SUMMARY
Recent advances in proteomic technologies provide tremendous opportunities for biomarker-related clinical applications; however, the unique characteristics of human biofluids such as the high dynamic range in protein abundances, and extreme complexity of the proteomes present tremendous challenges. In this review we summarize recent advances in LC-MS based proteomic profiling and its applications in clinical proteomics, as well as discuss the major challenges associated with implementing these technologies for more effective candidate biomarker discovery. Developments in immunoaffinity depletion and various fractionation approaches in combination with substantial improvements in LC-MS platforms have enabled the plasma proteome to be profiled with considerably greater dynamic range of coverage, allowing many proteins at low ng/mL levels to be confidently identified. Despite these significant advances and efforts, major challenges associated with the dynamic range of measurements and extent of proteome coverage, confidence of peptide/protein identifications, quantitation accuracy, analysis throughput, and the robustness of present instrumentation must be addressed before a proteomic profiling platform suitable for efficient clinical applications can be routinely implemented.
Keywords: LC-MS, Proteomics, Biomarker, Clinical, Advance, Challenge, Human plasma, Human serum, False positive, AMT tag, Throughput, Ion mobility spectrometry
INTRODUCTION
Advances in mass spectrometry (MS) technologies, high resolution liquid phase separations, and informatics/bioinformatics for large scale data analysis have made MS-based proteomics an indispensable research tool with the potential to broadly impact biology and laboratory medicine1. In particular, proteomic technologies have been increasingly applied to the study of disease-related clinical samples (e.g., human blood serum/plasma, proximal fluids, and disease tissues) for the purposes of identifying novel disease-specific protein biomarkers, gaining better understandings of disease processes, and discovering novel protein targets for therapeutic interventions and drug developments2.
Proteomics-based candidate biomarker discovery efforts have recently gained significant attention due to the power of these technologies for analyzing complex protein mixtures and their potential for identifying novel markers indicative of disease. It is widely accepted that many complex human diseases, including cancers, might be effectively cured if specific disease biomarkers were available to enable detection and treatment at very early stages of disease3. Despite noteworthy efforts, only a handful of cancer biomarkers have been approved by the US Food and Drug Administration (FDA) for clinical use, with the majority of these being protein biomarkers 4. While existing markers play a significant role in screening, monitoring, and staging, effective biomarkers are not currently available for most cancers, and are generally nonexistent for early detection3. Therefore, there is a clear need for applying advanced technologies such as proteomics in the quest for novel candidate clinical biomarkers.
Although widely speculated that advances in genomics and proteomics would alter the landscape of clinical biomarker discovery and validation, the declining trend of new FDA-approved biomarkers reported over the last decade5 highlights the magnitude of the challenges associated with human clinical samples and validation of candidate biomarkers. Contributing to these challenges are the substantial complexity of the human proteome itself and the tremendous heterogeneity of the human population, both of which make the search for biomarkers from either biofluids or disease tissues a daunting task. As a result of this heterogeneous nature of humans and the complexity of diseases like cancer, a panel of biomarkers rather than a single marker may be required to achieve the high sensitivity and specificity required for clinical applications3. Proteomics technologies offer significant potential for discovering such panels of markers.
Many different technologies have been applied for biomarker discovery and other clinical applications, including two-dimensional (2D) gel-electrophoresis6, liquid chromatography coupled with mass spectrometry (LC-MS), and protein- and antibody-based microarrays7-9. LCMS or tandem MS (MS/MS) based proteomic technologies offer highly sensitive analytical capabilities and a relatively large dynamic range of detection, and have increasingly become the method of choice for in-depth profiling of complex protein mixtures1. In addition, the relatively high throughput of LC-MS technologies is amenable to clinical applications that involve human biofluids and disease tissues. The application of LC-MS/MS for human biofluid protein profiling initiated with the first global shotgun proteomic study of human plasma/serum published in 2002 by Adkins et al.10 An explosion of LC-MS based applications in human plasma/serum and various biofluids soon followed due to the tremendous interest in identifying disease related proteins11, 12. Various depletion/fractionation/ enrichment techniques have been developed along the way and coupled to LC-MS to increase coverage of the biofluid proteomes13.
