Abstract
Systematic mapping of protein–protein interactions has become a central task of functional genomics. To map the human interactome, several strategies have recently been pursued. The generated interaction datasets are valuable resources for scientists in biology and medicine. However, comparison reveals limited overlap between different interaction networks. This divergence obstructs usability, as researchers have to interrogate numerous heterogeneous datasets to identify potential interaction partners for proteins of interest. To facilitate direct access through a single entry gate, we have started to integrate currently available human protein interaction data in an easily accessible online database. It is called UniHI (Unified Human Interactome) and is available at http://www.mdc-berlin.de/unihi. At present, it is based on 10 major interaction maps derived by computational and experimental methods. It includes more than 150 000 distinct interactions between more than 17 000 unique human proteins. UniHI provides researchers with a flexible integrated tool for finding and using comprehensive information about the human interactome.
INTRODUCTION
Protein-protein interactions (PPIs) are central to many if not all cellular processes. Their importance has provoked broad interest in their analysis, which in turn has led to the construction of various large-scale interaction maps. The first PPI datasets were generated for model organisms such as Saccharomyces cerevisiae, Drosophila melanogaster and Caenorhabditis elegans (1–5). Recently, the focus has shifted towards the systematic mapping of human PPIs. Both computationally and experimentally derived interaction datasets have been produced. They are mostly based on review of literature (6–8), extrapolation from interactions between orthologous proteins observed in other organisms (9–11) or application of high-throughput yeast two-hybrid (Y2H) assays (12,13).
Although these maps will certainly have profound impact on biological research, major limitations are lack of overlap, completeness and integration. Scientists are required to interrogate numerous databases if they seek comprehensive information on potential interaction partners for specific human proteins. This generally involves time-consuming searches as various query formats and identifiers have to be used in different interaction databases. Some datasets are even stored in simple flat files. To overcome these obstacles, we have constructed the UniHI database for the integration of large-scale human PPI maps. UniHI offers a search platform that combines and gives access to ten different large-scale human PPI datasets. It includes over 150 000 interactions between more than 17 000 proteins. UniHI is intended to reduce unnecessary duplication of data, while incorporating the strength of single databases regarding careful curation and annotation of PPIs.
HIGH DIVERGENCE OF HUMAN PPI DATASETS
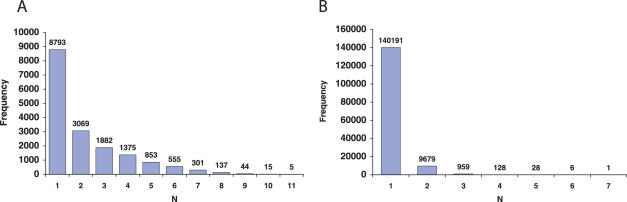
The construction of UniHI was motivated by the observation that human interaction maps tend to be highly divergent (14,15). This is also the case for the interaction maps integrated in UniHI (Table 1). We observed that <10% of all interactions occur in multiple maps, indicating a low degree of saturation (Figure 1B and Supplementary Data). The small number of shared interactions is remarkable considering the large number of proteins common to different datasets. More than 50% of all proteins are included in two or more maps (Figure 1A). Thus, current PPI datasets are highly complementary sharing few interactions between many common proteins.
Table 1.
PPI datasets currently integrated in UniHI
| Dataset | Proteins | Interactions | Method | References | Database location |
|---|---|---|---|---|---|
| MDC-Y2H | 1703 | 3186 | Y2H screen | (12) | www.mdc-berlin.de/neuroprot |
| CCSB-Y2H | 1549 | 2754 | Y2H screen | (13) | vidal.dfci.harvard.edu (flat file only) |
| CCSB-LIT | 2192 | 4067 | Text mining | (13) | vidal.dfci.harvard.edu (flat file only) |
| HPRD-BIN | 5908 | 15 508 | Literature | (8) | www.hprd.org |
| HPRD-COMP | 1277 | 4468 | Literature | (8) | www.hprd.org |
| DIP | 1033 | 1303 | Literature | (7) | dip.doe-mbi.ucla.edu |
| BIND | 4273 | 5863 | Literature | (6) | www.bind.ca |
| COCIT | 3737 | 6580 | Text mining | (10) | bioinformatics.icmb.utexas.edu/idserve |
| REACTOME | 679 | 12 639 | Literature | (16) | www.reactome.org |
| ORTHO | 6225 | 71 466 | Orthology | (11) | www.sanger.ac.uk/PostGenomics/signaltransduction/interactionmap |
| HOMOMINT | 4127 | 10 174 | Orthology | (17) | mint.bio.uniroma2.it |
| OPHID | 4785 | 24 991 | Orthology | (9) | ophid.utoronto.ca |
Number of proteins and interactions in each dataset as well as construction approach are given.
