Abstract
Knowing gene structure is vital to understanding gene function, and accurate genome annotation is essential for understanding cellular function. To this end, we have developed a genome-wide assay for mapping introns in Saccharomyces cerevisiae. Using high-density tiling arrays, we compared wild-type yeast to a mutant deficient for intron degradation. Our method identified 76% of the known introns, confirmed 18 previously predicted introns, and revealed 9 formerly undiscovered introns. Furthermore, we discovered that all 13 meiosis-specific intronic yeast genes undergo regulated splicing, which provides posttranscriptional regulation of the genes involved in yeast cell differentiation. Moreover, we found that ≈16% of intronic genes in yeast are incompletely spliced during exponential growth in rich medium, which suggests that meiosis is not the only biological process regulated by splicing. Our tiling-array assay provides a snapshot of the spliced transcriptome in yeast. This robust methodology can be used to explore environmentally distinct splicing responses and should be readily adaptable to the study of other organisms, including humans.
Keywords: meiosis, regulated splicing, Saccharomyces cerevisiae
Intronic sequences provide numerous functional elements that direct pre-mRNA processing and alternative splicing. In the relatively simple eukaryote Saccharomyces cerevisiae, introns direct splicing (1), can increase gene expression (2), and, in specific cases, may contain small nucleolar RNAs (3). Additionally, introns in yeast can modulate translation posttranscriptionally through a process known as regulated splicing (4–7). During regulated splicing, yeast cells under certain conditions can limit intron splicing in specific genes, which, in turn, disrupts translation through frame-shifting and/or introduction of nonsense codons (4–7). Accurate mapping of introns is an essential first step to understanding RNA splicing and function.
S. cerevisiae is an easily manipulatable eukaryote with a relatively small extensively studied genome that shares many core spliceosome functions with humans (1). Only 5% of S. cerevisiae genes are interrupted by introns (8, 9), and all introns are constitutively removed before translation (10). Because its genome is relatively small and well characterized, yeast serves as an ideal model organism for new technologies.
Tiling DNA microarrays, comprised of overlapping, end-to-end, or closely spaced DNA probes, have been used to map cellular transcription in a variety of organisms. Tiling-array data have improved gene annotation and revealed extensive transcription of noncoding RNAs (11–13). We used a high-density yeast-tiling array with overlapping probes, which provides a per-strand resolution of eight nucleotides, to research pre-mRNA processing in yeast. The closely spaced probes allowed for accurate measurement of small transcriptional features, such as single exons and small introns.
In this paper, we describe the results of a high-resolution microarray investigation of yeast splicing during exponential growth in rich medium. We show that, even for the extensively investigated, curated, and refined S. cerevisiae genome, we were able to verify 18 previously predicted introns and identify nine previously uncharacterized introns, representing a 10% increase over the number of intron-containing genes currently annotated by the Saccharomyces Genome Database (SGD) (8, 9). Additionally, we show that 13 of 13 meiosis-specific intron-containing genes are subject to regulated splicing, such that splicing is repressed during vegetative growth and induced during sporulation. Note that we define meiosis-specific genes as those that function primarily during the biological process of meiosis, as defined by the SGD (8, 9). Because the 13 meiotic genes represent a small subset of the 45 inefficiently spliced introns identified, we suggest that regulated splicing may be more pervasive in yeast than initially suspected. In summary, we developed a methodology for whole-genome intron identification and analysis. This study improves gene annotation and extends our understanding of regulated splicing in S. cerevisiae.
Results
High-Density Tiling Arrays Identify the Majority of Known Introns in Yeast.
To identify and map intronic sequences, we compared the transcriptional content of wild-type yeast with a mutant strain deficient for degradation of processed intron lariats (dbr1Δ/dbr1Δ) (14). Intron lariats are formed during RNA splicing when the 2′ hydroxyl from the branchpoint adenosine nucleophilically attacks the 5′-splice site. Lariats are released from the mRNA during exon ligation when the 5′-exon nucleophilically attacks the 3′-splice site. This reaction creates a 2′-hydroxyl-5′-phosphate bond within the processed intron, forming a lariat structure. RNA nucleases within the cell can digest the 3′ tail of the intron lariat (14), but the loop structure is impervious to digestion until Dbr1, a specialized debranching enzyme, severs the 2′-hydroxyl-5′-phosphate bond (14). When DBR1, a nonessential gene, is deleted from the genome, intron lariats build up in the cell (15). We postulated that subtractive comparison of the transcriptomes from wild-type and dbr1Δ/dbr1Δ yeast would reveal expressed and processed intronic sequences.
Affymetrix Tiling Analysis Software (16) was used to analyze data from three replicate pairs of wild-type and mutant samples. We identified all regions of differing expression, termed “intervals” between the strains. Interval identification was optimized by using the known introns as positive- and exon sequences as a negative-training set (described in detail in Methods). From our optimized analysis, we identified a list of candidate intervals. We defined an intron as being detected if a candidate interval overlapped the intronic sequence. Using this approach, we successfully identified the majority of known introns.
