Abstract
Background
Meningiomas are the most common intracranial neoplasias, representing a clinically and histopathologically heterogeneous group of tumors. The neurofibromatosis type 2 (NF2) tumor suppressor is the only gene known to be frequently involved in early development of meningiomas. The objective of this study was to identify genetic and/or epigenetic factors contributing to the development of these tumors. A large set of sporadic meningiomas were analyzed for presence of 22q macro-mutations using array-CGH in order to identify tumors carrying gene dosage aberrations not encompassing NF2. The NF2 locus was also comprehensively studied for point mutations within coding and conserved non-coding sequences. Furthermore, CpG methylation within the NF2 promoter region was thoroughly analyzed.
Results
Monosomy 22 was the predominant finding, detected in 47% of meningiomas. Thirteen percent of the tumors contained interstitial/terminal deletions and gains, present singly or in combinations. We defined at least two minimal overlapping regions outside the NF2 locus that are small enough (~550 kb and ~250 kb) to allow analysis of a limited number of candidate genes. Bialleinactivationo the NF2 gne was detected in 36% of meningiomas. Among the monosomy 22 cases, no additional NF2 mutations could be identified in 35% (17 out of 49) of tumors. Furthermore, the majority of tumors (9 out of 12) with interstitial/terminal deletions did not have any detectable NF2 mutations. Methylation within the NF2 promoter region was only identified at a single CpG site in one tumor sample.
Conclusion
We confirmed previous findings of pronounced differences in mutation frequency between different histopathological subtypes. There is a higher frequency of biallelic NF2 inactivation in fibroblastic (52%) compared to meningothelial (18%) tumors. The presence of macro-mutations on 22q also shows marked differences between fibroblastic (86%) and meningothelial (39%) subtypes. Thus, inactivation of NF2, often combined with the presence of macro-mutation on 22q, is likely not as important for the development of the meningothelial subtype, as opposed to the fibroblastic form. Analysis of 40 CpG sites distributed within 750 bp of the promoter region suggests that NF2 promoter methylation does not play a major role in meningioma development.
Background
Meningiomas are the most frequently occurring intracranial tumors. Clinically treated meningiomas comprise approximately 13–26% of all primary brain tumors [1]. Epidemiological studies indicate that ~90% of all meningiomas are asymptomatic [2]. The majority are benign, slowly growing, solitary and sporadic tumors, but atypical or malignant meningioma constitutes approximately 10% of all cases. Meningiomas are thought to be derived from the arachnoid cap cells of the leptomeninges, which is the covering of the brain and the spinal cord [1,3]. They usually occur in the cranial meninges, but approximately 10% of tumors develop in the meninges covering the spine. From histopathological and clinical points of view, meningiomas are a heterogeneous group of tumors, which are classified into 15 histopathological types with WHO malignancy grades I–III [4].
Meningiomas was the first solid tumor to be characterized as containing a specific genetic aberration, which is monosomy 22 [5]. Since then, loss of genetic material from chromosome 22 has been the most consistent aberration, observed in up to 70% of tumors [6,7]. Meningiomas are closely associated with the genetic syndrome neurofibromatosis type-2 (NF2). Approximately 50% of NF2 patients suffer from meningiomas, making them the second most frequent neoplasm associated with this tumor syndrome [8-10]. In 1993, the neurofibromatosis type 2 (NF2) tumor suppressor gene was characterized from chromosome 22 [11,12]. Sporadic meningiomas were therefore screened for mutations in the NF2 gene, which was found to be frequently inactivated. There is, however, a wide discrepancy in the reported mutation frequency (20–60%) [7,13-18]. In fact, it has been shown that meningiomas may be grouped into tumors with and without mutations in the NF2 gene. These groups can also be correlated with histopathological subtypes [15,17]. Tumors with mutations in the NF2 gene are often of the transitional or fibrous subtype, commonly with allelic losses on chromosome 22. Tumors without NF2 gene mutations are predominantly of the meningothelial subtype, and allelic losses on chromosome 22 are less commonly observed in this subtype. Furthermore, the meningothelial meningiomas are more often located in the anterior parts of the skull base [19]. Transcriptional silencing of the NF2 tumor suppressor by CpG methylation in meningiomas was recently tested by two research groups, but the results were conflicting. Van Tilborg et al. reported methylation in 1 out of 21 tumors, whereas Lomas and colleagues reported a higher methylation frequency (23 out of 88 cases) [20,21].
Despite almost 40 years of cytogenetic and molecular genetic research, the NF2 tumor suppressor is still the only specific gene which has been shown to be frequently involved early in the development of meningiomas. There is, however, a general consensus that other not yet characterized loci are important for the development and/or progression of these tumors [22-27]. Considering the clinical and histopathological heterogeneity of meningiomas, it is possible that alternative genetic mechanisms underlay its tumorigenesis. There are reports of the loss of chromosome 22 as the primary consistent aberration in meningiomas, in the absence of mutations in the NF2 gene. Moreover, young patients with constitutional ring chromosomes and multifocal meningiomas have been characterized. This highlights the possibility of an alternative genetic etiology related to other genes located on chromosome 22 [23,28-30]. In addition, other sites in the genome have been implicated in meningioma development or progression [23]. Among these, aberrations of chromosome 1 have been established as the second major genetic factor for the initiation/progression of meningiomas and correlate with increased morbidity [31,32]. Furthermore, the NF2 gene has also been excluded by linkage analysis in one family with multiple meningothelial meningiomas and ependymomas [33]. These various reports suggest that there exist additional loci contributing to the formation of meningiomas.
The rationale of this study was to perform a multilevel analysis of mutations that include gene dosage aberrations on chromosome 22, analysis of point mutations in the NF2 locus, and profiling of the methylation status within the NF2 gene promoter. We also intended to relate our findings from chromosome 22 to the high resolution DNA copy number variation in meningiomas of chromosome 1, which was recently reported [34].
Results and discussion
We analyzed a comprehensive series of sporadic meningiomas on multiple levels, i.e. for presence of "macro-mutations" involving large scale aberrations of 22q using an array-based approach and for "micro-mutations" within the NF2 tumor suppressor gene. The latter involved analysis of point mutations within the mRNA encoding part of this gene and within the conserved non-genic (CNGs) sequences, which we have previously defined [35]. We also comprehensively studied the promoter region of the NF2 gene for CpG methylation.
Chromosome 22 DNA copy number profiling using array-CGH
We studied 126 sporadic meningioma samples, composing a representative series of tumors operated on at a single hospital in Sweden. All tumor samples studied were analyzed previously using a lower resolution approach [7,22,23,31,36]. The main rationale in this part of the project was to identify tumors possessing gene dosage aberrations of 22q which did not involve the NF2 locus. We carried out DNA copy number profiling using array-CGH on a genomic microarray covering chromosome 22. This array, containing 480 measurement points across the sequenced part of this autosome, was developed and validated previously [37]. Among the samples used for validation of this array was meningioma T24 DNA, which is a well characterized tumor containing a large terminal deletion of 22q and an additional approximately 150 kb biallelic deletion involving EWSR1, GAR22, RRP22, AP1B1, RFPL1 and NEFH genes [37,38]. On average, each array-CGH experiment generated ~400 positively scored data points from 22q.
Table 1 summarizes results obtained from DNA copy number profiling of chromosome 22 and other results, as well as relevant clinical information pertaining to the studied patients. The predominant profile detected in 59 tumors was monosomy 22, which accounted for 47% of meningiomas, and this result is consistent with previous studies [7,22,23,31,36]. The biological consequence of this very common aberration for the affected tumor cells is one copy loss of all genes from 22q. The second category comprised of16 tumors (13%) that were affected by terminal deletions, interstitial deletions and gains, present singly or in combinations (Figure 1A). This number is also consistent with previous analysis using a loss of heterozygosity (LOH) approach [7,22,23,31,36]. However, due to a much higher density of data points used in the current analysis, we were able to detect additional aberrations as well as exactly define the breakpoints of all rearrangements in samples which were characterized previously using LOH. Figure 1A shows all 16 cases in detail, and Figs. 2 and 3 display array-CGH profiles of 7 tumors providing new information. Tumor M64 (Figs. 1A and 2A) disclosed a previously unknown biallelic deletion affecting the NF2 gene. This was the only locus affected by a homozygous deletion in the studied series of tumors, which confirms the importance of the NF2 gene in meningioma development. This fibroblastic meningioma has an overall 22q profile consistent with a very large (> 30 Mb) terminal deletion of the long arm of chromosome 22. The second deletion affects only one clone (ID 151) [GenBank: AC005529] and specifically targets the NF2 gene, thus inactivating its second allele in tumor cells. Another interesting example is fibroblastic meningioma M11, since it displays a terminal and multiple interstitial deletions, all of which are located outside the NF2 locus (Figs. 1A and 2E). Three of these deletions, located towards the telomere of 22q, were previously characterized [39]. However, current analysis revealed an additional, smaller (~550 kb) deletion, encompassing 5 contiguous clones (ID 85–89) [GenBank: U07000–AP000348], located on the centromeric side of the NF2 locus (Figures 1A, 1B and 2E). This tumor is intriguing since no additional genetic/epigenetic aberration in the NF2 gene could be detected in this sample (see below). The vast majority of copy number aberrations of 22q in meningiomas point to inactivation or a decreased gene dosage. However, in two tumors (M109 and M96, Figs 1A, 2C and 3A) we were able to detect a low copy number gain. For instance, tumor M109 displays a combination of low copy number gain in the centromeric part of 22q and a large terminal deletion (Figure 2A). A minimum overlapping region of gain is approximately 3.8 Mb (clone ID 11–48) [GenBank: AC006285 – AC007957] and is located in 22q11. The importance of these gains for meningioma tumorigenesis is difficult to judge, as they are not frequent events. One possible explanation is that these gains reflect cell-specific chromosomal instability of 22q11, since this region is rich in intra-chromosomal segmental duplications and is prone to rearrangements leading to several genetic disorders.