Human blood serum/plasma remains the most commonly used clinical sample to date for proteomic applications since it has the potential to contain specific biomarkers for virtually all human diseases due to its either direct or indirect interaction with the entire cell complement of the body, i.e., tissue-specific proteins may be released into the blood stream upon cell damage or cell death. Additionally, serum/plasma can be readily obtained by clinical sampling. However, the magnitude of the previously mentioned challenges associated with human clinical samples coupled with the anticipation that potential biomarkers of interest could be present at extremely low concentrations in plasma has raised doubts as to whether disease biomarkers can be accurately detected or identified from plasma by using a proteomic approach. As a result, analysis of various other biofluids/tissues has gained increasing attention. Due to their proximity to the source of disease or perturbation in the body, tissues14 and various biofluids such as cerebrospinal fluid15, bronchoalveolar lavage fluid16, synovial fluid17, nipple aspirate fluid18, saliva19, and urine20 are believed to provide a more focused pool of potential biomarkers of interest. In addition, tumor interstitial fluids (TIF) have also been reported as a novel source for proteomic biomarker and therapeutic target discovery21, offering a unique alternative to direct tissue analysis.
In the following review, we highlight LC-MS based proteomic profiling for clinical applications by summarizing recent advances, as well as the major challenges facing this technology for more effective candidate biomarker discovery.
Challenges and Requirements for Designing a Robust LC-MS Discovery Platform
The unique nature of human biofluid proteomes, in particular, the serum/plasma proteome, presents significant challenges for current analytical technologies aimed at quantitative protein profiling and biomarker discovery. First, the serum/plasma protein content is dominated by several very abundant proteins (i.e., the 22 most abundant proteins represent approximately 99% of the total protein mass in plasma), yet at the same time presents an extraordinary dynamic range (>10 orders of magnitude) in protein concentrations that begins with serum albumin at ∼45 mg/mL and extends to cytokines (and potentially many disease-related proteins) at around 1-10 pg/mL or lower5. Second, the serum/plasma proteome presents tremendous biological complexity as a result of tissue “leakage” proteins from the entire body, complex post-translational plasma protein modifications such as glycosylation, as well as the existence of various forms (i.e., splicing variants, proteolytic products, and the tremendous variability in the immunoglobulin class) for each expressed gene. Finally, the substantial genetic and non-genetic biological variability of human clinical samples contributes significantly to the overall analytical challenge.
In spite of significant recent advances, major challenges remain to prevent routine implementation of a LC-MS protein profiling platform suitable for efficient biomarker discovery (Table 1). To effectively address these challenges, a protein profiling platform suitable for biomarker discovery and clinical applications must provide at the very minimum: (1) overall high dynamic range of measurements and extensive coverage of the proteome for effective detection of low abundance proteins, (2) highly confident and specific protein identifications, (3) accurate quantitation of relative protein abundances across many clinical samples, and (4) high throughput capable of analyzing large numbers of clinical samples to provide sufficient statistical power needed to address biological variability. In addition, the platform, including both sample processing and LC-MS instrumentation, must be robust and include efficient informatics software capabilities for data mining and statistical analyses. Currently, there is a broad consensus that no existing platform meets all of these requirements for effective biomarker discovery.
Table 1.
Challenges and limitations of current LC-MS based proteomic technologies applied to biomarker discovery.