The S. cerevisiae nuclear genome consists of 5,762 nondubious, verified, or uncharacterized ORFs, of which 259 are known to contain introns (8, 9). The software identified 180 (70%) of the 259 known intronic genes (i-genes), and informed manual assessment of the array data identified an additional 17 introns. Additionally, an intron contained within a “dubious” ORF (YPR170W-B) (8, 9) was revealed and verified by the array data. In total, 198 of 260 (76%) intron-containing genes were identified by using this simple approach (Table 1).
Table 1.
Summary of introns identified with the yeast tiling array
| Introns | i-genes |
Splicing of undetected introns |
Percent detected |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Total | Detected | Undetected | 0% | <50% | >50% | 100% | Total, % | Spliced, % | |
| Known | 260 | 198 | 62 | 20 | 9 | 9 | 24 | 76 | 83 |
| Predicted | 20 | 18 | 2 | 2 | 0 | 0 | 0 | 90 | 100 |
Known introns were gathered from the SGD (8, 9), and predicted introns were collected from the literature (17–19). The numbers of intronic genes (‘Total,’ ‘Detected,’ and ‘Undetected’) are listed under i-genes. All undetected introns, 62 known and 2 predicted, are recompiled under ‘Splicing of undetected introns;’ their respective splicing efficiencies are presented as percentiles (0%, <50%, >50%, and 100%). Under ‘Percent detected,’ ‘Total’ is the percent of introns detected compared to total i-genes [Total(% detected) = 100 × (Detected(i-gene)/Total(i-gene))]. ‘Spliced’ is the percent of introns detected compared to the set of introns that splice [Spliced = 100 × (Detected(i-gene)/(Total(i-gene) − ‘0%’))].
Closer inspection of the array data showed that signal intensity for introns was concentrated toward the 5′ end of the introns. This signal corresponded well with the intron-lariat loops, located between the 5′-splice site and the branchpoint adenosine (Fig. 1). Alternatively, the lariat tails showed little signal intensity, verifying that the tails are susceptible to nuclease degradation (14). Additionally, we observed that the expression level of the intron lariats generally correlated well with gene expression, such that highly expressed genes produced more intronic signal (data not shown). These patterns of expression allowed us to identify introns, as well as to map 5′-splice sites and branchpoint sites with a high degree of confidence.
Fig. 1.
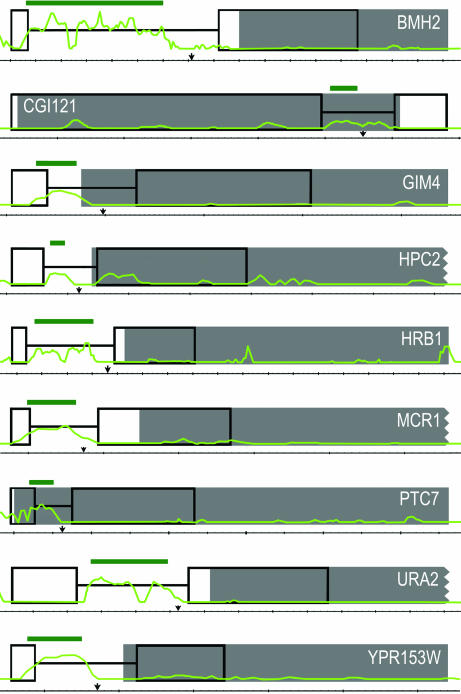
Nine introns were discovered by using a high-density tiling array. Intronless gene annotations from the SGD are shown as solid gray boxes (8, 9). For large genes, the uninformative 3′ ends are not shown; truncations are indicated by a jagged edge on the gray boxes. Newly annotated introns are depicted as thin black lines flanked by open boxes, which designate the position of high-quality exonic sequence. Light-green lines graph the intensity that results when wild-type data are subtracted from dbr1Δ/dbr1Δ data; peaks are indicative of increasing levels of signal from intron lariats. Dark-green bars show intronic intervals automatically identified by the software. Black arrows designate the position of conserved branchpoint sequences. Genomic distances are specified along the thin black line beneath the gene annotations; large tick marks demarcate 100 bp. Gene names are specified in white lettering.
Eighteen Previously Predicted Introns Are Independently Verified by Using Tiling Arrays.
Several groups of RNA biologists have made efforts to identify and catalog all known and predicted intronic genes (17–19). In total, 20 potential intronic genes identified by these researchers have not yet been included in the SGD (8, 9). Our array-directed method for intron detection was able to identify 18 of 20 of these proposed introns [supporting information (SI) Table 2]. To authenticate the array results, we used 5′-end RACE and RT-PCR to amplify and sequence all 20 proposed intronic cDNAs. We validated all 18 introns identified with the tiling arrays. Notably, using three different methods (tiling arrays, 5′-end RACE sequencing, and directed RT-PCR), we were not able to accumulate any evidence for the presence of introns in two proposed intronic genes (MTR2 and SNT1). It remains a possibility that MTR2 and SNT1 contain introns that are not excised by the spliceosome under the growth conditions we tested. In summary, we have accumulated exceptionally strong evidence verifying the existence of 18 previously predicted introns that are not currently annotated by the SGD.
Nine Recently Discovered Introns Are Revealed by Using High-Density Yeast-Tiling Arrays.
