Abstract
The variation resources within the University of California Santa Cruz Genome Browser include polymorphism data drawn from public collections and analyses of these data, along with their display in the context of other genomic annotations. Primary data from dbSNP is included for many organisms, with added information including genomic alleles and orthologous alleles for closely related organisms. Display filtering and coloring is available by variant type, functional class or other annotations. Annotation of potential errors is highlighted and a genomic alignment of the variant's flanking sequence is displayed. HapMap allele frequencies and linkage disequilibrium (LD) are available for each HapMap population, along with non-human primate alleles. The browsing and analysis tools, downloadable data files and links to documentation and other information can be found at http://genome.ucsc.edu/.
INTRODUCTION
The development of high throughput platforms to study genomic variation has provided new datasets that have deepened our understanding of genome structure and evolution. The single nucleotide polymorphism (SNP) discovery and genotyping platforms (http://www.affymetrix.com/index.affx; http://www.perlegen.com/; http://www.illumina.com/) allow the cost effective generation of deep, dense datasets of human variation. Although many such datasets are publicly available through individual web sites, the disconnected nature of these resources makes it difficult for many biologists to use. The variation resources section of the Genome Browser collects much of these data in a common format in a single location, provides additional variation annotations and allows examination of the data in a genomic context with additional types of information.
The primary resource for small scale variation data is dbSNP [http://www.ncbi.nlm.nih.gov/snp/; (1)] which is a repository that collects data from all public projects. These data are presented in the Genome Browser through a display that simplifies comparison with other genomic annotations. In addition, filtering on this track allows increased focus on specific subsets of the data, such as non-synonymous mutations or variants in conserved regions.
Several of the public efforts—HapMap, Affymetrix, Perlegen and SeattleSNPs—are complete enough to warrant additional focus at UC Santa Cruz. Additional analyses of these data can shed new light on genome evolution and help target functional regions, including the identification of disease susceptibility loci. Calculations of linkage disequilibrium (LD) and Tajima's D′ have been performed, and orthologous alleles have been identified for human biallelic SNPs. In addition, external groups have contributed other analyses, such as the recombination rates and hotspots and the structural variants. These are all available for integrated display, analysis and download.
RESULTS
Data types and data
dbSNP
We provide mappings of all variant data from dbSNP to the current assembly of our supported species, including human, chimpanzee, mouse, rat, dog and chicken. When a new assembly is released for a given species, dbSNP (1) maps all of the submitted variant data to the new coordinates and releases their build within a few months. Sometimes there are enough new submissions to warrant the development and release of a new build for an existing assembly. In either case, we then process the new dbSNP build through our pipeline for public release within weeks. In addition to displaying data extracted from dbSNP, the orthologous states from closely related species (e.g. chimpanzee and macaque for the human genome) are identified using liftOver (2). Also, comparison with the reference sequence shows the direction of insertion/deletion (indel) events (i.e. was there a deletion of bases in the reference sequence, or were new bases inserted?). The pipeline extracts the most relevant characteristics of the variant and checks for consistency. These include the variant class (SNP, indel, microsatellite, etc.), how it maps to the genomic sequence (single base, between two bases or a range of bases), the variant's function (splice site, non-synonymous, etc.), validation status, map weight (a quality measure for the alignment between the variant's flanking sequence and the genomic sequence) and the allele represented in the current genome assembly. The error checking includes several types of error checking and annotation: data format inconsistencies, missing data, disagreement between expected and observed allele lengths, poor quality alignments of flanking sequences, alignment of a variant to multiple genomic locations, inconsistency in functional classification and disagreement between the genomic alleles as identified by dbSNP and the reference assembly. Considering all errors and other annotations together, ∼5% of the dbSNP build for human warrants additional scrutiny.
Genotyping array SNPs
The growth of common genotyping platforms is making genome-wide association studies more accessible, but integration of the genotyping results with other genomic features may be challenging for some users. We have added tracks for the large, fixed SNP sets to assist in the visualization and analysis of these data. Currently, we support the Affymetrix GeneChip Human Mapping 500K Array Set and the Illumina HumanHap300 BeadChip genotyping platforms, which are available in the ‘SNP Arrays’ track in the Genome Browser. This is built in the flexible composite track format which simplifies the process of adding new platforms as they are released; we intend to support the Affymetrix ‘Million-SNP’ chip when it is released in early 2007.