Figure 1.
Numbers of proteins (A) and interactions (B) common to multiple maps. The histograms display frequency of proteins and interactions that are included in N different maps. Comparisons were performed after mapping of proteins to their corresponding Entrez Gene IDs.
INTEGRATION OF PPI DATASETS
We have started to integrate available large-scale human PPI maps in UniHI. In its initial version, UniHI is based on the unification of the following interaction datasets recently generated: MDC-Y2H, CCSB, HPRD, DIP, BIND, COCIT, REACTOME, ORTHO, HOMOMINT and OPHID (Table 1). These maps have been derived from manually curated databases (6–8,16), computational approaches employing text-mining (13,17), predictions based on orthology, (9–11) and from large Y2H screenings (12,13). For details see Supplementary Data. Matching of protein identifiers, which is essential for standardization, was performed using information from Ensmart and HGNC (18,19). For the combined map, we could assign 150 992 interactions between 17 064 unique proteins.
For user friendliness, some modifications of the integrated datasets were carried out. First, we wanted to indicate whether interactions are binary or complex. Most of the included interactions are binary, while REACTOME comprises only complex interactions and HPRD comprises both binary and complex PPIs. To enable users to distinguish easily between the two types, we have split interaction data from HPRD into two sets (HPRD-BIN, HPRD-COMP).
Secondly, differentiation between PPIs identified with different strategies was facilitated as choice of mapping approach has considerable impact on the PPIs detected. Maps based on multiple approaches were divided according to the methods used. CCSB data were divided into Y2H- and literature-based interaction maps (CCSB-Y2H, CCSB-LIT). OPHID comprises orthology-based PPIs as well as interactions imported from other databases. We included only orthology derived PPIs.
DATABASE STRUCTURE AND IMPLEMENTATION
The structure of the UniHI database has been designed to integrate PPI data obtained from different sources. UniHI is implemented as relational database using an open source MySQL database management system. It consists of six key tables: Protein, ProteinAliases, ProteinDistribution, InteractionDistribution, InteractionProperties and InteractionScore. It links the proteins with information about their properties, their interactions and their distribution and in the different PPI datasets (Supplementary Figure S1). A full description of the UniHI database structure and its implementation can be found in the Supplementary Data.
DATA ACCESS
Our aim was to provide easy and intuitive, but nevertheless efficient and comprehensive access to the integrated data. UniHI is accessible via a web-server at http://www.mdc-berlin.de/unihi. A search interface based on Java programming language offers two different search options: In a single protein search, users input a single protein to query for its direct interaction partners. In a network-oriented multiple protein search users can supply a list of proteins. Proteins can be entered by their corresponding gene symbol, Entrez Gene ID, Uniprot ID, Unigene ID, OMIM ID, NCBI Geneinfo ID or Ensembl ID.
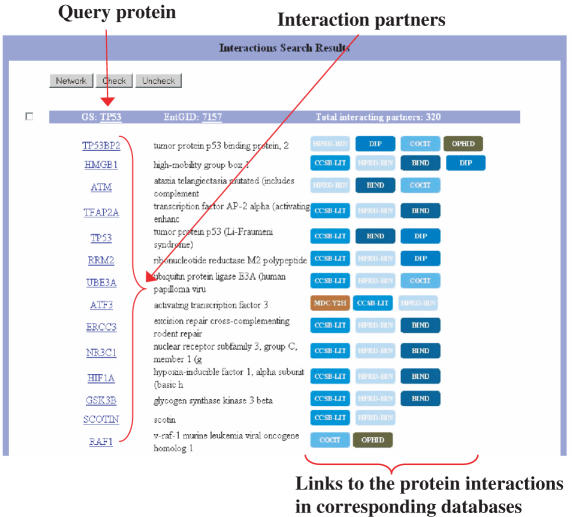
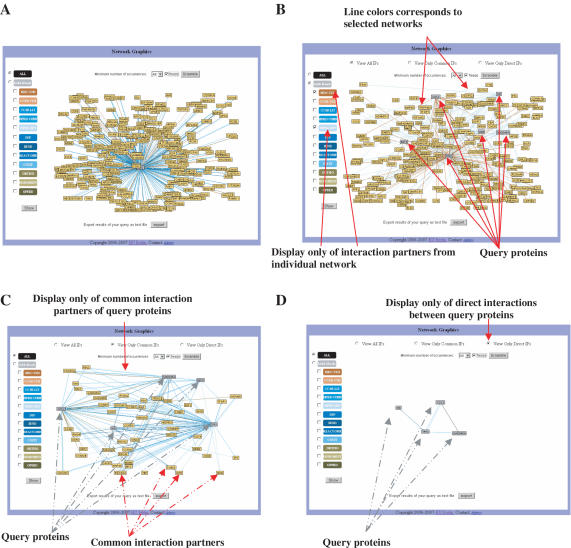
A visualization tool for interaction data with various features has been implemented. We utilized and extended a pre-existing Java applet for graphical presentation of interaction networks (20). Retrieved interactions can be displayed either in textual (Figure 2) or graphical form (Figure 3). For both types of views, interactions are directly hyperlinked to the maps from which they originate, with the exception of OPHID, due to technical reasons, and CCBS, which is only available as a text file. To facilitate the interpretation of results, characteristic sets of colors were used distinguishing maps as well as mapping approaches.