After we optimized our high-density tiling-array analysis for the verification of known introns, we redirected the analysis toward the discovery of uncharacterized introns. We postulated that, even for the extensively studied and thoroughly annotated S. cerevisiae genome, there may be unidentified introns that could be easily revealed only by using a genomic approach. All intervals identified with the Affymetrix software were ranked based on intensity, size, proximity to known genes, and splice-site sequence conservation. We examined the highest-ranking intervals and identified 384 candidate i-genes for RT-PCR and sequencing analysis. Using this approach, we identified nine introns in nine genes: BMH2, CGC121, GIM4, HPC2, HRB1, MCR1, PTC7, URA2, and YPR153W (Fig. 1). Each intron was validated with RT-PCR gel electrophoresis, sequencing, and splice-site signal identification (5′-splice, branchpoint, and 3′-splice sites).
An examination of the intronic sequences of the nine introns finds only one previously undescribed splice signal (SI Table 2). Sequencing of the cDNAs for the intron-containing genes was used to determine the exact 5′- and 3′-splice signals. Branchpoint sequences were deduced by comparing sequence data with array data and finding the branchpoint most closely associated with the 3′-end of the interval. Eight of the nine introns contain one of two highly conserved 5′-splice site motifs (GTATGT or GTACGT), whereas GIM4 contains a less-common version of the motif (GTATGC), which it shares with nine other S. cerevisiae introns (20). We saw more variability in the branchpoint site sequences. Six of the introns contain either of the top two most-conserved branchpoint motifs (TACTAAC or GACTAAC). The branchpoint site in HRB1 (TACTAAT) is one of only two found in S. cerevisiae, whereas the branchpoint site in YPR153W is unique in S. cerevisiae, although it occurs in Pichia angusta, another hemiascomycetous yeast (20). The only previously undescribed splice signal we identified was the branchpoint site in HPC2 (GATTAAC), which differs at one position from the second-most common site (GACTAAC). We had expected that the introns would share some sequences with the conserved splice-site homologies, but we were surprised at the extent of the sequence conservation, and that these introns had not previously been revealed by using bioinformatic approaches. Another interesting characteristic of the introns is that HPC2, GIM4, PTC7, and YPR153W are inefficiently spliced (splicing rates are 85%, 72%, 55%, and 50%, respectively), which could explain why they have been overlooked by molecular approaches until now. It is our proposition from these data that the SGD should be updated to include 27 additional introns, the nine new introns, plus the 18 previously predicted introns described above. Accordingly, we have submitted these data to the SGD for consideration.
Inefficient Splicing and Low Expression Impede Intron Identification.
We next asked why our array technology, which confirmed 76% of the known introns, failed to detect the other 24%. Because it is possible that some of the undetected i-genes were misannotated, we sought to verify the presence of introns within all 62 undetected i-genes by using RT-PCR and gel electrophoresis. We used RT-PCR, because the assay can detect even minute levels of transcriptional activity. We found that the transcripts of all 62 undetected i-genes (spliced or unspliced) were identifiable by RT-PCR analysis, which suggested all i-genes are expressed at some level. Notably, the observed constant basal level of gene transcription is not a result of genomic DNA contamination (see for example genes) and is not unique to intronic genes (L. David, personal communication). Because several wild-type yeast genes are known to be inefficiently spliced (4–7), we investigated the splicing efficiencies of the undetected introns. We found that >60% of the undetected introns were incompletely spliced. Specifically, 20 of the 62 undetected i-genes did not splice in rich medium, 9 spliced at a rate of <50%, and another 9 spliced between 50% and 75% (Table 1). In contrast, when we tested a subset of the detected known i-genes, we found that 13 of 15 spliced completely; and the two outliers, RAD14 and REC114, spliced relatively efficiently at 75% and 89%, respectively. Our array-based assay can detect only introns that are spliced. We detected 83% (198 of 240) of the introns that spliced in rich medium. We acknowledge it may be unrealistic to expect arrays to detect i-genes that are partially spliced, especially if they are minimally expressed. Thus, we suggest that 83% represents a conservative estimate of the performance of our methodology.
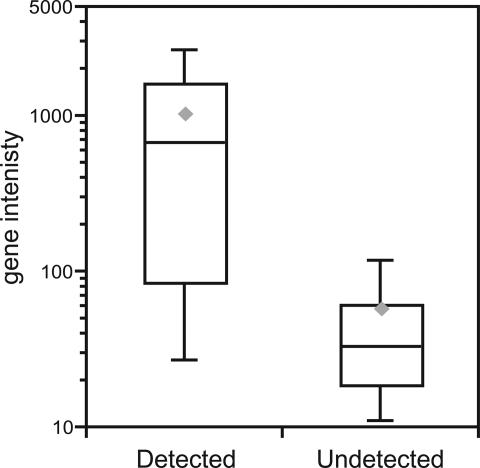
Because expression levels also correlate with array detection, we asked how expression levels of the 198 detected i-genes compared with the 42 undetected spliced i-genes. To evaluate gene expression, we calculated the average probe intensity across the exons of each i-gene and compared the averaged intensities of the undetected with the identified i-genes. We observed a significantly lower level of expression for the undetected i-genes (Fig. 2). The median expression of undetected i-genes was 16-fold lower than for detected i-genes. In addition to inefficient splicing and low gene expression, poor probe hybridization, repetitive sequences, and short introns also can contribute to the difficulty of detecting expressed and spliced introns. Further improvement of microarray design and data analysis should minimize some of these impediments.