| Challenge | Current techniques for addressing the challenge | Limitations |
|---|---|---|
| Dynamic range of measurements | immunoaffinity depletion and multidimensional fractionation coupled with high resolution LC-MS or MS/MS instrumentation | Low throughput |
| Require relatively large sample sizes | ||
| Sensitivity | Small inner diameter LC column (50 μm or less) coupled with nanoflow electrospray ionization and advanced MS instrumentation (i.e., FTICR, LTQ-FT). | Issues in robustness and expense |
| Reproducibility and Quantitation | Platform automation (including sample processing) | Variations from multi-step sample processing |
| “Label-free” direct quantitation and isotope labeling based quantitation | Ionization suppression and instrument variations | |
| Labeling efficiencies | ||
| Throughput | Automated fast LC and gas phase ion mobility separations | Limited Dynamic range or coverage |
| False positive identifications | Improved database searching algorithms and statistical models | Lack of consensus |
Figure 1 shows a component-based diagram of an LC-MS protein profiling platform. Note that such a platform is not based on a single instrument, but rather on a compilation of current technologies to achieve high dynamic range quantitative proteome profiling for clinical samples. A key performance factor of any such platform is the overall dynamic range of detection and extent of proteome coverage, which in turn dictates its ability to detect low abundance proteins. Many disease specific proteins in plasma/serum are anticipated to be present at very low levels (ng/mL or even lower), e.g., within the same range as current FDA approved markers such as prostate specific antigen (PSA, 0.01-100 ng/mL) and Troponin-T (0.02-100 ng/mL). This is particularly obvious for cancer markers of early detection, where tumor size is very small (millimeter size) and cancer specific proteins in plasma may present at pg/mL or lower levels. This overall dynamic range presents a tremendous challenge for any MS-based technology. The achievable dynamic range or proteome coverage for a platform depends on the peak capacity (the number of chromatographic peaks that can be fit into the length of separation) of the on-line LC separations prior to MS measurements, the dynamic range of the MS instrumentation, and the efficiency of sample enrichment or fractionation steps at both protein and peptide levels prior to LC-MS analyses. Analysis throughput inevitably determines the size of any clinical study sample set and largely depends on factors such as automation of each platform component, LC-MS analysis duty cycle, and the extent of pre-fractionation prior to LC-MS analysis. While the application of more extensive fractionation can lead to a higher dynamic range of detection, the overall throughput can be severely reduced. Other key performance factors are the confidence of protein identifications and the quantitative accuracy, which determine the ability of the platform to confidently identify a potential biomarker based on the abundance differences between healthy and diseased conditions. Both the reproducibility of sample processing/fractionation prior to LC-MS and the LC-MS instrumentation will contribute to the accuracy of quantitation.
Figure 1.
A component diagram of a LC-MS protein profiling platform.
Advances in LC-MS Technologies
A high resolution LC (or LC/LC) separation coupled on-line with MS is the central component of many proteomics platforms. Over the past decade, there have been significant advances in LC separations, as well as in MS instrumentation and electrospray ionization (ESI). To date, the “bottom-up” proteomic strategy that combines high efficiency separations with MS to characterize highly complex peptide mixtures still accounts for the majority of proteomics measurements. This strategy relies on the identification of peptides sufficiently unique for protein identification. Protein mixtures from cellular lysates or biofluids are typically digested by trypsin (or other proteases) into polypeptides, which are then separated by capillary LC and analyzed by MS on-line via an electrospray ionization (ESI) interface. Peptide sequences are identified by using automated database searching algorithms such as SEQUEST 22, MASCOT 23, or X!Tandem24 to correlate experimental MS/MS spectra to theoretical mass spectra based on sequences in a given protein database for a specific organism. With the recent development of high speed 2D linear ion-trap instruments, i.e., LTQ, the protein profiling coverage has been greatly enhanced compared to traditional 3D ion-trap systems25. When coupled with SCX fractionation either on-line or offline,26, 27 LC-MS/MS technologies now routinely allow for identification of thousands of proteins from complex mammalian tissues and cells. Although routinely used for peptide/protein identifications, data-dependent LC-MS/MS still has an inherent “under-sampling” limitation whereby only a portion of the species observed in the survey MS scan is selected for fragmentation28.