Linkage disequilibrium
LD describes the association of alleles at separate loci in the genome, usually on the same chromosome. It is useful for understanding the associations between genetic variants throughout the genome, and can be helpful in selecting SNPs for genotyping. LD measures the difference between the observed allele frequency for a pair of alleles—one at each of two loci—as compared to the expected joint allele frequency, which is the product of the two individual allele frequencies. When LD is low, the two loci tend to be inherited independently because recombination decouples them. Regions of high LD reflect lack of recent recombination between the two loci within the population, causing the inheritance of their alleles to be linked.
Three pairwise measures of LD are commonly used. Between two alleles, r2 is the square of the correlation coefficient, D′ is the covariance normalized to 1, and the LOD score is the log odds score (3). These values are calculated separately for three HapMap populations—the Yoruba people in Ibadan, Nigeria (YRI), the European samples from the Centre d'Etude du Polymorphisme Humain (CEU), and the combined populations of Japanese from Tokyo and Han Chinese from Beijing (JPT + CHB) (4).
Calculation of LD from diploid data first requires breaking each diploid genotype into its underlying genotypes and assigning each haploid genotype to one of the two chromosomes in the individual, often called phasing. This process of defining the phase of the mutations determines which alleles are paired on the same chromosome. Inferring the phasing between heterozygous genotypes from two nearby loci in the same individual is complicated and addressed elsewhere (5). Given the diploid genotypes A/a and B/b at two loci for one individual, probabilistic methods are used to infer which of the four possible combinations—AB/ab, Ab/aB, aB/Ab and ab/AB—are most likely at these loci for that individual. All genotype data at all loci from the population is used together in this process. The result is that all diploid genotypes for each individual are then partitioned into two sets representing the individual's two inherited chromosomes. This phased data then forms the basis for the calculation of LD as described above. Although not currently displayed in the Browser, this phased data also forms the basis for partitioning of the SNPs into commonly co-inherited sets, or haplotype blocks.
A unique feature of this resource is its data compression, which is an internal engineering trick used to decrease the time of data retrieval and image rendering in the Genome Browser. First, LD values were precomputed and binned, allowing storage of an intensity index in a single byte rather than as a real number. Second, the data were stored in a single denormalized table for each population such that all LD values downstream of a given SNP were stored in a single record; this requires fewer table joins and disk hits, allowing tens of megabases of LD data to be drawn within a few seconds.
To build the current ‘HapMap LD’ track, Haploview (3) was used to infer phasing and calculate LD values for pairs of genotypes separated by 250 kb or less from HapMap release 20 (http://www.hapmap.org/genotypes/2006-01/non-redundant). This approach infers phase using a standard EM algorithm with a partition–ligation approach for blocks with >10 markers. The dataset is currently being updated to use the results from another phasing algorithm, PHASE (6,7), as it typically produces lower error rates in several measures used to evaluate phasing methods (5).
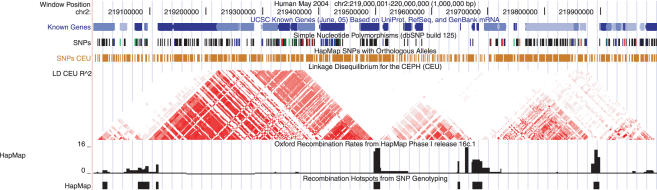
An example of the HapMap LD track display is available in Figure 1. As LD data describes the relationship of a pair of alleles, the display must also reflect this complexity. To understand this plot, consider a pair of SNPs at two points on a chromosome, then follow the lines up from those points until they meet. This point defines the top of a trapezoid, whose color represents the amount of LD observed between the two SNPs.
Figure 1.
SNPs, Linkage Disequilibrium (LD) and Recombination Display across a 1 Mb region of human chromosome 2 in the Genome Browser. Genes and their polymorphisms from dbSNP are shown in the first two tracks, where SNPs are colored by their function; red: non-synonymous or splice site, green: synonymous, blue: untranslated, black: intron. The HapMap SNPs track shows all of the polymorphic SNPs that were used to calculate LD, which is displayed in the HapMap LD track. Regions of high LD (red) tend to be separated by areas of high recombination rate and recombination hotspots.