Fig. 2.
Undetected introns are predominantly located in minimally expressed genes. The y axis specifies gene intensity, which is calculated by averaging probe intensities across the exons of each i-gene. ‘Detected’ introns were identified by using our array-based methodology; ‘Undetected’ introns were not identified. On the bar-and-whiskers diagram, the lowest, second-lowest, middle, second-highest, and highest lines represent the 10th percentile, 25th percentile, median, 75th percentile, and 90th percentile, respectively. The gray diamonds designate the mean.
Meiotic Intronic Genes Appear to Be Posttranscriptionally Regulated at the Level of Splicing.
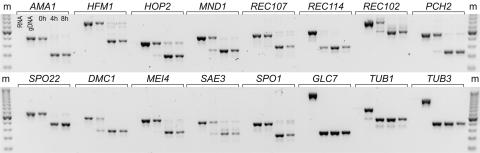
We noticed that genes involved in meiosis were enriched within the set of undetected introns. In fact, our intron-array analysis identified only four (GLC7, REC114, TUB1, and TUB3) of 16 i-genes that the SGD identifies as functioning during meiosis (8, 9). Only one of these genes (REC114) is meiosis-specific, in that it functions primarily, if not exclusively, during meiosis; the other three participate in important biological processes beyond meiosis (8, 9) and are thus not considered meiosis-specific. We suspected deficient detection of meiotic introns could be the result of inefficient splicing caused by regulated splicing, given three meiotic genes have previously been reported as undergoing regulated splicing (AMA1, REC107, and HFM1) (4, 6, 21). Using RT-PCR and gel electrophoresis, we found that all 13 meiosis-specific introns were incompletely spliced during exponential growth in rich media (Fig. 3, lane “0h”). The 12 undetected introns were spliced very inefficiently with splicing rates that ranged between 0% and 51%, whereas the only meiosis-specific intron identified by the array (REC114) spliced much more efficiently at 89%. We supposed that splicing might be induced in the meiotic genes during sporulation. Using SK1, a strain of S. cerevisiae that sporulates efficiently, we studied the consequences of sporulation on splicing. We synchronized SK1 cells; induced sporulation; collected cells after 0, 4, and 8 hours; extracted and reverse-transcribed the RNA; and used PCR and electrophoresis to assess the degree of splicing (Fig. 3). The result was dramatic; all 13 meiosis-specific i-genes spliced more efficiently (>84%) during sporulation. Even REC114, which spliced well during vegetative growth, saw an increase in splicing from 89% to 94% during sporulation. Previously, 3 of the 13 meiotic genes had been identified as having regulated splicing (AMA1, HFM1, and REC107); of the three, AMA1 and REC107 have shown sporulation-dependent splicing (4, 21). We have now demonstrated that all 13 meiosis-specific i-genes splice in a sporulation-dependent manner. Our data suggest that, during vegetative growth, yeast require potent repression of meiotic gene expression, and that regulated splicing functions to minimize meiotic gene expression posttranscriptionally.
Fig. 3.
Meiosis-specific genes are regulated posttranscriptionally by splicing. PCR products, from primers that surround intronic sequences in 16 meiotic genes (designated above), were separated by using gel electrophoresis on an agarose gel stained with SYBR green. The image color has been inverted for clarity; bands of DNA appear dark on a light background. For each gene, five PCRs were carried out under identical conditions by using five different SK1-derived templates: total RNA (RNA) as a control for genomic contamination, genomic DNA (gDNA), cDNA from cells grown in rich media after 0 hours of sporulation (0h), cDNA from cells harvested after 4 (4h) or 8 (8h) hours of sporulation. The larger PCR products result from genomic DNA in the gDNA lanes or unspliced pre-mRNA in the 0h, 4h, and 8h lanes. The smaller products result from spliced mRNA. The marker (m) is a 50-bp marker. All 13 meiosis-specific genes perform regulated splicing. Only GLC7, TUB1, and TUB3, which have important cellular functions outside of meiosis, are completely spliced in rich media and do not display regulated splicing activity.
Discussion
We have developed a strategy to measure splicing patterns in S. cerevisiae by using DNA microarrays. By comparing wild-type yeast with a mutant deficient in intron-lariat debranching (dbr1Δ/dbr1Δ), we were able to identify 83% (198 of 240) of intronic genes shown to splice in rich media. The 42 spliced introns not identified were often inefficiently spliced and, on average, had lower levels of expression. Both of these characteristics contributed to the difficulty in monitoring the splicing of these 42 genes using DNA arrays. Interestingly, our assay did allow us to identify introns within some very low-expressing genes, possibly because the mRNAs in question were subject to high rates of turnover. The intron lariats would not be subject to the same rapid degradation as the mRNAs and thus would build up to levels detectable on the array. If this supposition is true, this assay could possibly be extended to analyze RNA turnover by comparing exon expression with lariat buildup.