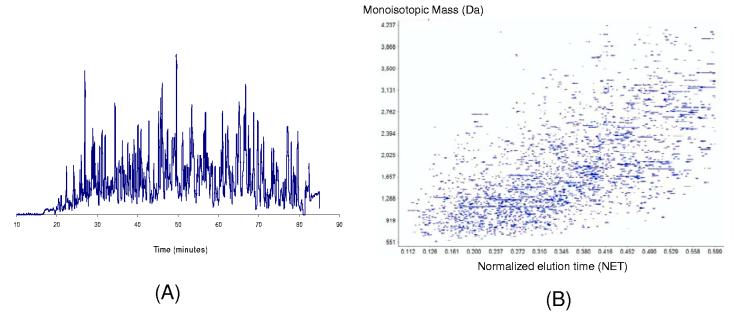
To overcome the under-sampling issue, our laboratory developed an accurate mass and time (AMT) tag approach that utilizes highly accurate mass measurements from a high resolution mass spectrometer (e.g., Fourier transform ion cyclotron resonance (FTICR) or time-of-flight (TOF) mass spectrometer) in conjunction with accurate elution time measurements from high resolution capillary LC separations to achieve high throughput proteome profiling without routine MS/MS measurements29, 30. The concept of this AMT tag approach is based on the principle that the accurate mass and time measurements will allow reliable peptide identifications by correlating the mass and time of detected peaks to a pre-established peptide AMT tag reference library for a particular biological system (e.g., plasma). With this approach, LCMS/MS proteome analyses coupled with extensive fractionation only need to be performed once to create an effective reference database of peptide markers defined by accurate masses and elution times, i.e., AMT tags, for a particular biological system, such as serum/plasma. The AMT tag database then serves as a comprehensive “look-up table” for subsequent higher throughput LC-MS analyses, allowing many peptides in each spectrum to be identified without MS/MS. Figure 2 exemplifies an LC chromatogram and 2D display of ∼2,800 peptides identified using the AMT tag strategy resulting from a single LC-FTICR analysis of a ProteomeLab™ IgY-12 depleted human plasma sample.
Figure 2.
A typical LC-FTICR analysis of an IgY-12 depleted human plasma sample. (A) The base peak chromatogram. (B) A 2D-display of ∼2800 identified species at the mass and normalized elution time (NET) space. The analysis was performed using a Bruker 9.4 Tesla FTICR instrument coupled with an LC system equipped with a 150 μm i.d. and 65 cm long capillary column operated at 5000 psi.
The fact that application of the AMT tag approach obviates the need for routine MS/MS is particularly attractive in high throughput repeated analyses of similar samples (e.g., serum/plasma) in clinical proteomic studies. We have recently demonstrated the application of the AMT tag approach coupled with 18O labeling for quantitative profiling of the human plasma proteome in response to lipopolysaccharide administration31. The availability of commercial high performance mass spectrometers (e.g., ThermoElectron Finnigan LTQ-FT, LTQ-Orbitrap) will likely lead to an even broader range of applications based on this LC-MS only approach for higher throughput peptide identifications.
As mentioned previously, the achievable dynamic range for the LC-MS platform depends significantly on the peak capacity of the on-line gradient reversed phase separations, the dynamic range of the MS system, and the efficiency and stability of the ESI interface. A single MS spectrum can provide a dynamic range of up to 103 for a high resolution instrument (e.g., FTICR), and one would expect to achieve a dynamic range of at least 105 by coupling this instrument to an on-line high resolution LC separation that provides a peak capacity of ∼1000. However, the observed dynamic range of measurements can be significantly reduced for complex biological samples such as human plasma due to the charge competition of co-eluting high abundance species, which leads to ion suppression of the relatively low abundance species. Ion suppression is a particular issue when analyzing human biofluid samples, as these samples are dominated by a handful of highly abundant proteins. Significant ion suppression will occur when peptides originating from low abundance proteins of interest co-elute with peptides originating from high abundance proteins, which leads to the inability to detect the co-eluting low abundance peptides.