Tajima's D′
Tajima's D′ (8) is one of many classic tests of the neutral model. It is a statistic used to compare two measures of nucleotide diversity under the assumptions that all polymorphisms are selectively neutral and the population has constant size. The neutral model expects correlation between the expected number of polymorphic sites (θs) and the average number of nucleotide differences (π). Tajima's D′ is the normalized difference between these two estimates (8):
The theoretical distribution of Tajima's D′ (95% confidence interval between −2 and +2) assumes that polymorphism ascertainment is independent of allele frequency. High values of Tajima's D′ suggest an excess of common variation in a region, which can be consistent with balancing selection or population contraction. Negative values of Tajima's D′, on the other hand, indicate an excess of rare variation, consistent with population growth or positive selection. Previous analyses have suggested that using the empiric distribution of Tajima's D′ from a collection of regions across the genome provides advantages in assessing whether selection or demography might explain an observed deviation from expectation (9,10).
It is important to note here that the observed allele frequency spectrum is dependent on the depth of coverage during SNP detection (ascertainment). If this coverage is uneven or if part of the frequency spectrum has been ignored, as is the case in the Perlegen dataset where a minor allele frequency cutoff was used, this can lead to ascertainment bias. Because of this bias toward common polymorphism in the Perlegen data set and the difficulty of modeling such bias, positive Tajima's D′ values are difficult to interpret. The ascertainment bias raises the mean of the distribution, so high values have reduced significance. However, extreme negative values in extended regions can be useful in qualitatively identifying interesting regions for full resequencing and more rigorous theoretical analysis of nucleotide diversity. As less biased datasets become available, this track will become more useful.
The University of Calfornia Santa Cruz (UCSC) Tajima's D′ track shows estimates from the three human populations in the Perlegen data set (11), which includes genotypes for 1 586 383 SNPs in 71 Americans of European, African and Asian ancestry. A separate track of SNPs that were used in this study is also available.
Recombination rates and recombination hotspots
A better understanding of the genomic landscape of human recombination rate variation would facilitate the efficient design and analysis of disease association studies and greatly improve inferences from polymorphism data about selection and human demographic history. Recombination rate estimates also provide a new route to understanding the molecular mechanisms underlying human recombination. Observations from sperm studies (12) and patterns of genetic variation (13,14) show that recombination rates in the human genome vary extensively over kilobase scales and that much recombination is observed in recombination hotspots, providing an explanation for the apparent block-like structure of LD (15,16).
Estimates of recombination rates and the locations of recombination hotspots were contributed by Gil McVean and Simon Myers (13) for the HapMap release 16a (17) and the Perlegen (11) datasets. They are available in the SNP Recombination tracks in the Genome Browser.
Structural variants
During the finishing of the human genome, it became obvious that there were major differences between individuals, causing problems in reconciling the different sequences for a given region. Some of these problems turned out to be caused by copy number polymorphism (CNP) and structural variation (SV) (18). These describe related types of variation, both of which are important in disease (19). Typically, CNPs refer to a single large region (>100 kb) being duplicated multiple times (20). SVs describe intermediate sized regions, larger than are included in dbSNP and smaller than CNPs, which complete the spectrum of observed indel sizes.
Array-based comparative genomic hybridization [array CGH; (21)] can identify variants between 20 and 400 kb by hybridizing genomic DNA to arrays containing BAC clones (22,23). Representational oligonucleotide microarray analysis (ROMA) follows a similar approach, but uses oligonucleotide arrays and has found variants that range in size from several 100 bp up to 1.6 Mb (20). Paired-end sequencing of fosmid clones detects variants in approximately the same range, or 700 bp to 2 Mb (24). Using the diploid HapMap data to look for failures of Mendelian inheritance has shown deletions up to 1 Mb (25,26). Hybridizations of haploid samples found deletions up to 8 kb (27). Further details about populations and individual genotypes are available in the ‘Structural Var’ track in the Genome Browser.