We used our intron identification assay to search for previously unidentified introns within S. cerevisiae. We discovered nine introns in nine genes and verified 18 previously predicted introns. The addition of these 27 introns to the SGD brings the total number of i-genes to 287, which represents a 10% increase in the total number of nuclear encoded intronic genes identified in S. cerevisiae. This method of intron identification could be adapted to study cultured human cells, because the DBR1 gene, which is conserved in humans (hDBR1), maintains the same function in human cells as in yeast (22). If we extrapolate from our studies on yeast, which has ≈300 introns, to human, which has >140,000 (23), our method could possibly reveal tens of thousands of new human introns. Furthermore, this assay could make important contributions to our understanding of alternative splicing in human, as it did for regulated splicing in yeast.
Another powerful aspect of our assay is its ability to capture a snapshot of yeast splicing under a specific growth condition. It was this characteristic that revealed the extensive amount of regulated splicing occurring during the transition from vegetative growth to sporulation. Strikingly, all 13 meiosis-specific intronic genes appear to be regulated posttranscriptionally by splicing. Rigorous regulation of the transition from mitosis to meiosis is essential for yeast to maintain vigorous growth in rich media (24). The transcription of many genes appears to be leaky (L. David, personal communication); therefore, sporulation-specific splicing is likely used to further repress protein synthesis and prevent premature commencement of meiosis or ill-timed chromosome recombination, which could lead to DNA damage during vegetative growth (25). That all 13 meiosis-specific i-genes are subject to regulated splicing suggests there may be one protein or protein complex controlling meiotic splicing. Two genes, MER1 and NAM8, have demonstrated the ability to regulate splicing for specific subsets of meiosis-specific i-genes (4, 6, 21), but neither has proved capable of regulating all 13 i-genes (26–28). In addition to finding the protein components responsible for the observed splicing regulation, it would also be of great interest to identify the RNA sequences that regulate splicing of meiotic i-genes.
It has been shown that regulated splicing is not limited to meiosis (5, 7). If the splicing inefficiencies we see are any indication, it is possible that regulated splicing in S. cerevisiae is more pervasive than expected. We show that 16% of yeast i-genes splice inefficiently (45 of 287). Thirteen of the inefficiently spliced i-genes are meiotic, 16 others demonstrate some degree of splicing, and the remaining 16 do not splice while growing in rich media. The extent to which regulated splicing is utilized by yeast is unknown, and it is unclear whether all observed instances of inefficient splicing are truly regulated (29, 30). What is evident is that regulated splicing is an important way for yeast to modify gene expression posttranscriptionally.
This study was conducted to understand the splicing patterns of the S. cerevisiae transcriptome. On doing so, we discovered previously undescribed introns, validated predicted introns, and revealed an extensive network of meiosis-specific regulated splicing.
Methods
Summary of Methodology.
The methodology used included the following steps: wild-type and dbr1Δ/dbr1Δ cultures were grown in rich media (yeast extract/peptone/dextrose). RNA was isolated, and cDNA samples were prepared and labeled. Arrays were hybridized and computationally compared to identify intervals. Intervals were ranked based on splice site homologies and gene proximity. The highest-ranking intervals were validated with RT-PCR and sequencing.
Growth Conditions and Yeast Strains.
Standard yeast extract/peptone/dextrose media and 30°C growth conditions were used (31). We isolated total RNA from three strains: the isogenic S288c strain BY4743 (MATa/α, his3Δ1/his3Δ1, leu2Δ0/leu2Δ0, lys2Δ0/LYS2, MET15/met15Δ0, ura3Δ0/ura3Δ0; 4741/4742) (Open Biosystems, Huntsville, AL), the homozygous double-deletion strain dbr1Δ/dbr1Δ, which was constructed as part of the yeast deletion collection (32) (Open Biosystems, catalog no. YSC1021-664479), and an isogenic SK1 strain (MATa/α, flo8Δ0/flo8Δ0, his3Δ0/HIS3, and ura3Δ0/URA3).
Sample Preparation.
RNA was extracted with hot phenol from log-phase yeast growing in yeast extract/peptone/dextrose, as described (33). Total RNA was treated for 10 min at 37°C with RNase-free DNase I (GE Healthcare Life Sciences, Giles, U.K.; catalog no. 27-0514-01). RNA was repurified to remove the DNase I by using the RNeasy Mini Kit (Qiagen, Valencia, CA; catalog no. 74104). The RNeasy protocol was altered to optimize retention of small RNAs (<40 nt). Briefly, the RNA was treated with Qiagen's denaturing RLT buffer and vortexed, 3.5 volumes of 100% ethanol were added to facilitate binding of small RNAs, and the sample was washed twice with Qiagen's RPE buffer and eluted in 1 × TE buffer, pH 8.0 (10 mM Tris·HCl/1 mM EDTA).