Table 1.
Genetic alterations in one hundred meningiomas.
| Sample ID | Subtype1 | CH3 | NF2 gene mutations | DNA copy number profiles4 | ||||
| Position | Mutation2 | Assumed effect3 | Chromosome 22 | Chromosome 1 | Loss of non-22 genes5 | |||
| M26 | A | Ex 2 | 227_240del | Lys76fs | Monosomy | Monosomy | D1, S1A, S1B | |
| M71 | A | Ex8 | 810+2T>C | Abn. Spl. | Monosomy | 1p, 120.8 Mb TD | D1, P2A, S1A, S1B | |
| M38 | A | Ex6 | 573del; 575_581del | Trp191fs | Monosomy | 1p 107.9 Mb TD | P1B, P2A, S1A, S1B | |
| M43 | A | Ex12 | 1185del | Lys396fs | Monosomy | 1p 120.3 Mb TD, 4.4 Mb A | ||
| M46 | A | Ex8 | 810+2T>C | Abn. Spl. | NAD | NAD | ||
| M33 | A | NAD | 30.6 Mb TD [AC009516-AC002055] | 1p, 90.2 Mb TD, 45.8 Mb ID | ||||
| M9 | A | NAD | NAD | NAD | ||||
| M20 | A | NAD | NAD | NAD | ||||
| M13 | F | Ex 2 | 240+2T>C | Abn. Spl. | Monosomy | 1p, 142.9 Mb TD | P2A, S1A, S1B | |
| M88 | F | Ex4 | 364-1G>A | Abn. Spl. | Monosomy | 1p 84.1 Mb TD; 1q 4.6, 7.1 Mb IDs, 12.5 Mb TD | P2A | |
| M3 | F | Ex 2 | 240+2T>C | Abn. Spl. | Monosomy | 1p 119.8 Mb TD | ||
| M14 | F | Ex3 | 249del | His84fs | Monosomy | 1p 120.2 Mb TD | ||
| M15 | F | Ex8 | 683del | Lys228fs | Monosomy | 1p 116.0 Mb TD | ||
| M39 | F | Ex8 | 810+1G>A | Abn. Spl. | Monosomy | NS | ||
| M42 | F | Ex10 | 995del | Lys332fs | Monosomy | NS | ||
| M70 | F | Ex6 | 599+1G>A | Abn. Spl. | Monosomy | NS | ||
| M91 | F | Ex 2 | 154del | Arg52fs | Monosomy | NS | ||
| M102 | F | Ex9 | 865A>T | Lys289X | Monosomy | NS | ||
| M115 | F | Ex7 | 634C>T | Gln212X | NAD | 1p 119.6 Mb TD | ||
| M646 | F | NAD | 30.4 Mb TD [AC008079-AC002055], NF2 biallelic del [AC005529] | 1p 98.8 Mb ID; 1q 24.2 Mb ID | P1B, P2A | |||
| M66 | F | NAD | Monosomy | 1p 118.1 Mb TD | P2A | |||
| M117 | F | NAD | 0.55, 2.8, 4.5 Mb IDs [U07000-AP00034, AC005004-Z69907, AL022311-Z83851]; 5.9 Mb TD [Z93244-AC002055] | 1p 116.8 Mb TD, 3.6 Mb A | ||||
| M32 | F | NAD | 5.3 Mb ID [AL022329-AL022336], 15.2 Mb TD [AL009049-AC002055] | NS | D1 | |||
| M4 | F | NAD | Monosomy | NAD | ||||
| M29 | F | NAD | Monosomy | NS | ||||
| M41 | F | NAD | Monosomy | NS | ||||
| M92 | F | NAD | Monosomy | NS | ||||
| M114 | F | NAD | 18.0 Mb ID [AP000525-Z68324] | NS | ||||
| M119 | F | NAD | NAD | NS | ||||
| M10 | M | Ex13 | 1341-2A>G | Abn. Spl. | Monosomy | 1p 120.3 Mb TD | P2A | |
| M23 | M | Ex3 | 241-2A>C | Abn. Spl. | Monosomy | 1p 120.9 Mb TD | P2A | |
| M67 | M | Ex4 | 432C>A | Tyr144X | Monosomy | 1p 120.8 Mb TD | P2A | |
| M98 | M | Ex14 | 1519_1520del | Phe507fs | Monosomy | NAD | D1, P2A, S1A, S1B | |
| M57 | M | Ex 2 | 133_144 del | Asp45fs | Monosomy | NS | ||
| M72 | M | Ex7 | 634C>T | Gln212X | Monosomy | NS | ||
| M40 | M | NAD | Monosomy | 1p 164.9 Mb TD | P1B, P2A | |||
| M76 | M | NAD | Monosomy | 1p 142.7 Mb TD | P2A | |||
| M105 | M | NAD | Monosomy | 1p 116.3 Mb TD | P1B, P2A | |||
| M348 | M | NAD | 20.0 Mb TD [AC004542-AC002055] | 1p 102.8 Mb ID | ||||
| M44 | M | NAD | Monosomy | 1p 85.4 Mb TD | ||||
| M101 | M | NAD | 25.1 Mb ID [AP000525-AL035450], 7.9 Mb TD [AL049758-AC002055] | Monosomy, 1p 2.1 Mb R | ||||
| M30 | M | NAD | Monosomy | NAD | D1, P2A | |||
| M112 | M | NAD | NAD | 1p 114.2 Mb TD | ||||
| M2 | M | NAD | NAD | NAD | ||||
| M5 | M | NAD | NAD | NAD | ||||
| M77 | M | NAD | NAD | NAD | ||||
| M82 | M | NAD | NAD | NAD | ||||
| M45 | M | NAD | NAD | NS | ||||
| M51 | M | NAD | NAD | NS | ||||
| M54 | M | NAD | NAD | NS | ||||
| M61 | M | NAD | NAD | NS | ||||
| M63 | M | NAD | NAD | NS | ||||
| M65 | M | NAD | NAD | NS | ||||
| M69 | M | NAD | NAD | NS | ||||
| M86 | M | NAD | NAD | NS | ||||
| M95 | M | NAD | NAD | NS | ||||
| M99 | M | NAD | NAD | NS | ||||
| M107 | M | NAD | NAD | NS | ||||
| M108 | M | NAD | NAD | NS | ||||
| M111 | M | NAD | NAD | NS | ||||
| M116 | M | NAD | NAD | NS | ||||
| M120 | M | NAD | NAD | NS | ||||
| M93 | P | Ex8 | 810G>T | Glu270Asp | Monosomy | 1p 114.3 Mb ID | P2A | |
| M229 | P | Ex13, inter1 | 1390del; 28309611 C>A | Ala464fs, unknown | Monosomy | NS | ||
| M62 | P | NAD | Monosomy | NS | ||||
| M21 | P | NAD | NAD | NS | ||||
| M60 | T | Ex14 | 1519_1528del | Phe507fs | Monosomy | 1p 118.2 Mb TD | P2A | |
| M1 | T | Ex7 | 650del | Tyr217X | Monosomy | 1p 65.0 Mb TD | ||
| M31 | T | Ex13 | 1325del | Ala451fs | Monosomy | 1p 53.2, 36.3 Mb IDs | ||
| M58 | T | Ex 2 | 115-3G>C | Abn. Spl. | 30.1 Mb TD [AC007050-AC002055] | 1p 120.2 Mb TD | ||
| M90 | T | Ex3 | 241-2A>G | Abn. Spl. | 20.1Mb ID [AP000525-AL079295], 13.4 Mb TD [AL049850-AC002055] | 1p 83.3 Mb TD | ||
| M96 | T | Ex12 | 1175G>T | Glu392X | 2.0 Mb ID [AP000525-AC007666], 5.4 Mb A [AC006285-AC000102], 27.5 Mb TD [U07000-AC002055] | 1p 43.3 Mb TD, 1q 69.9 Mb TD | ||
| M52 | T | Ex4 | 431_432insAA | Tyr144X | Monosomy | NS | P1B | |
| M113 | T | Ex4 | 377del | Leu127X | Monosomy | NAD | D1, P1B, P2A, S1A, S1B | |
| M18 | T | Ex12 | 1198C>T | Gln400X | Monosomy | NS | ||
| M37 | T | Ex8 | 745_755del | Arg249fs | Monosomy | NS | ||
| M48 | T | Ex 1 | 36_37del | Leu14fs | Monosomy | NS | ||
| M75 | T | Ex8 | 750del | Thr251fs | Monosomy | NS | ||
| M87 | T | Ex4 | 436G>A | Val146Ile | NAD | NS | ||
| M56 | T | NAD | Monosomy | 1p 116.5 Mb TD | P2A | |||
| M85 | T | NAD | Monosomy | 1p 108.2 Mb TD | P2A | |||
| M109 | T | NAD | 5.7 Mb A [AC006548-AC007957], 30.6 Mb TD [AC009516-AC002055] | 1p 120.2 Mb TD | ||||
| M27 | T | NAD | 15.5Mb ID [AP000525-AC005005], 14.1 Mb TD [AL049749-AC002055] | NS | ||||
| M59 | T | NAD | Monosomy | NS | ||||
| M68 | T | NAD | Monosomy | NS | ||||
| M94 | T | NAD | Monosomy | NS | ||||
| M97 | T | NAD | Monosomy | NS | ||||
| M25 | T | NAD | NAD | 1p 98.7 Mb TD | ||||
| M117 | T | NAD | NAD | 1p 36.1 Mb TD | ||||
| M10410 | T | + | NAD | NAD | NS | |||
| M8 | T | NAD | NAD | NS | ||||
| M12 | T | NAD | NAD | NS | ||||
| M17 | T | NAD | NAD | NS | ||||
| M35 | T | NAD | NAD | NS | ||||
| M55 | T | NAD | NAD | NS | ||||
| M89 | T | NAD | NAD | NS | ||||
| M103 | T | NAD | NAD | NS | ||||
| M78 | U | Ex10 | 999+1G>T | Abn. Spl. | Monosomy | 1p 3.8 Mb TD | ||
| M79 | U | NAD | NAD | 1p 39.1Mb ID, 1q 41.3Mb TD | ||||
| M83 | U | NAD | NAD | NS | ||||
1 Tumors are grouped according to histopathological subtypes. Abbreviations: A, Anaplastic; F, Fibroblastic; M, Meningothelial; P, Psammomatous; T, Transitional and U, Unclassified.