Table 2 provides a summary of the relative proteome coverage and estimated dynamic ranges achieved by coupling high resolution reversed phase capillary LC separations with either MS/MS using an LTQ instrument or MS using a 9.4 tesla FTICR instrument. The enhanced coverage and dynamic ranges obtained by the removal of high abundance proteins and SCX fractionation are illustrated. All results shown in this table are based on triplicate experiments that involved a pooled plasma sample from healthy subjects. The number of peptide identifications are reported with >95% confidence based on either a reversed database evaluation for MS/MS data32 or a shifted database evaluation for the LC-FTICR data (Petyuk et al., manuscript submitted), with all proteins identified using a minimum of two different peptides. As shown, the single LC-MS/MS analysis only identifies ∼100 proteins with high confidence and provides a dynamic range of ∼103. With the removal of either the top-6 (MARS) or top-12 (IgY-12) abundant proteins, the overall dynamic range is enhanced to ∼105. LC-FTICR shows greater coverage for both peptide and protein identifications compared to LC-MS/MS, and the dynamic range is estimated to be similar to that observed for LC-MS/MS. When IgY-12 depletion and SCX fractionation are combined with LC-MS/MS, a dynamic range of 106-107 can be achieved, allowing identification of nearly 500 proteins in plasma with high confidence, including many at the low ng/mL level. Note that this dynamic range still falls three orders of magnitude short for detecting pg/mL protein concentrations. In addition, it should be noted that not all the proteins within the estimated dynamic range will be detected due to the differences in digestion efficiency and ion-suppression effects for different proteins/peptides within the complex sample.
Table 2.
The proteome coverage and estimated dynamic range offered by current LC-MS technologies*.
| Methods | Replicate 1 | Replicate 2 | Replicate 3 | Overlap | Identified low abundance proteins | Estimated dynamic range of coverage* | |
|---|---|---|---|---|---|---|---|
| Non-depleted plasma and 1D-LC-MS/MS | Peptides | 1398 | 1213 | 1466 | 972 | ALS, 25 μg/mL | |
| Proteins | 99 | 97 | 102 | 96 | Factor XII, 30 μg/mL | ∼ 10 3 | |
| APOC2, 35 μg/mL | |||||||
| MARS depletion and 1D-LC-MS/MS | Peptides | 1723 | 1732 | 1692 | 1250 | B2M, 1.1 μg/mL | |
| Proteins | 119 | 118 | 115 | 111 | VWF, 1.3 μg/mL | ∼ 10 4 | |
| SAA, 10 μg/mL | |||||||
| IgY-12 depletion and 1D-LC-MS/MS | Peptides | 1869 | 1912 | 1999 | 1309 | Myoglobin, 90 ng/mL | |
| Proteins | 130 | 141 | 130 | 122 | CRP, 500 ng/mL | ∼ 10 5 | |
| HGFA, 500 ng/mL | |||||||
| CD14, 1.4 μg/mL | |||||||
| IgY-12 depletion and 1D-LC-FTICR | Peptides | 2800 | 2840 | 2630 | 2070 | Myoglobin, 90 ng/mL | |
| Proteins | 174 | 172 | 167 | 162 | CRP, 500 ng/mL | ∼ 10 5 | |
| HGFA, 500 ng/mL | |||||||
| CD14, 1.4 μg/mL | |||||||
| IgY-12 depletion and SCX-LC-MS/MS | Peptides | 5196 | 6148 | 5687 | 3391 | MSF, 1 ng/mL | |
| Proteins | 498 | 474 | 476 | 369 | Leptin, 5 ng/mL | ∼ 106 – 107 | |
| NAP1L1, 7 ng/mL | |||||||
| MMP2, 9 ng/mL | |||||||
| Cathepsin D, 9 ng/mL | |||||||
| EGFR, 11 ng/mL |
A pooled reference plasma sample from healthy individuals was used for this evaluation. A prepacked 4.6 mm × 50 mm (loading capacity: 15 μL plasma) MARS affinity column (Agilent, Palo Alto, CA) and a 7 mm × 52 mm (loading capacity: 25 μL plasma) ProteomeLab IgY-12 affinity column (Beckman Coulter, Fullerton, CA) were used for the depletion of high abundance proteins. For each method, the samples were processed in triplicate and individually analyzed using a 150 μm i.d. and 65 cm long column coupled either with a Finnigan LTQ system (MS/MS) or a Bruker 9.4 Tesla FTICR instrument. 10 μg and 5 μg of peptide samples were loaded for each LC-MS/MS and LC-FTICR analyses, respectively. 300 μg peptides were used for each SCX fractionation. The LC and SCX operations were the same as previously described31. Peptides were filtered with a confidence level >95% based on reversed database evaluation32 and proteins were identified with at least two different peptides.