Figure 2.
Textual representation of a query result for protein interactions in UniHI. For each interaction partner found, a hyperlink is provided to the database from which the interaction originates. Multiple links indicate inclusion in multiple maps. For easy discrimination between maps, specific colors have been assigned. Shades of blue have been used for datasets derived by literature search, shades of green for orthology-based maps, shades of red for maps derived from Y2H screens.
Figure 3.
Graphical representation of PPIs. After retrieval, users of UniHI can visualize the interactions as graphs with interactions displayed as lines. (A) Output of the query for interaction partners of TP53. (B–D) Output for a query with multiple proteins (TP53, CDC2, E2F4, HD, A2M and GADD45A). Several features allow quick assessment of results. Line color indicates the map from which the interaction is derived. Multiple lines between proteins signify presence in several maps. Queried proteins are symbolized by gray rectangles, their interacting partners by yellow rectangles. To assist users in the evaluation of results, several tools are offered to restrict interaction sets displayed according to selection of maps, multiple occurrences, common neighbourhood of proteins (C), direct interactions between query proteins (D).
To permit users a highly targeted search, UniHI offers several tools to specify the displayed interactions: (i) Display only interactions from selected maps. This option can be used to exclude certain mapping approaches. (ii) Display only proteins that are common interaction partners to multiple proteins in the query. Such procedure can narrow down the context of a chosen set of proteins and can help to identify putative modifiers of physiological processes (12). (iii) Display only interactions that occur in multiple maps. This approach may be used to gain confidence in interactions retrieved (21). (iv) Display only direct interactions between query proteins. This option can be used for the identification of protein complexes.
SCOPE OF UniHI AND FUTURE DIRECTIONS
The aim of UniHI is to provide a unified set of protein interactions included in the major human PPI maps that are publicly available. As these are constantly extended, this demands ongoing integration of additional interaction data. UniHI has been designed with an open structure permitting future integration of further human interactome datasets. Links to already included maps will be updated every three months. Currently, Perl scripts with integrated SQL commands are used to preprocess and import interaction data after manual download from the corresponding web-pages. For future versions of UniHI, we aim to automate this process. Detailed information about the updating procedure can be found in the Supplementary Data.
To examine the constitution of UniHI, extensive statistical analysis was performed regarding network structure and functional annotation of integrated datasets. We also scrutinized the reliability of interaction maps using independent expression data and annotation (see Supplementary Data). Since the scope of UniHI can be expected to be continuously expanding, these analyses will be regularly repeated and presented on the UniHI webpage. This allows users a critical assessment of the single maps included in UniHI as well as of UniHI itself. To assess the quality of the interaction data, information on co-expression and co-annotation is presented for each interaction pair. We also list how protein interactions were validated in each dataset. Additionally, UniHI provides available links to the original PubMed articles that were used for curation in literature-based interactions maps.
CONCLUSIONS
Increasing numbers of human PPI datasets provide enormous amounts of valuable, but frequently unconnected information whose application in biology and medicine is still limited (22–24). Lack of integration and overlap need to be addressed more strongly with experimental and bioinformatical strategies.
UniHI constitutes a highly practical integrated platform that allows simultaneous querying of the major human protein-protein interaction maps. It does not replace already available interaction maps, but facilitates single portal access to the larger part of the human interactome analyzed so far. UniHI enables the assembly of comprehensive lists of protein interactions and flexible network-orientated searching. It allows identification of network structures which would not be detectable if single maps were analyzed separately. UniHI is a flexible tool for the systematic utilization of human interactome data in biomedical research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Acknowledgments
We would like to thank S. Schnögl for critical reading and suggestions and to acknowledge the support of the German BMBF (NGFN2, KB-P04T03, 01GR0471) and the Deutsche Forschungsgemeinschaft (DFG) by the SFB 618 grant. Funding to pay the Open Access publication charges for this article was provided by SFB 618.