Single-stranded cDNA was synthesized in 200-μl reactions containing 0.25 μg/μl total RNA, 12.5 ng/μl random primers (Invitrogen, Carlsbad, CA; catalog 48190-01), 12.5 ng/μl Oligo(dT)12–18 primer (Invitrogen, catalog no. 18418-012), 15 units/μl SuperScript II (Invitrogen, catalog no. 18064-014), 1 × First Strand Buffer, 10 mM DTT, and 10 mM dNTPs (Invitrogen, catalog no. 18427013). After the RNA and primers were denatured for 10 min at 70°C, the remaining reagents were added, and the reaction was incubated at 25°C for 10 min, 37°C for 60 min, 42°C for 60 min, and 70°C for 10 min. After cDNA synthesis, the RNA was degraded with 1/3 volume of 1 M NaOH incubated for 30 min, and an addition of 1/3 volume of 1 M HCl was used to neutralize the solution before cleanup. The cDNA was cleaned up by using the columns from the MinElute Reaction Cleanup Kit (Qiagen, catalog no. 28204) and the buffers and protocol from QIAquick Nucleotide Removal Kit (Qiagen, catalog no. 28304).
cDNA (7 μg) was fragmented with 2.1 units/μl DNase I (Amersham, Santa Clara, CA; catalog no. E70194Y) in 1× One-Phor-All Buffer PLUS (Amersham; catalog no. 27-0901-02) for 10 min at 37°C and quenched by incubating at 98°C for 15 min. The fragmented cDNA (7 μg) was labeled by incubating in a 50-μl reaction containing 0.3 mM GeneChip DNA Labeling Reagent (Affymetrix; catalog no. 900542), 1× Terminal Transfer Reaction Buffer, and 2 μl of Terminal Deoxynucleotidyl Transferase (Promega, Madison, WI; catalog no. M1871) for 60 min at 37°C.
Array Hybridization.
Labeled cDNA (7 μg) was mixed with 100 mM MES (Sigma–Aldrich, St. Louis, MO; catalog nos. M5287 and 5057), 1 M Na+ (from NaCl and MES sodium salt), 20 mM EDTA (Invitrogen, catalog no. 15575-020), 0.01% Tween-20 (Sigma–Aldrich, catalog no. P8942), 50 pM control oligonucleotide B2 (Affymetrix, catalog no. 900301), 0.1 mg/ml herring sperm DNA (Promega, catalog no. D1811), and 0.5 mg/ml BSA (Invitrogen, catalog no. 15561-020) in a total volume of 330 μl, from which 220 μl was hybridized per array. Arrays were hybridized for 16 h at 45°C with a rotation rate of 60 rpm (Affymetrix, Santa Clara, CA; GeneChip Hybridization Oven 640).
Analysis of Array Data.
High-density tiling arrays were analyzed, normalized, and compared by using Affymetrix Tiling Analysis Software (TAS) (16). Intronic intervals were identified by creating a difference map from normalized sets of three arrays hybridized with either wild-type (BY4743) or dbr1Δ/dbr1Δ cDNA. We identified all regions of differing expression (termed “intervals”) between the strains. Within the TAS software, the user can define values for variables that direct interval calling (16). These variables include minimum intensity, maximum P value, bandwidth (distance in base pairs for which data were grouped for statistical evaluation), minimum run (minimum size in base pairs of a detected interval), and maximum gap (maximum tolerated gap between signals in an interval). As a first step, we optimized each variable individually to identify a minimum range of values that overlapped with the most known introns (positive training set) and the fewest exons (negative control). We then carried out a second optimization of these variables by calculating all of the intervals for a 4 × 3 × 3 × 2 matrix of variable combinations (maximum gap, minimum run, bandwidth, and P value, respectively). We calculated two sets of intervals, one with a maximum P value of 0.01 and one with a minimum intensity of five, while holding maximum gap, minimum run, and bandwidth constant (numerical values 9, 16, and 17, respectively). We combined the two sets of intervals and picked 384 candidate i-genes, which were tested by using RT-PCR and sequencing.
Sequencing.
Genes with introns identified by using the yeast-tiling array were confirmed by sequencing. DNased total RNA (1 μg) was reverse-transcribed into 5′-RACE Ready cDNA by using the SMART RACE cDNA Amplification Kit (BD Biosciences, Franklin Lakes, NJ; catalog no. 634914). For sequencing, cDNAs were PCR-amplified by using gene-specific reverse primers, 5′-RACE universal forward primers, and Platinum PCR SuperMix (Invitrogen, catalog no. 11306-016). PCRs were incubated with 0.1 units/μl SAP (USB, Cleveland, OH; catalog no. 70092X) and 0.1 units/μl Exonuclease I (USB, catalog no. 70073X) for 60 min at 37°C and 15 min at 80°C, before sequencing. Sequence reactions were carried out by using Big Dye Terminator version 3.0 Ready Reaction Cycle Sequencing Kit (Applied Biosystems, Foster City, CA; catalog no. 4390244) and included 1–3 μl of gene specific PCR, 0.7–1.0× of 2.5X Big Dye Terminator, 7–10 μM primer, and 0.7–1.0× of 5× sequencing buffer. Sequencing reactions were cleaned up with Sephadex G-50 Fine (Amersham Biosciences, 17-0573-02) in −HV 0.45-μm Filter Plates (Millipore, Billerica, CA; catalog no. MAHVN4550). Reactions were run on a 3730xl DNA Analyzer (Applied Biosystems).