A common database for these types of results is still in the planning phase at NCBI. We provide a unified view of these data gathered in a single ‘composite’ track (28), which groups together many datasets of the same type, allowing common control of the display while reducing track clutter and simplifying access. This approach also makes it straightforward to add new datasets as they become available.
Orthologous alleles
In addition to variation within an organism, a key component of genome analysis is comparison with other related genomes. Fixed differences between species can be used either alone or in combination with other polymorphism data such as the HKA test (29) to find genomic regions that are under selection. For variant sites within a genome, the direction of the mutation can be determined by inferring the ancestral state and identifying the new (derived) allele. When deep genotyping information exists for a population and the derived allele can be inferred, the derived allele frequency (DAF) can be calculated. The theoretical basis for a shift in DAF between neutral and functional regions is presented in detail elsewhere (30), showing that the DAF spectrum is independent of population history.
We have used the blastz/chain/net/liftOver process (2) to identify the chimpanzee and macaque states for each of the biallelic SNPs in dbSNP (1), in HapMap release 21 (http://www.hapmap.org/genotypes/2006-07/rsstrand/nonredunant) and in the Seattle SNP datasets (http://pga.gs.washington.edu, http://egp.gs.washington.edu) (31,32). When one of the two human alleles is the same as the chimpanzee state, that allele is most likely to be ancestral, so the other allele represents a new mutation and is called the derived allele. This simple parsimony approach can lead to errors in up to 2% of the derived allele calls, due to mutation on the chimpanzee lineage that may still be polymorphic or could be fixed, so the macaque allele is also provided to be used for confirmation. As more advanced approaches to inferring the ancestral state are developed, we intend to provide these results as well. The orthologous alleles for the variant data are available in the ‘HapMap SNPs’ track and the fixed differences are available in the chimpSimpleDiff table in the Genome Browser Database.
Analysis of the derived alleles was used to address the question of whether sequences that have been constrained throughout mammalian evolution are currently under selection in the human population. The DAF spectrum shift for non-coding elements between constrained and non-constrained regions showed that selection on the non-coding constrained elements is currently active in the human population. Furthermore, it is approximately as strong as the pressure on protein coding regions, indicating that the non-coding constrained regions are likely to be functional (33). Thus, examining the DAF spectrum can identify candidate functional regions for further scrutiny during the follow-up of association studies and help us develop a complete understanding of locus-specific biology.
Self alignments and repeats
Duplications and repeats play important roles in both genomic disease and gene evolution, as replication slippage leads to copy number differences and retrotransposition-mediated duplications allow the birth of new gene variants. In addition, repetitive complexity can lead to misassignment and misassembly of genome sequence (34), which may confound interpretation within these regions. The Genome Browser provides results from several approaches to look for these types of features that are biologically interesting but can be a source of error for some analyses.
The ‘Self Chain’ track shows alignments of the human genome with itself, using a scoring system that allows longer gaps than traditional affine scoring systems. These can be used to identify genomic intervals that are not unique in the genome. Similarly, the ‘Segmental Dups’ are experimentally validated blocks of duplicated genomic DNA, typically ranging in size from 1 to 200 kb (35). They often contain sequence features such as high copy repeats and gene sequences with intron–exon structure. Arian Smit's RepeatMasker (http://www.repeatmasker.org) program (36,37) screens DNA sequences for interspersed repeats, low complexity DNA sequences and other repeat elements. These results are available in the ‘RepeatMasker’ track. Similarly, simple tandem repeats identified by the Tandem Repeat Finder (38) are available in the ‘Simple Repeats’ track. These repeat regions can lead to genotyping errors as cross-hybridization is more likely.
DATA DISPLAY AND AVAILABILITY
As described elsewhere, the Genome Browser (28,39–41) and the Table Browser (42) are useful tools for the display, analysis and retrieval of genome scale biological data. All of these data are available for display in the Genome Browser at http://genome.ucsc.edu, for interactive analysis through the Table Browser at http://genome.ucsc.edu/cgi-bin/hgTables, and via direct access to text files on our download site at http://hgdownload.cse.ucsc.edu/.
WEB SITE REFERENCES
http://genome.ucsc.edu; UCSC Genome Browser. http://genome.ucsc.edu/cgi-bin/hgTables; UCSC Table Browser. http://hgdownload.cse.ucsc.edu/; UCSC Database Downloads.