2 Position of the mutations according to numbering of the cDNA sequence, nucleotide number one is the A in the ATG translation initiation codon [GenBank:AF369658]. Abbreviations: substitution, >; deletion, del; insertion, ins; No Aberration Detected, NAD. Donor splice site mutations indicated by the last nucleotide of the preceding exon, a plus sign and position in the intron (e.g. 240+2T>C). Acceptor splice site mutations denoted by the first nucleotide of the following exon, a minus sign and position in the intron (e.g. 241-2A>C).
3 Amino acid number one is the translation initiator Methionine. Abbreviations: translation termination codon, X; frame shift, fs; abnormal splicing, Abn. Spl.
4 Data from copy number imbalances on chromosome 1 were previously published [34]. Abbreviations: Terminal Deletion, TD; Interstitial Deletion, ID; Amplification/gain, A; Not Studied, NS; No Aberration Detected, NAD; Retained segment R. Genbank accession numbers in squared brackets.
5 Loss of non-chromosome 22 clones spanning the DAL1, D1; PTCH2A, P2A; PTCH1B, P1B; SUFU1A, S1A; and SUFU1B, S1B genes.
6 M64 displays a homozygous deletion of the NF2 gene.
7 M11 demonstrates multiple deletions not affecting the NF2 gene.
8 M34 displays a terminal deletion not affecting the NF2 locus.
9 M22 presents two mutations in the NF2 locus, in exon 13 and in the CNG inter1. The position of the inter1 mutation is according to numbering on chromosome 22 (UCSC Homo sapiens Genome Browser, assembly July 2003). Ten bases on each side of the mutation: agcaaagagaCaaattaccca.
10 Methylation of a single CpG site in the intron 1 of the NF2 gene in ~40% of DNA molecules.
Figure 1.
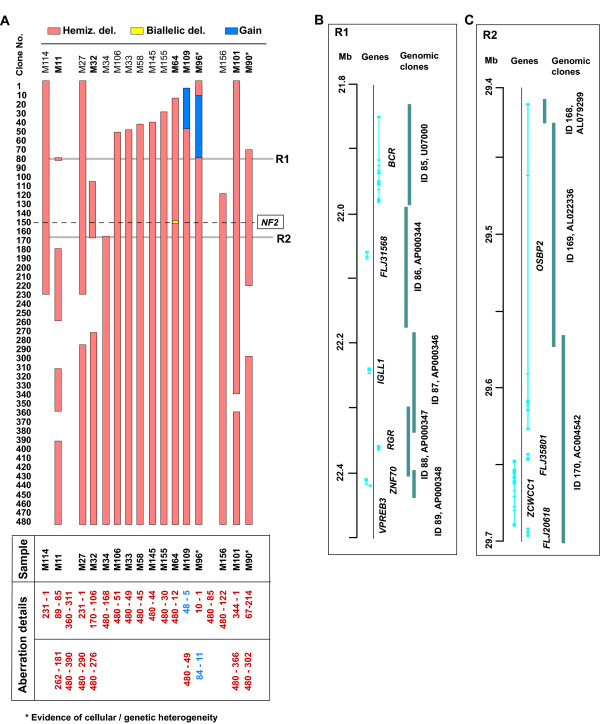
Summary of array-CGH profiles for chromosome 22 insporadic meningioma. A) Detailed summary of all complex aberrations. Deletions and gains are mapped according to the clones present on the chromosome 22 minimal tiling path array. Chromosome 22 clones are grouped in bins of 10 along the y-axis. The panel of tumors which displayed complex aberrations of chromosome 22 is shown from left to right. The red, yellow and blue boxes denote hemizygous deletion, biallelic deletion and gain, respectively. The horizontal dashed line displays the approximate position of the NF2 gene in relation to the measurement points on the chromosome 22 minimal tiling path array. The table below details the clone ID's at which the deletion/gain starts and finishes. Smallest regions of overlapping deletions are highlighted with grey lines (R1 and R2) and genes present within these regions are shown to the left. Tumor names highlighted in bold are displayed in more detail in other figures. B) Detailed view of candidate region 1 (R1) with gene and clone information presented according to the positions on chromosome 22 sequence (UCSC Homo sapiens Genome Browser, assembly July 2003). C) Detailed view of candidate region 2 (R2) with gene and clone information presented according to the positions on chromosome 22 sequence (UCSC Homo sapiens Genome Browser, assembly July 2003).
Figure 2.
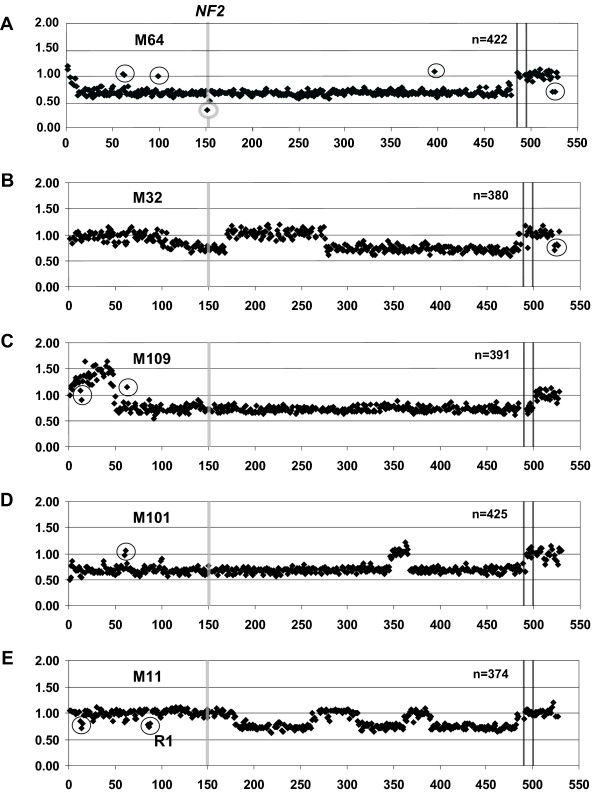
Array-CGH profiles of chromosome 22 in tumors displaying complex aberrations. The chromosome 22 array-CGH plots consist of 480 chromosome 22 clones ordered from centromere to telomere and 9 chromosome X controls between the vertical lines, as well as 31 non-chromosome 22 controls plotted on the right hand side. The y-axis displays the raw fluorescent intensity ratios for each clone on the genomic array. The position of the NF2 gene is highlighted with the vertical grey bar. A) Array-CGH profile of case M64 displaying a single copy loss of the majority of the chromosome with a biallelic deletion encompassing a single measurement point (clone ID 151) [GenBank:AC005529], specifically targeting the NF2 gene (highlighted by a grey circle). Encircled clones centromeric of NF2 display fluorescent ratio consistent with two copies; clone IDs 61–63 [GenBank:D87000–D87003] (previously reported Copy Number Polymorphism, CNP) and clone IDs 99–100 [GenBank:AL049759–AL050312]. Clone ID 400 [GenBank:Z84478] positioned towards the telomere, also exhibit two gene copies (highlighted by a circle). Control clone derived from chromosome 10 encompassing SUFU1B gene presents with one copy loss (highlighted by a circle). B) Array-CGH profile of case M32 displaying an interstitial deletion encompassing the NF2 gene and a terminal deletion. Clones spanning the DAL-1 gene on chromosome 18 and clones spanning the PTCH1B locus on chromosome 9 display a fluorescent ratio consistent with a one copy loss are highlighted by a circle. C) Case M109 displaying a gain spanning clone IDs 5–48 [Genbank:AC006548–AC007957] followed by a large terminal deletion covering clones 49–480 [Genbank:AC009516–AC002055]. Two CNPs are positioned at clone IDs 13–15 and 63 [Genbank: AC008079–AC008132 and D87003] and are indicated by circles. D) Meningioma M101 displaying a large one copy loss for the majority of the chromosome apart from a region encompassing clone IDs 348–366 [GenBank:AL080242–Z82176] and a CNP spanning clone IDs 60–61 [GenBank:D88269–D87000]. E) Detection of multiple interstitial deletions outside the NF2 region, spanning clone IDs 85–89 (R1, highlighted by a circle), 181–262 and 311–360 [GenBank: U07000–AP000348, AC005004–Z69907 and AL022311–Z83851] and a terminal deletion covering clone IDs 390–480 [GenBank:Z93244–AC002055] in case M11. A CNP spanning clone IDs 14–15 [GenBank:AC008132–AC008103] is highlighted by a circle.