Publicly available software was used to analyze the sequence data; bases were called with Phred, sequences were aligned with Phrap, and data were visualized and manually manipulated by using Consed (34–36). The presence of a splice junction within a sequenced PCR product was identified by using BLAST and SIM4 (37, 38); BLAST was used to find the sequence within the yeast genome, SIM4 was used to align the sequence to a 20-kb genomic fragment centered on the blast match start. Using a 20-kb fragment (rather than the entire chromosome) for alignment allowed for the identification of small, ≈10 bp, exons that are often present at the 5′ termini of the sequences. We searched gap sequences for intron-specific motifs to further substantiate the existence of the intron.
Quantifying Splicing Efficiencies with Gel Electrophoresis.
RT-PCR was carried out on total RNA (10 ng/μl), genomic DNA (50 pg/μl), and cDNA (50 pg/μl) from various sources. RNA was extracted with the hot-phenol method described above and DNased twice; RNA was treated for 30 min at 37°C with RNase-free DNase I (GE Healthcare Life Sciences, catalog no. 27-0514-01) before being repurified and re-DNased using the RNeasy Mini Kit and RNase-free DNase Set (Qiagen, catalog nos. 74104 and 79254). Genomic DNA was extracted from yeast by using the Yeastar Genomic Kit (Zymo Research, Orange, CA; catalog no. D2002). The cDNA was synthesized as described above. In Fig. 3, the concentration of RNA that was PCR amplified was 2-fold higher than the residual RNA present in the unpurified cDNA samples that were PCR-amplified. This concentration difference ensured that, if genomic contamination was present in our cDNA, it would be readily PCR-amplified and visualized in the RNA controls. Primers were designed to surround intronic sequences and were used at a concentration of 600 nM, in 1× Platinum PCR SuperMix (Invitrogen, catalog no. 11306-016). PCRs were cycled 32 times (94°C for 30 s, 56°C for 30 s, and 72°C for 60 s) before 8 μl of the resulting reactions was loaded onto 2% agarose gels stained with 1.5× SYBR Safe DNA Gel Stain (Invitrogen, catalog no. S33102). Bands were identified and quantified automatically by using Labworks Image Acquisition and Analysis Software (UPV, Upland, CA). Values were normalized with respect to DNA length and reported as percentiles of spliced over spliced plus unspliced by using the following equation: percent spliced = 100 × ((intensity of spliced/base pairs of spliced PCR)/(((intensity of spliced/base pairs of spliced PCR) + (intensity of unspliced))/(base pairs of unspliced PCR))).
Measuring Meiotic Splicing.
The SK1 strain of S. cerevisiae was used for all of the meiosis experiments. Cell synchronization and sporulation were carried out as described (39). Aliquots of cells (1.5 ml) were collected at a constant cell density of 1.0 OD, spun down, and frozen in dry ice after the supernatant was removed. RNA was extracted from the cells by using the Masterpure Yeast RNA Purification Kit (Epicentre Technologies, Madison, WI; catalog no. MPY03100). The methods used for RT-PCR, gel electrophoresis, splicing quantitation, and cDNA preparation from doubly DNased RNA are described above.
Supplementary Material
Acknowledgments
We thank Lior David, Alex M. Plocik, and Corey Nislow for experimental advice; Michelle Nguyen for technical assistance; Keith Anderson and Michael Jensen for oligonucleotide synthesis; Marilyn Fukushima for comprehensive editing of the manuscript; and Jed Dean, Robert St. Onge, and Michael Primig for helpful comments on the manuscript. This work was supported by National Institutes of Health Grant RR020000 (to K.J. and R.W.D.).
Abbreviations
- i-gene
intronic gene
- SGD
Saccharomyces Genome Database.
Note Added in Proof.
Miura et al. (40) recently reported similar results from a large scale cDNA sequencing effort. They identified all 18 of the previously predicted introns we found and five of nine of the new introns we discovered.
Footnotes
The authors declare no conflict of interest.
Data deposition: The array data reported in this paper have been deposited in the ArrayExpress database (accession no. E-MEXP-919). The cDNA sequences reported in this paper have been deposited in the GenBank database (accession nos. EF123123–EF123149).
This article contains supporting information online at www.pnas.org/cgi/content/full/0610354104/DC1.
