami S, Terashima Y, Suzuki O, Nakagawa S, Senoh A, Mizoguchi H, Goto Y, Shimizu F, Wakebe H, Hishigaki H, Watanabe T, Sugiyama A, Takemoto M, Kawakami B, Yamazaki M, Watanabe K, Kumagai A, Itakura S, Fukuzumi Y, Fujimori Y, Komiyama M, Tashiro H, Tanigami A, Fujiwara T, Ono T, Yamada K, Fujii Y, Ozaki K, Hirao M, Ohmori Y, Kawabata A, Hikiji T, Kobatake N, Inagaki H, Ikema Y, Okamoto S, Okitani R, Kawakami T, Noguchi S, Itoh T, Shigeta K, Senba T, Matsumura K, Nakajima Y, Mizuno T, Morinaga M, Sasaki M, Togashi T, Oyama M, Hata H, Watanabe M, Komatsu T, Mizushima-Sugano J, Satoh T, Shirai Y, Takahashi Y, Nakagawa K, Okumura K, Nagase T, Nomura N, Kikuchi H, Masuho Y, Yamashita R, Nakai K, Yada T, Nakamura Y, Ohara O, Isogai T, Sugano S. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- Tommerup N, Aagaard L, Lund CL, Boel E, Baxendale S, Bates GP, Lehrach H, Vissing H. A zinc-finger gene ZNF141 mapping at 4p16.3/D4S90 is a candidate gene for the Wolf-Hirschhorn (4p-) syndrome. Hum Mol Genet. 1993;2:1571–1575. doi: 10.1093/hmg/2.10.1571. [DOI] [PubMed] [Google Scholar]
- del Castillo I, Cohen-Salmon M, Blanchard S, Lutfalla G, Petit C. Structure of the X-linked Kallmann syndrome gene and its homologous pseudogene on the Y chromosome. Nat Genet. 1992;2:305–310. doi: 10.1038/ng1292-305. [DOI] [PubMed] [Google Scholar]
- Soussi-Yanicostas N, de Castro F, Julliard AK, Perfettini I, Chedotal A, Petit C. Anosmin-1, defective in the X-linked form of Kallmann syndrome, promotes axonal branch formation from olfactory bulb output neurons. Cell. 2002;109:217–228. doi: 10.1016/s0092-8674(02)00713-4. [DOI] [PubMed] [Google Scholar]
- Hardelin JP, Julliard AK, Moniot B, Soussi-Yanicostas N, Verney C, Schwanzel-Fukuda M, Ayer-Le Lievre C, Petit C. Anosmin-1 is a regionally restricted component of basement membranes and interstitial matrices during organogenesis: implications for the developmental anomalies of X chromosome-linked Kallmann syndrome. Dev Dyn. 1999;215:26–44. doi: 10.1002/(SICI)1097-0177(199905)215:1<26::AID-DVDY4>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Wang Q, McEwen DG, Ornitz DM. Subcellular and developmental expression of alternatively spliced forms of fibroblast growth factor 14. Mech Dev. 2000;90:283–287. doi: 10.1016/s0925-4773(99)00241-5. [DOI] [PubMed] [Google Scholar]
- van Swieten JC, Brusse E, de Graaf BM, Krieger E, van de Graaf R, de Koning I, Maat-Kievit A, Leegwater P, Dooijes D, Oostra BA, Heutink P. A mutation in the fibroblast growth factor 14 gene is associated with autosomal dominant cerebellar ataxia. Am J Hum Genet. 2003;72:191–199. doi: 10.1086/345488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren P, Silberg DG, Sirica AE. Expression of an intestine-specific transcription factor (CDX1) in intestinal metaplasia and in subsequently developed intestinal type of cholangiocarcinoma in rat liver. Am J Pathol. 2000;156:621–627. doi: 10.1016/S0002-9440(10)64766-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberg DG, Furth EE, Taylor JK, Schuck T, Chiou T, Traber PG. CDX1 protein expression in normal, metaplastic, and neoplastic human alimentary tract epithelium. Gastroenterology. 1997;113:478–486. doi: 10.1053/gast.1997.v113.pm9247467. [DOI] [PubMed] [Google Scholar]
- Yuasa Y. Control of gut differentiation and intestinal-type gastric carcinogenesis. Nat Rev Cancer. 2003;3:592–600. doi: 10.1038/nrc1141. [DOI] [PubMed] [Google Scholar]
- de Smet C, Loriot A, Boon T. Promoter-dependent mechanism leading to selective hypomethylation within the 5′ region of gene MAGE-A1 in tumor cells. Mol Cell Biol. 2004;24:4781–4790. doi: 10.1128/MCB.24.11.4781-4790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JZ, Stirzaker C, Harrison J, Melki JR, Clark SJ. Hypermethylation trigger of the glutathione-S-transferase gene (GSTP1) in prostate cancer cells. Oncogene. 2002;21:1048–1061. doi: 10.1038/sj.onc.1205153. [DOI] [PubMed] [Google Scholar]
- Kuniyasu H, Yasui W, Yokozaki H, Tahara E. Helicobacter pylori infection and carcinogenesis of the stomach. Langenbecks Arch Surg. 2000;385:69–74. doi: 10.1007/s004230050248. [DOI] [PubMed] [Google Scholar]
- Kawachi H, Takizawa T, Eishi Y, Shimizu S, Kumagai J, Funata N, Koike M. Absence of either gastric or intestinal phenotype in microscopic differentiated gastric carcinomas. J Pathol. 2003;199:436–446. doi: 10.1002/path.1323. [DOI] [PubMed] [Google Scholar]
- Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van Engeland M, Toyota M, Tokino T, Hinoda Y, Imai K, Herman JG, Baylin SB. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet. 2004;36:417–422. doi: 10.1038/ng1330. [DOI] [PubMed] [Google Scholar]
- Tycko B. Genetic and epigenetic mosaicism in cancer precursor tissues. Ann NY Acad Sci. 2003;983:43–54. doi: 10.1111/j.1749-6632.2003.tb05961.x. [DOI] [PubMed] [Google Scholar]