Figure 3.
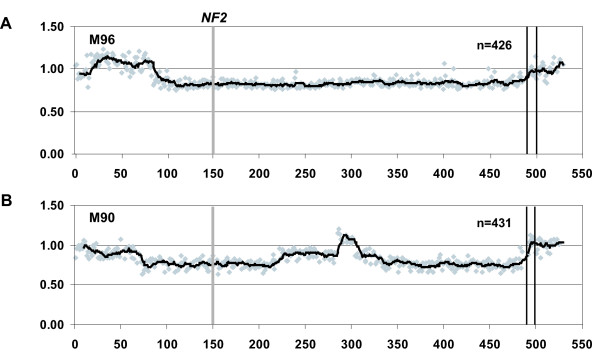
Detection of cellular or genetic heterogeneity in meningioma samples. The array-CGH plots follow the same general scheme as for Figure 3. However, the data points are shaded in grey and a trend line (with a moving average of 10 data points) is shown, which highlights the difference in fluorescent ratio levels along the chromosome. A) Identification of genetic heterogeneity in meningioma sample M96 which displays 3 different fluorescent ratio values across chromosome 22. The clone IDs and Average Normalized Inter Locus Fluorescence Ratio (ANILFR) for each of these are 1–15, 0.95; 16–84, 1.1 and 85–480, 0.83, respectively. B) Array-CGH profile of a complex set of aberrations on chromosome 22, which suggest genetic/cellular heterogeneity within the tumor M90. The regional shifts in fluorescent ratio across chromosome 22 are as summarized with the clone IDs and their ANILFR value; 1–23, 0.95; 24–66, 0.90; 67–214, 0.76; 215–284, 0.88; 285–303, 1.09; 304–326, 0.87; 327–480, 0.76. The non-chromosome 22 autosomal controls have an ANILFR value of 1.00, since this value was used to normalize all values across the genomic array.
All samples displaying aberrations other than monosomy 22 are graphically summarized in Fig 1A. This analysis enables the discrimination of several minimal overlapping deleted regions which should be studied further. Two of these regions, named as R1 (~550 kb) and R2 (~250 kb) are small enough to allow for detailed analysis of a limited number of candidate genes (Figs. 1B and 1C). The breakpoint cluster region (BCR) gene is one interesting candidate located in R1. This gene is located at the site of the chromosome 22 breakpoint generating the Philadelphia chromosome (9;22) translocation that creates the BCR/ABL fusion gene found in chronic myeloid leukemia and acute myeloid leukemia. The normal BCR gene was reported to have a serine/threonine kinase activity [40]. Another candidate gene located within R1 is the RGR oncogene. This gene was originally identified as a fusion protein which can promote tumor development in nude mice, but has also displayed oncogenic properties when not in the fusion form [41]. The RGR gene has frequently been found to be altered in human T-cell malignancies [42]. The zinc finger protein encoding gene ZCWCC1, located within R2, may also be a candidate for further investigation. The relative weakness of the R1 and R2 regions is that they are defined by rare cases with small deletions. There is therefore a possibility that the latter chromosome 22 aberrations are neutral "passenger mutations", with little consequence to tumorigenesis. Yet another apparent, large candidate region is located towards the telomere of 22q, starting from clone ID 390. This segment is affected by hemizygous deletion in all but one (M114) of the studied tumors which disclosed gene copy number imbalances of chromosome 22.
The third major category of tumors (51 cases, 40%) was composed of samples that did not display detectable changes in gene dosage on 22q (Table 1, "No Aberration Detected"; NAD) despite the high average resolution (~75 kb per data point). There may be several possible explanations for these results. One possibility is that additional genes on 22q (other than NF2 tumor suppressor) are important for meningioma development and these are mutated by alternative mechanisms, such as point mutation or epigenetic silencing. Another explanation might be that this subset of samples involves an alternative tumorigenic pthway which does not involve chromosome 22-located genes.
The current version of the chromosome 22 array has been supplemented with additional non- 22q-derived genomic clones from other chromosomes, which encompass genes (e.g. PTCH2A, PTCH1B, SUFU and DAL1 genes) of potential interest in the fields of meningiomas and brain tumors in general. The complete list of these additional clones is provided in additional file 1. Table 1 shows the samples in which loss of DNA copy number was detected for some of these loci. The highest rate of loss detected was at the PTCH2A locus on 1p34.1, which displayed a rate of loss of 18%. The other loci, namely DAL-1, PTCH1B, SUFU1A and SUFU1B, displayed a level of DNA copy number loss of 6%, 5%, 6% and 6%, respectively. We also detected DNA copy number losses in a number of non-chromosome 22-derived control clones. The most frequently deleted control clones were those derived from chromosomes 1 and 3, suggesting the likely importance of other regions of the genome in meningioma development/progression.
Mutation analysis of NF2 exons and evolutionarily conserved non-genic sequences (CNGs)
Inactivating micro-mutations in the NF2 tumor suppressor gene are found in meningiomas of all histopathological subtypes and malignancy grades and are thought to be an early event in tumorigenesis [43]. However, large differences (20–60%) of mutation frequency in sporadic meningiomas were previously reported [7,13-18]. Our rationale was to reassess the mutation frequency of NF2, especially when combined with very high resolution array-CGH analysis of these tumors. The rationale was also to test the hypothesis that mutations might be located in evolutionarily well conserved non-genic sequences within introns or in the 5' promoter region of the NF2 gene, in addition to mutations in the exons. Comparisons of the orthologous genomic sequence from the NF2 locus derived from five species (human, baboon, mouse, rat and pufferfish) were previously performed in order to identify CNGs that might be of functional importance for the NF2 gene [35]. The exonic sequence of the NF2 gene constitutes ~2.7% of this locus. When applying a threshold of 70% sequence identity over >100 bp of gap-free alignment in comparisons between human and mouse, the resulting level of extra-exonic conservation was 3.8%. We selected CNGs for mutational analysis that displayed a similar level of evolutionarily conservation as the protein coding sequence. In total, 17.5 kb was sequenced in 100 tumors, which included all 17 exons, the highly conserved exon flanking sequences and seven CNGs not contiguous with exons (Figure 4A). Four of the CNGs were positioned upstream of exon 1 and the remaining three CNGs were located in introns 1, 4 and 10.
Figure 4.
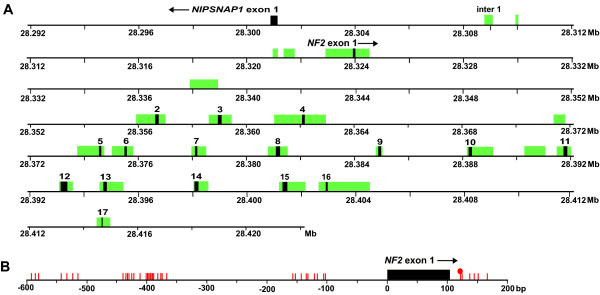
Overview of the NF2 locus analyzed for point mutations and methylation of CpG dinucleotides. A) Illustration of the NF2 gene displaying exons (black boxes) and conserved non-genic sequences (CNGs) (green boxes) that were analyzed for point mutations. Gene orientation is displayed with an arrow next to the gene name. The scale is according to sequence position on chromosome 22 in million base pairs (Mb) (UCSC Homo sapiens Genome Browser, assembly July 2003). B) Schematic view of the CpG sites in the 5' part of the NF2 gene that were analyzed for methylation. The 40 sequenced CpG sites were located between positions -591 and +166 bp (position +1 refers to transcriptional start site). A single methylated CpG site was detected in tumor M104 at position +120 in ~40% of DNA molecules (displayed as a red filled circle).
Thirty-nine alterations were identified in 100 analyzed samples (Tables 1, 2 and 3). Fifteen mutations caused frame-shifts, twelve altered the acceptor- or donor-splice sites, nine were nonsense, two were missense and one mutation occurred in the inter1 CNG. None of the identified mutations were present in the corresponding peripheral blood DNA. Thirty-five of these mutations were accompanied by a macro-mutation on the other allele, either through monosomy or terminal/interstitial deletions. Overall, we could detect mutations consistent with biallelic inactivation of the NF2 tumor suppressor gene in 36% of meningiomas (Table 3). However, in three cases (M46, M115 and M87) no second inactivating mutation could be identified. Considering what is known to date about the NF2 gene mutational spectrum in different tumors, it can be assumed that at least two of the latter three mutations (M46 – aberrant splicing and M115 – nonsense mutation) are pathogenic. These types of mutations are, however, usually combined with the second inactivating hit on the remaining normal allele of the gene [13,44-47]. The third aberration is a missense mutation (tumor M87, exon 4, 436 G>A, Val-146-Ile, Table 1), which is possibly introducing a dysfunction to the N-terminal FERM domain (residues 1–302 of the protein, plasma membrane binding domain) of the protein. Merlin/schwannomin provides a link between the cytoskeleton and plasma membrane and interacts with the actin cytoskeleton through binding sites in its FERM domain [48,49]. Thus, there exists a possibility of this missense mutation exerting a sufficiently strong dominant negative effect on the protein function, which does not necessitate the presence of the second hit NF2 gene inactivating event in this tumor. The three tumors discussed above (M46, M115 and M87) allow us to estimate our false negative detection rate of mutations in the NF2 gene. We can speculate that, in at least two and a maximum of three of the studied tumors, we could expect the second hit NF2 gene mutation to be present, but we were unable to detect it. Assuming the above reasoning to be correct, our false negative rate of micro-mutation detection is between 5.1 – 7.6%.