References
- 1.Burge CB, Tuschl T, Sharp PA. In: Cold Spring Harbor Monograph Series. Gesteland RF, Cech T, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1999. pp. 525–560. [Google Scholar]
- 2.Juneau K, Miranda M, Hillenmeyer ME, Nislow C, Davis RW. Genetics. 2006;174:511–518. doi: 10.1534/genetics.106.058560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherry JM, Ball C, Weng S, Juvik G, Schmidt R, Adler C, Dunn B, Dwight S, Riles L, Mortimer RK, et al. Nature. 1997;387:67–73. [PMC free article] [PubMed] [Google Scholar]
- 4.Engebrecht JA, Voelkel-Meiman K, Roeder GS. Cell. 1991;66:1257–1268. doi: 10.1016/0092-8674(91)90047-3. [DOI] [PubMed] [Google Scholar]
- 5.Li B, Vilardell J, Warner JR. Proc Natl Acad Sci USA. 1996;93:1596–1600. doi: 10.1073/pnas.93.4.1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakagawa T, Ogawa H. EMBO J. 1999;18:5714–5723. doi: 10.1093/emboj/18.20.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Preker PJ, Kim KS, Guthrie C. RNA. 2002;8:969–980. doi: 10.1017/s1355838202020046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong EL, Balakrishnan R, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hirschman JE, Livstone MS, Nash R, et al. Saccharomyces Genome Database. 2006 Oct 5; [Google Scholar]
- 9.Hirschman JE, Balakrishnan R, Christie KR, Costanzo MC, Dwight SS, Engel SR, Fisk DG, Hong EL, Livstone MS, Nash R, et al. Nucleic Acids Res. 2006;34:D442–D445. doi: 10.1093/nar/gkj117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ast G. Nat Rev Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 11.Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al. Science. 2005;308:1149–1154. doi: 10.1126/science.1108625. [DOI] [PubMed] [Google Scholar]
- 12.David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al. Science. 2003;302:842–846. doi: 10.1126/science.1088305. [DOI] [PubMed] [Google Scholar]
- 14.Chapman KB, Boeke JD. Cell. 1991;65:483–492. doi: 10.1016/0092-8674(91)90466-c. [DOI] [PubMed] [Google Scholar]
- 15.Clark TA, Sugnet CW, Ares M., Jr Science. 2002;296:907–910. doi: 10.1126/science.1069415. [DOI] [PubMed] [Google Scholar]
- 16.Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, Drenkow J, Piccolboni A, Bekiranov S, Helt G, et al. Genome Res. 2004;14:331–342. doi: 10.1101/gr.2094104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lopez PJ, Seraphin B. Nucleic Acids Res. 2000;28:85–86. doi: 10.1093/nar/28.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Planta RJ, Mager WH. Yeast. 1998;14:471–477. doi: 10.1002/(SICI)1097-0061(19980330)14:5<471::AID-YEA241>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 19.Spingola M, Grate L, Haussler D, Ares M., Jr RNA. 1999;5:221–234. doi: 10.1017/s1355838299981682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bon E, Casaregola S, Blandin G, Llorente B, Neuveglise C, Munsterkotter M, Guldener U, Mewes HW, Van Helden J, Dujon B, et al. Nucleic Acids Res. 2003;31:1121–1135. doi: 10.1093/nar/gkg213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis CA, Grate L, Spingola M, Ares M., Jr Nucleic Acids Res. 2000;28:1700–1706. doi: 10.1093/nar/28.8.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim JW, Kim HC, Kim GM, Yang JM, Boeke JD, Nam K. Nucleic Acids Res. 2000;28:3666–3673. doi: 10.1093/nar/28.18.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International Consortium H G S C. Nature. 2004;431:931–945. [Google Scholar]
- 24.Harigaya Y, Tanaka H, Yamanaka S, Tanaka K, Watanabe Y, Tsutsumi C, Chikashige Y, Hiraoka Y, Yamashita A, Yamamoto M. Nature. 2006;442:45–50. doi: 10.1038/nature04881. [DOI] [PubMed] [Google Scholar]
- 25.Hochwagen A, Amon A. Curr Biol. 2006;16:R217–R228. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 26.Leu JY, Roeder GS. Mol Cell Biol. 1999;19:7933–7943. doi: 10.1128/mcb.19.12.7933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Menees TM, Ross-MacDonald PB, Roeder GS. Mol Cell Biol. 1992;12:1340–1351. doi: 10.1128/mcb.12.3.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Malone RE, Pittman DL, Nau JJ. Mol Gen Genet. 1997;255:410–419. doi: 10.1007/s004380050513. [DOI] [PubMed] [Google Scholar]
- 29.Goguel V, Rosbash M. Cell. 1993;72:893–901. doi: 10.1016/0092-8674(93)90578-e. [DOI] [PubMed] [Google Scholar]
- 30.Warner JR, Mitra G, Schwindinger WF, Studeny M, Fried HM. Mol Cell Biol. 1985;5:1512–1521. doi: 10.1128/mcb.5.6.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guthrie C, Fink GR. Guide to Yeast Genetics and Molecular Biology. New York: Academic; 1991. [Google Scholar]
- 32.Shoemaker DD, Lashkari DA, Morris D, Mittmann M, Davis RW. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 33.Schmitt ME, Brown TA, Trumpower BL. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ewing B, Green P. Genome Res. 1998;8:186–194. [PubMed] [Google Scholar]
- 35.Ewing B, Hillier L, Wendl MC, Green P. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 36.Gordon D, Abajian C, Green P. Genome Res. 1998;8:195–202. doi: 10.1101/gr.8.3.195. [DOI] [PubMed] [Google Scholar]
- 37.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W. Genome Res. 1998;8:967–974. doi: 10.1101/gr.8.9.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Primig M, Williams RM, Winzeler EA, Tevzadze GG, Conway AR, Hwang SY, Davis RW, Esposito RE. Nat Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 40.Miura F, Kawaguchi N, Sese J, Toyoda A, Hattori M, Morishita S. Proc Natl Acad Sci USA. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.