Acknowledgments
We acknowledge support from the National Human Genome Research Institute (NHGRI; for D. J. Thomas, B. L. Rhead, R. M. Kuhn, W. J. Kent and H. Trumbower), the Howard Hughes Medical Institute (HHMI; for D. Haussler), the National Cancer Institute (NCI; for H. Trumbower) and A. D. Kern's NIH Kirschstein-NRSA postdoctoral fellowship. We would like to thank the many collaborators who have contributed annotation data to our project, as well as our users for their feedback and support. We would also like to thank the dedicated system administrators who have provided an excellent computing environment: Jorge Garcia, Patrick Gavin, Chester Manuel, Victoria Lin and Paul Tatarsky. Funding to pay the Open Access publication charges for this article was provided by NHGRI.
Conflict of interest statement. D. J. Thomas, H. Trumbower, B. L. Rhead, R. M. Kuhn, D. Haussler, and W. J. Kent receive royalties from the sale of UCSC Genome Browser source code licenses to commercial entities.
REFERENCES
- 1.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kent W.J., Baertsch R., Hinrichs A., Miller W., Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett J.C., Fry B., Maller J., Daly M.J. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 4.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marchini J., Cutler D., Patterson N., Stephens M., Eskin E., Halperin E., Lin S., Qin Z., Munro H., Abecasis G., et al. A comparison of phasing algorithms for trios and unrelated individuals. Am. J. Hum. Genet. 2006;78:437–450. doi: 10.1086/500808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stephens M., Smith N.J., Donnelly P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephens M., Donnelly P. A comparison of Bayesian methods for haplotype reconstruction from population genotype data. Am. J. Hum. Genet. 2003;73:1162–1169. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Akey J.M., Eberle M.A., Rieder M.J., Carlson C.S., Shriver M.D., Nickerson D.A., Kruglyak L. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2004;2:e286. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stajich J.E., Hahn M.W. Disentangling the effects of demography and selection in human history. Mol. Biol. Evol. 2005;22:63–73. doi: 10.1093/molbev/msh252. [DOI] [PubMed] [Google Scholar]
- 11.Hinds D.A., Stuve L.L., Nilsen G.B., Halperin E., Eskin E., Ballinger D.G., Frazer K.A., Cox D.R. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–1079. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 12.Jeffreys A.J., Kauppi L., Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nature Genet. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- 13.McVean G.A., Myers S.R., Hunt S., Deloukas P., Bentley D.R., Donnelly P. The fine-scale structure of recombination rate variation in the human genome. Science. 2004;304:581–584. doi: 10.1126/science.1092500. [DOI] [PubMed] [Google Scholar]
- 14.Crawford D.C., Bhangale T., Li N., Hellenthal G., Rieder M.J., Nickerson D.A., Stephens M. Evidence for substantial fine-scale variation in recombination rates across the human genome. Nature Genet. 2004;36:700–706. doi: 10.1038/ng1376. [DOI] [PubMed] [Google Scholar]
- 15.Daly M.J., Rioux J.D., Schaffner S.F., Hudson T.J., Lander E.S. High-resolution haplotype structure in the human genome. Nature. 2001;29:229–232. doi: 10.1038/ng1001-229. [DOI] [PubMed] [Google Scholar]
- 16.Gabriel S.B., Schaffner S.F., Nguyen H., Moore J.M., Roy J., Blumenstiel B., Higgins J., DeFelice M., Lochner A., Faggart M., et al. The structure of haplotype blocks in the human genome. Science. 2002;296:2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- 17.Thorisson G.A., Smith A.V., Krishnan L., Stein L.D. The International HapMap Project Web site. Genome Res. 2005;15:1591–1593. doi: 10.1101/gr.4413105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feuk L., Carson A.R., Scherer S.W. Structural variation in the human genome. Nature Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 19.Botstein D., Risch N. Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nature Genet. 2003;33:228–237. doi: 10.1038/ng1090. [DOI] [PubMed] [Google Scholar]