Table 2.
Chromosome 22 DNA copy number status and NF2 gene mutations in a subset of 100 sporadic meningiomas.
| NF2 gene | |||
| Chr. 22 status | Point mutation | No point mutation | Total |
| Monosomy | 32 | 17 | 49 |
| NAD1 | 3 | 36 | 39 |
| Interstitial/terminal deletion | 3 | 9 | 12 |
| Total | 38 | 62 | 100 |
1 No Aberration Detected, NAD
Table 3.
NF2 gene mutation frequencies in meningiomas of different histological subtypes.
| Histological subtype | Total no. | NF2 gene mutation frequency1 | Chr. 22 deletions affecting the NF2 gene | Biallelic inactivation of the NF2 gene |
| Meningothelial | 33 | 6/33 (18%) | 13/33 (39%) | 6/33 (18%) |
| Transitional | 31 | 13/31 (42%) | 20/31(65%) | 12/31(39%) |
| Fibroblastic | 21 | 11/21 (52%) | 18/21 (86%)4 | 11/21 (52%)5 |
| Anaplastic | 8 | 5/8 (61%)2 | 5/8 (61%) | 4/8(50%) |
| Psammomatous | 4 | 2/4 (50%)3 | 3/4 (75%) | 2/4 (50%) |
| Unclassified | 3 | 1/3 (33%) | 1/3 (33%) | 1/3 (33%) |
| Overall | 100 | 39 | 60 | 36 |
1 The methylation status of NF2 gene promoter in tumor M104 is not included.
2 The two micro deletions (1 bp and 7 bp) detected in M169 within exon 12 are considered as one.
3 Two point mutations were detected in M22, only one is used for calculations.
4 Fifteen of the 18 fibroblastic meningiomas that display deletions on chromosome 22 are monosomy cases. The three remaining tumors (M64, M32, and M114) all disclose a minimum of 18 Mb deletion of 22q.
5 Case M64 (biallelic deletion affecting the NF2 gene) is not included in this calculation
In case M22 we identified two tumor-specific micro-mutations, one mutation causing a frame shift in exon 13 introducing a premature stop codon and the second being a mutation in the inter1 CNG element. Inter1 is situated approximately 15 kb upstream of the NF2 gene (Figure 4A). It displays sequence conservation between human and pufferfish, which are evolutionarily very distant species [35], suggesting that inter1 may possess functional properties. Details on the exact position of the inter1 mutation on chromosome the 22 sequence are shown in Table 1. The tumor-specific mutation in inter1 CNG is one of three changes present in M22, as this tumor also contains monosomy 22. Thus, we may speculate that three hits are affecting the NF2 gene. An alternative explanation is that one of these mutations has little consequence to the normal function of the NF2 gene.
Methylation profiling of the NF2 promoter region by bisulfite modification followed by pyrosequencing
Methylation of promoter associated CpG-islands is known as a possible mechanism for inactivation of tumor suppressor genes in a variety of human cancers. It has also been suggested as a mechanism of repressing transcription of the NF2 gene in schwannomas [50] and in meningiomas [21]. Methylation of other cancer related genes (GSTP1, THBS1, CDKN2A, CDH1, BRCA1, p14ARF and RB1) has also been reported to be associated with malignant progression of meningiomas [20,51,52]. We used an established method of bisulfite treatment of tumor DNA followed by Pyrosequencing (Biotage, Uppsala, Sweden) which allows detection and semi-quantitative estimation of the degree of methylation at the selected CpGs [53,54]. Two overlapping PCR products of 339 bp and 550 bp covered ~750 bp of the upstream promoter region, the first exon and approximately 70 bp of intron 1, immediately downstream of the first exon (Figure 4B). These two products were Pyrosequenced with 8 sequencing primers. In total, 40 CpG sites were sequenced, between positions -591 and +166 bp (position +1 refers to transcriptional start site). This analysis allowed interrogation of up to 11 CpG sites per assay, distributed over 79 bp and with a maximum of 60 dispensations of nucleotides (additional file 2). Methylation was identified in only one tumor sample (M104), at a single CpG site at position +120, in ~40% of DNA molecules in this tumor (Figure 4B). Kino et al. reported a 70 bp region (-591 to -522) essential for NF2 expression in schwannomas [50]. Three CpG sites within this region (at positions -591, -586 and -581) appeared to be of particular importance for silencing of the NF2 gene upon methylation in schwannomas. We paid particular attention to these CpG sites but no methylation was detected. In summary, upon comprehensive analysis of 40 CpG sites, we conclude that methylation does not play a major role in the silencing of the NF2 gene in meningiomas. These results are in accordance with a report by van Tilborg et al [20] although contradicting the results from Lomas et al. [21].
Summary of macro- and micro-mutations on chromosomes 22 and 1 in meningiomas
We observed pronounced differences of the NF2 gene mutation frequency between different histopathological subtypes of meningiomas (Table 3). There was a clear trend towards an increased frequency of the biallelic NF2 gene inactivation in meningothelial, transitional and fibroblastic subtypes which were 6 out of 33 tumors (18%), 12 out of 31 tumors (39%) and 11 out of 21 tumors (52%), respectively. Based on these results, we may speculate that the biallelic inactivation of the NF2 gene is not as important for the development of the meningothelial subtype of meningioma, when compared to the transitional and fibroblastic tumors. A related trend was even more pronounced, when the frequencies of large chromosome 22 deletions were correlated to the three above mentioned histopathological subtypes of meningioma. The presence of large hemizygous deletions showed marked differences between the meningothelial, transitional and fibroblastic subtypes, comprising 13 out of 33 cases (39%), 20 out of 31 cases (65%) and 19 out of 21 cases (90%), respectively. It should be stressed here that 15 out of 19 fibroblastic meningiomas, which displayed gene dosage imbalances on 22q, were monosomy 22 cases. The three remaining tumors (M64, M32, and M114) all showed a minimum loss of 18 Mb of 22q. It may thus be hypothesized that one copy loss of genes located within the 18 Mb on 22q is important for generation of the fibroblastic phenotype of cells.
More than half (61%) of the meningothelial cases were among the category of tumors with both chromosome 22 copies retained. One possible explanation for this finding is that meningothelial tumors more frequently harbor mutations in additional genes that are located outside chromosome 22. Our results correlate well with previous studies by Wellenreuther et al. [15] as well as with Evans and colleagues [17] that identified NF2 gene mutations in 4/26 (15%) and 1/20 (5%) of meningothelial tumors. The above results from DNA level analysis are supported by analysis of merlin/schwannomin protein expression, which show normal expression in the greater part of meningothelial tumors, but severely reduced expression levels in the majority of fibrous and transitional meningiomas [55-57]. Furthermore, correlation between tumor localization and histological subtype was previously reported. The majority of meningiomas with no chromosome 22 aberrations were localized in the anterior skull base and were predominantly meningothelial [19]. Taken together, these results might suggest that cellular origin plays a role in the genetic pathways which are operative during tumor formation of meningiomas. In the context of our results from chromosome 22, it is interesting to correlate this to the data obtained from chromosome 1, as genetic aberrations affecting these two autosomes are the most frequent genetic abnormalities in meningiomas [31,36]. Furthermore, aberrations of chromosome 1 are also related to recurrence-free survival from the disease [31]. We have recently completed a high resolution analysis of chromosome 1 using the same tumor series [34] and 49 of the samples were studied with similar resolution on both chromosomes (Table 1). There is a strong relationship between changes on both chromosomes, with 67% of samples showing concurrent aberrations. These results clearly indicate the need for a genome-wide and high-resolution array-based characterization of chromosomal aberrations in meningiomas.
Conclusion
One objective of this study was to identify the maximum number of tumors displaying copy number aberrations of 22q not encompassing the NF2 locus. From the category of tumors displaying terminal- and interstitial-deletions we identified at least two minimal overlapping candidate regions outside the NF2 locus. These regions allow analysis of a limited number of candidate genes. Interesting candidates in these regions include the BCR gene, implicated in chronic myeloid leukemia and acute myeloid leukemia; the RGR oncogene involved in RAS-signaling [58] and found to be altered in human T-cell malignancies [42]; and the zinc finger protein encoding gene ZCWCC1. One additional candidate region starts at clone ID 390 and extends toward the telomere (Fig 1A). This segment is hemizygously deleted in all but one (M114) of the studied tumors with 22q gene copy number imbalances.
We assessed the point mutation frequency of NF2 in combination with array-CGH of 22q. The rationale was to test the hypothesis that mutations also may be located in evolutionarily well conserved non-genic sequences within introns or in the 5' promoter region, in addition to mutations in the exons. We identified 39 point mutations in 100 tumors. In one single case (M22), there were three tumor-specific mutations detected; a point mutation in the CNG inter1, a nonsense mutation in exon 13 and monosomy 22. Inter1 displays sequence conservation between human and pufferfish, suggesting that it may possess functional properties. However, the fact that the inter1 mutation represented one out of three genetic alterations in this tumor, and that we only identified a single mutation in one CNG, suggests that this is not a major mechanism for inactivation of the NF2 gene. We also analyzed the methylation status of NF2 promoter associated CpGs in order to examine if this mechanism is associated with repressing NF2 transcription in sporadic meningiomas. We identified methylation in only one tumor sample (M104), at a single CpG site (Figure 4B). Thus, we conclude that methylation is unlikely to play a major role in silencing of the NF2 gene in meningioma.