Conflict of interest statement. None declared.
REFERENCES
- 1.Uetz P., Giot L., Cagney G., Mansfield T.A., Judson R.S., Knight J.R., Lockshon D., Narayan V., Srinivasan M., Pochart P., et al. A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–627. doi: 10.1038/35001009. [DOI] [PubMed] [Google Scholar]
- 2.Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gavin A.C., Bosche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 4.Giot L., Bader J.S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y.L., Ooi C.E., Godwin B., Vitols E., et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- 5.Li S., Armstrong C.M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P.O., Han J.D., Chesneau A., Hao T., et al. A map of the interactome network of the metazoan C. elegans. Science. 2004;303:540–543. doi: 10.1126/science.1091403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bader G.D., Donaldson I., Wolting C., Ouellette B.F., Pawson T., Hogue C.W. BIND—The Biomolecular Interaction Network Database. Nucleic Acids Res. 2001;29:242–245. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salwinski L., Miller C.S., Smith A.J., Pettit F.K., Bowie J.U., Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peri S., Navarro J.D., Amanchy R., Kristiansen T.Z., Jonnalagadda C.K., Surendranath V., Niranjan V., Muthusamy B., Gandhi T.K., Gronborg M., et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown K.R., Jurisica I. Online predicted human interaction database. Bioinformatics. 2005;21:2076–2082. doi: 10.1093/bioinformatics/bti273. [DOI] [PubMed] [Google Scholar]
- 10.Persico M., Ceol A., Gavrila C., Hoffmann R., Florio A., Cesareni G. HomoMINT: an inferred human network based on orthology mapping of protein interactions discovered in model organisms. BMC Bioinformatics. 2005;6:S21. doi: 10.1186/1471-2105-6-S4-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lehner B., Fraser A.G. A first-draft human protein–interaction map. Genome Biol. 2004;5:R63. doi: 10.1186/gb-2004-5-9-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F.H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., et al. A human protein–protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 13.Rual J.F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G.F., Gibbons F.D., Dreze M., Ayivi-Guedehoussou N., et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 14.Futschik M.E., Chaurasia G., Wanker E., Herzel H. Comparison of human protein–protein interaction maps. Lecture Notes Inform. 2006;P 83:21–32. [Google Scholar]
- 15.Chaurasia G., Herzel H., Wanker E., Futschik M.E. Systematic functional assessment of human protein–protein interaction maps. Genome Inform. 2006;17:36–45. [PubMed] [Google Scholar]
- 16.Joshi-Tope G., Gillespie M., Vastrik I., D'Eustachio P., Schmidt E., de Bono B., Jassal B., Gopinath G.R., Wu G.R., Matthews L., et al. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–D432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramani A.K., Bunescu R.C., Mooney R.J., Marcotte E.M. Consolidating the set of known human protein–protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol. 2005;6:R40. doi: 10.1186/gb-2005-6-5-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kasprzyk A., Keefe D., Smedley D., London D., Spooner W., Melsopp C., Hammond M., Rocca-Serra P., Cox T., Birney E. EnsMart: a generic system for fast and flexible access to biological data. Genome Res. 2004;14:160–169. doi: 10.1101/gr.1645104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eyre T.A., Ducluzeau F., Sneddon T.P., Povey S., Bruford E.A., Lush M.J. The HUGO Gene Nomenclature Database, 2006 updates. Nucleic Acids Res. 2006;34:D319–D321. doi: 10.1093/nar/gkj147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mrowka R. A Java applet for visualizing protein–protein interaction. Bioinformatics. 2001;17:669–671. doi: 10.1093/bioinformatics/17.7.669. [DOI] [PubMed] [Google Scholar]
- 21.von Mering C., Krause R., Snel B., Cornell M., Oliver S.G., Fields S., Bork P. Comparative assessment of large-scale data sets of protein–protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 22.Gunsalus K.C., Ge H., Schetter A.J., Goldberg D.S., Han J.D., Hao T., Berriz G.F., Bertin N., Huang J., Chuang L.S., et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;436:861–865. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- 23.Goehler H., Lalowski M., Stelzl U., Waelter S., Stroedicke M., Worm U., Droege A., Lindenberg K.S., Knoblich M., Haenig C., et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington's disease. Mol. Cell. 2004;15:853–865. doi: 10.1016/j.molcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 24.Lim J., Hao T., Shaw C., Patel A.J., Szabo G., Rual J.F., Fisk C.J., Li N., Smolyar A., Hill D.E., et al. A protein–protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]