- 20.Sebat J., Lakshmi B., Troge J., Alexander J., Young J., Lundin P., Månér S., Massa H., Walker M., Chi M., et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. doi: 10.1126/science.1098918. [DOI] [PubMed] [Google Scholar]
- 21.Albertson D.G., Pinkel D. Genomic microarrays in human genetic disease and cancer. Hum. Mol. Genet. 2003;12:R145–R152. doi: 10.1093/hmg/ddg261. [DOI] [PubMed] [Google Scholar]
- 22.Iafrate A.J., Feuk L., Rivera M.N., Listewnik M.L., Donahoe P.K., Qi Y., Scherer S.W., Lee C. Detection of large-scale variation in the human genome. Nature Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 23.Sharp A.J., Locke D.P., McGrath S.D., Cheng Z., Bailey J.A., Vallente R.U., Pertz L.M., Clark R.A., Schwartz S., Segraves R., et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 2005;77:78–88. doi: 10.1086/431652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tuzun E, Sharp A.J., Bailey J.A., Kaul R., Morrison V.A., Pertz L.M., Haugen E., Hayden H., Albertson D., Pinkel D., et al. Fine-scale structural variation of the human genome. Nature Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 25.McCarroll S., Hadnott T., Perry G., Sabeti P., Zody M., Barrett J., Dallaire S., Gabriel S., Lee C., Daly M., et al. Common deletion polymorphisms in the human genome. Nature Genet. 2006;38:86–92. doi: 10.1038/ng1696. [DOI] [PubMed] [Google Scholar]
- 26.Conrad D., Andrews T., Carter N., Hurles M., Pritchard J. A high-resolution survey of deletion polymorphism in the human genome. Nature Genet. 2006;38:75–81. doi: 10.1038/ng1697. [DOI] [PubMed] [Google Scholar]
- 27.Hinds D., Kloek A., Jen M., Chen X., Frazer K. Common deletions and SNPs are in linkage disequilibrium in the human genome. Nature Genet. 2006;38:82–85. doi: 10.1038/ng1695. [DOI] [PubMed] [Google Scholar]
- 28.Hinrichs A.S., Karolchik D., Baertsch R., Barber G.P., Bejerano G., Clawson H., Diekhans M., Furey T.S., Harte R.A., Hsu F., et al. The UCSC Genome Browser Database: Update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hudson R.R., Kreitman M., Aguade M.A. Test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fay J.C., Wyckoff G.J., Wu C.I. Positive and negative selection on the human genome. Genetics. 2001;158:1227–1234. doi: 10.1093/genetics/158.3.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.SeattleSNPs. NHLBI Program for Genomic Applications. Seattle, WA: SeattleSNPs; 2006. July, 2006. [Google Scholar]
- 32.NIEHS SNPs. NIEHS Environmental Genome Project. Seattle, WA: University of Washington; 2006. July, 2006. [Google Scholar]
- 33.Drake J.A., Bird C., Nemesh J., Thomas D.J., Newton-Cheh C., Reymond A., Excoffier L., Attar H., Antonarakis S.E., Dermitzakis E.T., et al. Conserved noncoding sequences are selectively constrained and not mutation cold spots. Nature Genet. 2006;38:223–227. doi: 10.1038/ng1710. [DOI] [PubMed] [Google Scholar]
- 34.Bailey J., Yavor A., Massa H., Trask B., Eichler E. Segmental duplications: organization and impact within the current human genome project assembly. Genome Res. 2001;11:1005–1017. doi: 10.1101/gr.187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.The International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature. 2003;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 36.Smit A.F.A., Green P. RepeatMasker. [Google Scholar]
- 37.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–420. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 38.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kent W.J., Sugnet C.W., Furey T.S., Roskin K.M., Pringle T.H., Zahler A.M., Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karolchik D., Kent W.J., Baertsch R., Furey T.S., Hinrichs A., Lu Y.T., Roskin K.M., Schwartz M., Sugnet C.W., Thomas D.J., et al. The UCSC Genome Browser Database. Nucleic Acids Res. 2003;31:51–54. doi: 10.1093/nar/gkg129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kuhn B., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., et al. The UCSC Genome Browser Database: Update 2007. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkl928. (in this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karolchik D., Hinrichs A.S., Furey T.S., Roskin K.M., Sugnet C.W., Haussler D., Kent W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]