Profound differences in mutation frequency were observed between various histopathological subtypes of tumors. There was a considerably higher frequency of biallelic NF2 inactivation in fibroblastic (52%) compared to meningothelial (18%) tumors. The presence of 22q copy number imbalances were also more common in the fibroblastic (86%) compared to the meningothelial (39%) subtype. Thus, inactivation of NF2 combined with the presence of macro-mutation on 22q, might not be as important for the development of the meningothelial subtype, as opposed to the fibroblastic form.
Chromosome 1 genetic abnormalities are frequently observed in meningiomas [31,36]. Forty-nine of the samples studied here were also previously analyzed for gene dosage aberrations on chromosome 1 by array-CGH [34]. We observed a strong correlation between changes on both chromosomes, with 67% of samples showing concurrent aberrations. A genome-wide and high-resolution copy number analysis could contribute to identify additional genetic alterations involved in tumor development, especially for the meningothelial subtype, which does not commonly display NF2 and 22q alterations.
Methods
Patient samples
Tumor tissue and matched samples of peripheral blood were collected and DNA was isolated using standard methods [59]. Samples were obtained from the Dept. of Neurosurgery, Karolinska Hospital, Stockholm, Sweden. All analyzed tumor samples were reported previously in various studies [7,22,23,31,34,36]. Ethical approval was granted from the Research Ethics Committee, Faculty of Medicine, Uppsala University. Copy number profiling of chromosome 22 was performed on 126 sporadic meningiomas and this sample set comprised 39 meningothelial, 38 transitional, 28 fibroblastic, 9 anaplastic, 6 psammomatous, 1 angiomatous and 5 tumors of unclassified histopathological subtype. For the analysis for point mutations and methylation in the NF2 gene promoter, we used a subset of 100 tumor DNA samples (Table 1 and additional file 3).
DNA copy number profiling using the chromosome 22 genomic microarray
The genomic microarray used to assess DNA copy number status of chromosome 22 has been described previously [37]. In brief, 480 genomic clones derived from the minimal tiling path of chromosome 22, 8 control clones from chromosome X and 30 non-chromosome 22 autosomal control clones were isolated and grown in bacterial cultures overnight. The clone DNA (cosmid, fosmid, BAC and PAC) was purified using Qiagen kits and quantified using the picoGreen assay (Molecular Probes, Inc., Eugene, OR). All clone DNA were verified using STS PCR. The prepared clone DNA stock was amplified by DOP-PCR [60]. A second round of PCR on the DOP-PCR products using an amino linked universal primer was performed and amplification products were printed in quadruplicate on CodeLink™ slides (GE Healthcare, UK). Protocols used for the hybridization and washing procedures have been previously described [37,61]. Image capture and analysis was performed using the axon 4000B Scanner and GenePix 6 image analysis software. For all clones on the array the average, standard deviation and coefficient of variation (CV) for each of the 4 replicas was calculated. Clones displaying a CV above 5% between a minimum of 3 replica spots were discarded from further analysis. Array-CGH profiles are plotted in Microsoft Excel with the clone IDs on the X-axis and the normalized fluorescent ratio intensities on the y-axis. The average fluorescent ratio value for a given set of continuous clones on the array is referred to as the Average Normalized Inter Locus Fluorescence Ratio or ANILFR value.
PCR and sequencing
Exons and conserved non-genic sequences (CNGs) in the NF2 locus were PCR amplified from genomic DNA derived from one hundred sporadic meningiomas, with customized primers using Ampli Taq Gold (Applied Biosystems) (primer list in Additional file 4). Peripheral blood DNA from phenotypically normal control persons was also PCR amplified and sequenced for CNGs in which mutations were observed. PCR fragments were purified by Exosapit (GE Healthcare, UK) digestion prior to sequencing. Sequencing of amplicons was performed with customized nested primers using ET Terminator (GE Healthcare, UK) and Big Dye v.3.1 (Applied Biosystems) sequencing chemistries, and ethanol precipitated according to standard protocol [59]. Electrophoresis and fluorescent detection was carried out on Megabace 1000 (GE Healthcare, UK) and ABI 3700 (Applied Biosystems) capillary sequencing machines. Processing and analysis of sequencing data was performed using the Staden package [62].
Methylation profiling by pyrosequencing of bisulfite treated DNA
Pyrosequencing (Biotage, Uppsala, Sweden) was used as method for detection of methylation of cytosines situated 5'of a guanine [53]. Briefly, the DNA was bisulfite treated using EZ-96 DNA methylation kit (Zymo research, Orange, CA, USA) for conversion of unmethylated cytosines into uracils. In the subsequent PCR uracil amplifies as thymidine. Methylated cytosines remain unchanged after treatment. Cytosines not followed by a guanine are not methylated and will therefore be converted to a thymidine after bisulfite treatment and PCR. These thymidines were used as internal controls for the completeness of bisulfite treatment. Human genomic DNA was methylated in vitro by the CpG methylase M. Sss I and used as positive controls for detection of methylation. Bisulfite treated DNA were amplified in a 50 μl reaction using Platinum Taq DNA polymerase (Invitrogen Life Technologies, Carlsbad, CA, USA) according to manufacturers protocol. Primers for amplification and sequencing were custom designed using PSQ assay design software (Biotage, Uppsala, Sweden). Data of primers used for amplification and sequencing, as well as the nucleotide sequence analyzed and the dispensation order of nucleotides during sequencing is shown in Additional file 2. One PCR primer in each pair was biotinylated for conversion of the PCR product to a single stranded template. The cycling condition used were: 94°C for 3 minutes, followed by 20 cycles of 94°C for 20 seconds, 65°C minus 0.5°C/cycle for 20 seconds, 72°C for 1 minute, followed by 40 cycles of 94°C for 20 seconds, 55°C for 20 seconds, 72°C for 1 minute and finally 72°C for 10 minutes. 45 μl of PCR product was used for the Pyrosequencing reaction. Sequencing was performed on a PSQ 96MA system with PSQ Gold reagent kit (Biotage, Uppsala, Sweden) according to manufacturer's protocol. Pyro Q-CpG software (Biotage, Uppsala, Sweden) was used for data analysis.
Authors' contributions
CMH: participated in the overall study design; performed mutation analysis experiments, methylation analysis experiments, array-CGH experiments; analyzed results and drafted the manuscript. PB: participated in the array-CGH study design, performed array-CGH experiments, data analysis and drafted the manuscript. GG: participated in the study design of the mutation screening part and analysis of sequencing data. AP: participated in the methylation analysis study design. ARH: performed mutation screening and analysis of sequencing data. KM: performed printing of the micro-array. CJ: participated in the preparations of the microarray. TM: provided with tumor and blood samples, provided clinical data of all patients. JPD: intellectual input in the overall study design, drafted the manuscript and generated financial support. All authors read and approved the final manuscript.
Supplementary Material
Table summarizing DNA copy number changes in the additional subset of 26 sporadic meningiomas not studied for NF2 gene point mutations or NF2 promoter methylation status.
Table summarizing primers used for PCR amplification and sequencing of the NF2 gene.
Table summarizing primer pairs used for amplification of bisulfite treated DNA, sequencing primers, analyzed sequences and dispensation order.
List of non-22q-derived genomic clones that encompass genes of potential interest in tumor formation/progression.
Acknowledgments
Acknowledgements
This work was supported by grants from the U.S. Army Medical Research and Materiel Command (award no. W81XWH-04-1-0269), the Wallenberg Consortium North, the Swedish Cancer Foundation, the Swedish Research Council and Uppsala University to JPD. Chris Partridge is greatly acknowledged for proofreading the manuscript.
Contributor Information
Caisa M Hansson, Email: Caisa.Hansson@gmail.com.
Patrick G Buckley, Email: Patrick.Buckley@gmail.com.
Giedre Grigelioniene, Email: Giedre.Grigelioniene@kbh.ki.se.
Arkadiusz Piotrowski, Email: apiotrowski@genetics.uab.edu.
Anders R Hellström, Email: hellstroem@gmail.com.
Kiran Mantripragada, Email: kiran.mantripragada@gmail.com.
Caroline Jarbo, Email: Caroline.Jarbo@amic.se.
Tiit Mathiesen, Email: mathiesen@karolinska.se.
Jan P Dumanski, Email: jdumanski@genetics.uab.edu.
References
- Russel DS, Rubinstein LJ. Pathology of the tumors of the nervous system. Fifth edition. London , Edward Arnold; 1989. [Google Scholar]
- Rausing A, Ybo W, Stenflo J. Intracranial meningioma--a population study of ten years. Acta Neurol Scand. 1970;46:102–110. doi: 10.1111/j.1600-0404.1970.tb05608.x. [DOI] [PubMed] [Google Scholar]
- Kepes JJ. Meningiomas; biology, pathology and differential diagnosis. New York , Masson Publishing, Inc. USA; 1982. [Google Scholar]
- Kleihues P, Cavenee WK. Pathology and Genetics. Tumors of the Nervous System. Lyon , IARC Press; 2000. [Google Scholar]
- Zang KD, Singer H. Chromosomal consitution of meningiomas. Nature. 1967;216:84–85. doi: 10.1038/216084a0. [DOI] [PubMed] [Google Scholar]
- Seizinger BR, de la Monte S, Atkins L, Gusella JF, Martuza RL. Molecular genetic approach to human meningioma: loss of genes on chromosome 22. Proc Natl Acad Sci U S A. 1987;84:5419–5423. doi: 10.1073/pnas.84.15.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruttledge MH, Sarrazin J, Rangaratnam S, Phelan CM, Twist E, Merel P, Delattre O, Thomas G, Nordenskjold M, Collins VP, Dumanski JP, Rouleau GA. Evidence for the complete inactivation of the NF2 gene in the majority of sporadic meningiomas. Nat Genet. 1994;6:180–184. doi: 10.1038/ng0294-180. [DOI] [PubMed] [Google Scholar]
- Evans DG, Huson SM, Donnai D, Neary W, Blair V, Teare D, Newton V, Strachan T, Ramsden R, Harris R. A genetic study of type 2 neurofibromatosis in the United Kingdom. I. Prevalence, mutation rate, fitness, and confirmation of maternal transmission effect on severity. J Med Genet. 1992;29:841–846. doi: 10.1136/jmg.29.12.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans DG, Huson SM, Donnai D, Neary W, Blair V, Newton V, Harris R. A clinical study of type 2 neurofibromatosis. Q J Med. 1992;84:603–618. [PubMed] [Google Scholar]
- Parry DM, Eldridge R, Kaiser-Kupfer MI, Bouzas EA, Pikus A, Patronas N. Neurofibromatosis 2 (NF2): clinical characteristics of 63 affected individuals and clinical evidence for heterogeneity. Am J Med Genet. 1994;52:450–461. doi: 10.1002/ajmg.1320520411. [DOI] [PubMed] [Google Scholar]
- Rouleau GA, Merel P, Lutchman M, Sanson M, Zucman J, Marineau C, Hoang-Xuan K, Demczuk S, Desmaze C, Plougastel B, Pulst SM, Lenoir G, Bijlsma E, Fashold R, Dumanski J, Jong P, Parry D, Eldrige R, Aurias A, Delattre O, Thomas G. Alteration in a new gene encoding a putative membrane-organizing protein causes neuro-fibromatosis type 2. Nature. 1993;363:515–521. doi: 10.1038/363515a0. [DOI] [PubMed] [Google Scholar]
- Trofatter JA, MacCollin MM, Rutter JL, Murrell JR, Duyao MP, Parry DM, Eldridge R, Kley N, Menon AG, Pulaski K, Haase VH, Ambrose CM, Munroe D, Bove C, Haines JL, Martuza RL, MacDonald ME, Seizinger BR, Short MP, Buckler AJ, Gusella JF. A novel moesin-, ezrin-, radixin-like gene is a candidate for the neurofibromatosis 2 tumor suppressor. Cell. 1993;75:826. doi: 10.1016/0092-8674(93)90501-G. [DOI] [PubMed] [Google Scholar]
- Lekanne Deprez RH, Bianchi AB, Groen NA, Seizinger BR, Hagemeijer A, van Drunen E, Bootsma D, Koper JW, Avezaat CJ, Kley N, Zwarthoff EC. Frequent NF2 gene transcript mutations in sporadic meningiomas and vestibular schwannomas. Am J Hum Genet. 1994;54:1022–1029. [PMC free article] [PubMed] [Google Scholar]
- Papi L, De Vitis LR, Vitelli F, Ammannati F, Mennonna P, Montali E, Bigozzi U. Somatic mutations in the neurofibromatosis type 2 gene in sporadic meningiomas. Hum Genet. 1995;95:347–351. doi: 10.1007/BF00225206. [DOI] [PubMed] [Google Scholar]
- Wellenreuther R, Kraus JA, Lenartz D, Menon AG, Schramm J, Louis DN, Ramesh V, Gusella JF, Wiestler OD, von Deimling A. Analysis of the neurofibromatosis 2 gene reveals molecular variants of meningioma. Am J Pathol. 1995;146:827–832. [PMC free article] [PubMed] [Google Scholar]
- De Vitis LR, Tedde A, Vitelli F, Ammannati F, Mennonna P, Bigozzi U, Montali E, Papi L. Screening for mutations in the neurofibromatosis type 2 (NF2) gene in sporadic meningiomas. Hum Genet. 1996;97:632–637. doi: 10.1007/BF02281874. [DOI] [PubMed] [Google Scholar]
- Evans JJ, Jeun SS, Lee JH, Harwalkar JA, Shoshan Y, Cowell JK, Golubic M. Molecular alterations in the neurofibromatosis type 2 gene and its protein rarely occurring in meningothelial meningiomas. J Neurosurg. 2001;94:111–117. doi: 10.3171/jns.2001.94.1.0111. [DOI] [PubMed] [Google Scholar]
- Evans DG, Watson C, King A, Wallace AJ, Baser ME. Multiple meningiomas: differential involvement of the NF2 gene in children and adults. J Med Genet. 2005;42:45–48. doi: 10.1136/jmg.2004.023705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kros J, de Greve K, van Tilborg A, Hop W, Pieterman H, Avezaat C, Lekanne Dit Deprez R, Zwarthoff E. NF2 status of meningiomas is associated with tumour localization and histology. J Pathol. 2001;194:367–372. doi: 10.1002/path.909. [DOI] [PubMed] [Google Scholar]
- van Tilborg AA, Morolli B, Giphart-Gassler M, de Vries A, van Geenen DA, Lurkin I, Kros JM, Zwarthoff EC. Lack of genetic and epigenetic changes in meningiomas without NF2 loss. J Pathol. 2005. [DOI] [PubMed]
- 21. Lomas J, Bello MJ, Arjona D, Alonso ME, Martinez-Glez V, Lopez-Marin I, Aminoso C, de Campos JM, Isla A, Vaquero J, Rey JA. Genetic and epigenetic alteration of the NF2 gene in sporadic meningiomas. Genes Chromosomes Cancer. 2005;42:314–319. doi: 10.1002/gcc.20141. [DOI] [PubMed] [Google Scholar]
- Dumanski JP, Carlbom E, Collins VP, Nordenskjold M. Deletion mapping of a locus on human chromosome 22 involved in the oncogenesis of meningioma. Proc Natl Acad Sci U S A. 1987;84:9275–9279. doi: 10.1073/pnas.84.24.9275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblom A, Ruttledge M, Collins VP, Nordenskjold M, Dumanski JP. Chromosomal deletions in anaplastic meningiomas suggest multiple regions outside chromosome 22 as important in tumor progression. Int J Cancer. 1994;56:354–357. doi: 10.1002/ijc.2910560310. [DOI] [PubMed] [Google Scholar]
- Cai DX, Banerjee R, Scheithauer BW, Lohse CM, Kleinschmidt-Demasters BK, Perry A. Chromosome 1p and 14q FISH analysis in clinicopathologic subsets of meningioma: diagnostic and prognostic implications. J Neuropathol Exp Neurol. 2001;60:628–636. doi: 10.1093/jnen/60.6.628. [DOI] [PubMed] [Google Scholar]
- Buschges R, Ichimura K, Weber RG, Reifenberger G, Collins VP. Allelic gain and amplification on the long arm of chromosome 17 in anaplastic meningiomas. Brain Pathol. 2002;12:145–153. doi: 10.1111/j.1750-3639.2002.tb00429.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry A, Banerjee R, Lohse CM, Kleinschmidt-DeMasters BK, Scheithauer BW. A role for chromosome 9p21 deletions in the malignant progression of meningiomas and the prognosis of anaplastic meningiomas. Brain Pathol. 2002;12:183–190. doi: 10.1111/j.1750-3639.2002.tb00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulman EP, White PS, Brodeur GM. Genomic annotation of the meningioma tumor suppressor locus on chromosome 1p34. Oncogene. 2004;23:1014–1020. doi: 10.1038/sj.onc.1206623. [DOI] [PubMed] [Google Scholar]
- Arinami T, Kondo I, Hamaguchi H, Nakajima S. Multifocal meningiomas in a patient with a constitutional ring chromosome 22. J Med Genet. 1986;23:178–180. doi: 10.1136/jmg.23.2.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella R, Levine S, Wilmot PL, Ashar KD, Casamassima AC, Shapiro LRR. Multiple meningiomas in a patient with constitutional ring chromosome 22. Am J Med Genet. 1993;47:184–186. doi: 10.1002/ajmg.1320470211. [DOI] [PubMed] [Google Scholar]
- Akagi K, Kurahashi H, Arita N, Hayakawa T, Monden M, Mori T, Takai S, Nishisho I. Deletion mapping of the long arm of chromosome 22 in human meningiomas. Int J Cancer. 1995;60:178–182. doi: 10.1002/ijc.2910600208. [DOI] [PubMed] [Google Scholar]
- Sulman EP, Dumanski JP, White PS, Zhao H, Maris JM, Mathiesen T, Bruder C, Cnaan A, Brodeur GM. Identification of a consistent region of allelic loss on 1p32 in meningiomas: correlation with increased morbidity. Cancer Res. 1998;58:3226–3230. [PubMed] [Google Scholar]
- Ishino S, Hashimoto N, Fushiki S, Date K, Mori T, Fujimoto M, Nakagawa Y, Ueda S, Abe T, Inazawa J. Loss of material from chromosome arm 1p during malignant progression of meningioma revealed by fluorescent in situ hybridization. Cancer. 1998;83:360–366. doi: 10.1002/(SICI)1097-0142(19980715)83:2<360::AID-CNCR21>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Pulst SM, Rouleau GA, Marineau C, Fain P, Sieb JP. Familial meningioma is not allelic to neurofibromatosis 2. Neurology. 1993;43:2096–2098. doi: 10.1212/wnl.43.10.2096. [DOI] [PubMed] [Google Scholar]
- Buckley PG, Jarbo C, Menzel U, Mathiesen T, Scott C, Gregory SG, Langford CF, Dumanski JP. Comprehensive DNA copy number profiling of meningioma using a chromosome 1 tiling path microarray identifies novel candidate tumor suppressor loci. Cancer Res. 2005;65:2653–2661. doi: 10.1158/0008-5472.CAN-04-3651. [DOI] [PubMed] [Google Scholar]
- Hansson CM, Ali H, Bruder CE, Fransson I, Kluge S, Andersson B, Roe BA, Menzel U, Dumanski JP. Strong conservation of the human NF2 locus based on sequence comparison in five species. Mamm Genome. 2003;14:526–536. doi: 10.1007/s00335-003-3011-3. [DOI] [PubMed] [Google Scholar]
- Dumanski JP, Rouleau GA, Nordenskjold M, Collins VP. Molecular genetic analysis of chromosome 22 in 81 cases of meningioma. Cancer Res. 1990;50:5863–5867. [PubMed] [Google Scholar]
- Buckley PG, Mantripragada KK, Benetkiewicz M, Tapia-Paez I, Diaz De Stahl T, Rosenquist M, Ali H, Jarbo C, De Bustos C, Hirvela C, Sinder Wilen B, Fransson I, Thyr C, Johnsson BI, Bruder CE, Menzel U, Hergersberg M, Mandahl N, Blennow E, Wedell A, Beare DM, Collins JE, Dunham I, Albertson D, Pinkel D, Bastian BC, Faruqi AF, Lasken RS, Ichimura K, Collins VP, Dumanski JP. A full-coverage, high-resolution human chromosome 22 genomic microarray for clinical and research applications. Hum Mol Genet. 2002;11:3221–3229. doi: 10.1093/hmg/11.25.3221. [DOI] [PubMed] [Google Scholar]
- Peyrard M, Fransson I, Xie YG, Han FY, Ruttledge MH, Swahn S, Collins JE, Dunham I, Collins VP, Dumanski JPP. Characterization of a new member of the human beta-adaptin gene family from chromosome 22q12, a candidate meningioma gene. Hum Mol Genet. 1994;3:1393–1399. doi: 10.1093/hmg/3.8.1393. [DOI] [PubMed] [Google Scholar]
- Ruttledge MH, Xie YG, Han FY, Peyrard M, Collins VP, Nordenskjold M, Dumanski JP. Deletions on chromosome 22 in sporadic meningioma. Genes Chromosomes Cancer. 1994;10:122–130. doi: 10.1002/gcc.2870100207. [DOI] [PubMed] [Google Scholar]
- Maru Y, Witte ON. The BCR gene encodes a novel serine/threonine kinase activity within a single exon. Cell. 1991;67:459–468. doi: 10.1016/0092-8674(91)90521-Y. [DOI] [PubMed] [Google Scholar]
- D'Adamo DR, Novick S, Kahn JM, Leonardi P, Pellicer A. rsc: a novel oncogene with structural and functional homology with the gene family of exchange factors for Ral. Oncogene. 1997;14:1295–1305. doi: 10.1038/sj.onc.1200950. [DOI] [PubMed] [Google Scholar]
- Leonardi P, Kassin E, Hernandez-Munoz I, Diaz R, Inghirami G, Pellicer A. Human rgr: transforming activity and alteration in T-cell malignancies. Oncogene. 2002;21:5108–5116. doi: 10.1038/sj.onc.1205694. [DOI] [PubMed] [Google Scholar]
- Perry A, Cai DX, Scheithauer BW, Swanson PE, Lohse CM, Newsham IF, Weaver A, Gutmann DH. Merlin, DAL-1, and progesterone receptor expression in clinicopathologic subsets of meningioma: a correlative immunohistochemical study of 175 cases. J Neuropathol Exp Neurol. 2000;59:872–879. doi: 10.1093/jnen/59.10.872. [DOI] [PubMed] [Google Scholar]
- Jacoby LB, MacCollin M, Barone R, Ramesh V, Gusella JF. Frequency and distribution of NF2 mutations in schwannomas. Genes, Chromosomes & Cancer. 1996;17:45–55. doi: 10.1002/(SICI)1098-2264(199609)17:1<45::AID-GCC7>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Merel P, Hoang-Xuan K, Sanson M, Moreau-Aubry A, Bijlsma EK, Lazaro C, Moisan JP, Resche F, Nishisho I, Estivill X, Delattre JY, Poisson M, Theillet C, Hulsebos T, Delattre O, Thomas G. Predominant occurrence of somatic mutations of the NF2 gene in meningiomas and schwannomas. Genes Chromosomes Cancer. 1995;13:211–216. doi: 10.1002/gcc.2870130311. [DOI] [PubMed] [Google Scholar]
- Twist EC, Ruttledge MH, Rousseau M, Sanson M, Papi L, Merel P, Delattre O, Thomas G, Rouleau GA. The neurofibromatosis type 2 gene is inactivated in schwannomas. Hum Mol Genet. 1994;3:147–151. doi: 10.1093/hmg/3.1.147. [DOI] [PubMed] [Google Scholar]
- De Vitis LR, Tedde A, Vitelli F, Ammannati F, Mennonna P, Bono P, Grammatico B, Grammatico P, Radice P, Bigozzi U, Montali E, Papi L. Analysis of the neurofibromatosis type 2 gene in different human tumors of neuroectodermal origin. Hum Genet. 1996;97:638–641. doi: 10.1007/BF02281875. [DOI] [PubMed] [Google Scholar]
- Xu HM, Gutmann DH. Merlin differentially associates with the microtubule and actin cytoskeleton. J Neurosci Res. 1998;51:403–415. doi: 10.1002/(SICI)1097-4547(19980201)51:3<403::AID-JNR13>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Gutmann DH, Haipek CA, Hoang Lu K. Neurofibromatosis 2 tumor suppressor protein, merlin, forms two functionally important intramolecular associations. J Neurosci Res. 1999;58:706–716. doi: 10.1002/(SICI)1097-4547(19991201)58:5<706::AID-JNR12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Kino T, Takeshima H, Nakao M, Nishi T, Yamamoto K, Kimura T, Saito Y, Kochi M, Kuratsu J, Saya H, Ushio Y. Identification of the cis-acting region in the NF2 gene promoter as a potential target for mutation and methylation-dependent silencing in schwannoma. Genes Cells. 2001;6:441–454. doi: 10.1046/j.1365-2443.2001.00432.x. [DOI] [PubMed] [Google Scholar]
- Bello MJ, Aminoso C, Lopez-Marin I, Arjona D, Gonzalez-Gomez P, Alonso ME, Lomas J, de Campos JM, Kusak ME, Vaquero J, Isla A, Gutierrez M, Sarasa JL, Rey JA. DNA methylation of multiple promoter-associated CpG islands in meningiomas: relationship with the allelic status at 1p and 22q. Acta Neuropathol (Berl) 2004;108:413–421. doi: 10.1007/s00401-004-0911-6. [DOI] [PubMed] [Google Scholar]
- Liu Y, Pang JC, Dong S, Mao B, Poon WS, Ng HK. Aberrant CpG island hypermethylation profile is associated with atypical and anaplastic meningiomas. Hum Pathol. 2005;36:416–425. doi: 10.1016/j.humpath.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Colella S, Shen L, Baggerly KA, Issa JP, Krahe R. Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
- Uhlmann K, Brinckmann A, Toliat MR, Ritter H, Nurnberg P. Evaluation of a potential epigenetic biomarker by quantitative methyl-single nucleotide polymorphism analysis. Electrophoresis. 2002;23:4072–4079. doi: 10.1002/elps.200290023. [DOI] [PubMed] [Google Scholar]
- Lee JH, Sundaram V, Stein DJ, Kinney SE, Stacey DW, Golubic M. Reduced expression of schwannomin/merlin in human sporadic meningiomas. Neurosurgery. 1997;40:578–587. doi: 10.1097/00006123-199703000-00031. [DOI] [PubMed] [Google Scholar]
- Hitotsumatsu T, Iwaki T, Kitamoto T, Mizoguchi M, Suzuki SO, Hamada Y, Fukui M, Tateishi J. Expression of neurofibromatosis 2 protein in human brain tumors: an immunohistochemical study. Acta Neuropathol (Berl) 1997;93:225–232. doi: 10.1007/s004010050608. [DOI] [PubMed] [Google Scholar]
- Den Bakker MA, van Tilborg AA, Kros JM, Zwarthoff EC. Truncated NF2 proteins are not detected in meningiomas and schwannomas. Neuropathology. 2001;21:168–173. doi: 10.1046/j.1440-1789.2001.00394.x. [DOI] [PubMed] [Google Scholar]
- Hernandez-Munoz I, Benet M, Calero M, Jimenez M, Diaz R, Pellicer A. rgr oncogene: activation by elimination of translational controls and mislocalization. Cancer Res. 2003;63:4188–4195. [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning; a laboratory manual. Second Edition. Cold Spring Harbor, New York , Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Fiegler H, Carr P, Douglas EJ, Burford DC, Hunt S, Scott CE, Smith J, Vetrie D, Gorman P, Tomlinson IP, Carter NP. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes Chromosomes Cancer. 2003;36:361–374. doi: 10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- http://puffer.genpat.uu.se/protocols
- Staden R. The Staden sequence analysis package. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table summarizing DNA copy number changes in the additional subset of 26 sporadic meningiomas not studied for NF2 gene point mutations or NF2 promoter methylation status.
Table summarizing primers used for PCR amplification and sequencing of the NF2 gene.
Table summarizing primer pairs used for amplification of bisulfite treated DNA, sequencing primers, analyzed sequences and dispensation order.
List of non-22q-derived genomic clones that encompass genes of potential interest in tumor formation/progression.