One key area of recent advances in LC-MS technologies is the improvement associated with capillary LC instrumentation that provides enhanced peak capacities and dynamic range of detection needed to analyze clinical samples. These improvements have been achieved primarily through the use of very high pressure (10-20 kpsi), very small porous particles (3 μm or less), smaller inner diameter (i.d.) columns (50 μm i.d. or less), nano-electrospray interfaces, and relatively long columns and long gradients for separations33-35. For example, high efficiency separations with peak capacities of ∼1000 have been achieved by using 15-75 μm i.d. and 85 cm long capillary columns packed with 3 μm C18-bonded silica particles operated at 10 kpsi. By employing smaller i.d. columns (e.g., 15 μm)34, the sensitivity of the system continues to increase inversely as the mobile phase flow rates drop to as low as 20 nL/min, which demonstrates the advantages of ESI-MS analyses at very low liquid flow rates36, 37. More recently, the use of 20 kpsi capillary LC columns packed with 1.4-3 μm porous C18-bonded silica particles has been demonstrated to provide chromatographic peak capacities of 1000-1500 for complex peptide and metabolite mixtures35. While these very high pressure systems present technical challenges for robust automated operations, the recently commercialized WATERS nanoACQUITY UPLC System that takes advantage of 1.7 μm sized particles and operates at >10 kpsi demonstrates the feasibility of such high performance systems for routine applications. With further improvements in robustness, these “ultra-performance” systems may become a powerful component for separating complex mixtures such as human biofluids while concurrently providing the high dynamic range needed for candidate biomarker discovery applications.
Multidimensional Fractionation Strategies Coupled with LC-MS for Improved Proteome Coverage
Given the tremendous dynamic range of protein abundances and the extraordinary complexity of human biofluid proteomes, many different fractionation techniques have been developed and applied in a multidimensional fashion to enhance dynamic range of detection and improve proteome coverage13. Multi-component immunoaffinity removal of highly abundant proteins in human plasma/serum38, 39 has increasingly become the method of choice for prefractionating human plasma samples due to the high specificity, efficacy, and the ease of coupling to other fractionation techniques. As shown in Table 2, coupling the immunoaffinity depletion step to LC-MS provides an additional one to two orders of magnitude increase in dynamic range, which allows for detection of more low abundance proteins by effectively increasing the sample loading; similar improvements were reported in other studies40, 41. Good reproducibility was demonstrated by performing immunoaffinity depletion with an automated LC system; however, some of the nontarget low abundance proteins have also been observed to bind to the columns, but in a reproducible fashion42. A possible approach to counter this effect is to analyze both the flow-through and bound fractions in more of a “partitioning” method instead of a pure “depletion” approach,39 with the accompanying trade-off of an increased number of required analyses. A further enhancement to the platform dynamic range will stem from the continuous improvement of antibody-based microbead technologies that will allow for removal of more highly- to moderately-abundant proteins.
Several different techniques for protein-level fractionation have been applied to human plasma/serum proteome profiling, including common gel-based techniques43, 44, PF2D automated chromotofocusing/reversed phase LC (RPLC)45 and other liquid chromatography-based separations46, free-flow electrophoresis41, 47, and isoelectric focusing (IEF) 46, 48-51. IEF is a common fractionation technique that has been applied to plasma profiling at both peptide and protein levels. Various forms of liquid-phase IEF techniques have been developed, including offgel electrophoresis48, Rotofor49 or MiniRotofor46, microscale solution IEF (ZOOM)50, and a preparative multi-channel electrolyte (MCE) system51. A common feature of these systems is the multiple tandem electrode chambers used to partition complex protein samples. Immobilized pH gradient (IPG) IEF followed by in-gel digestion has also been used for plasma protein fractionation prior to LC-MS/MS52. A number of recent large scale proteome profiling studies have combined different protein- and peptide-level fractionation techniques (e.g., PF2D53, SCX/RPLC54, FFE-IEF/RPLC47, ZOOM/SDS-PAGE50, and Rotofor/RPLC/SDS-PAGE49 protein fractionation) with peptide-level LC-MS/MS analyses to achieve more comprehensive coverage of the plasma proteome.
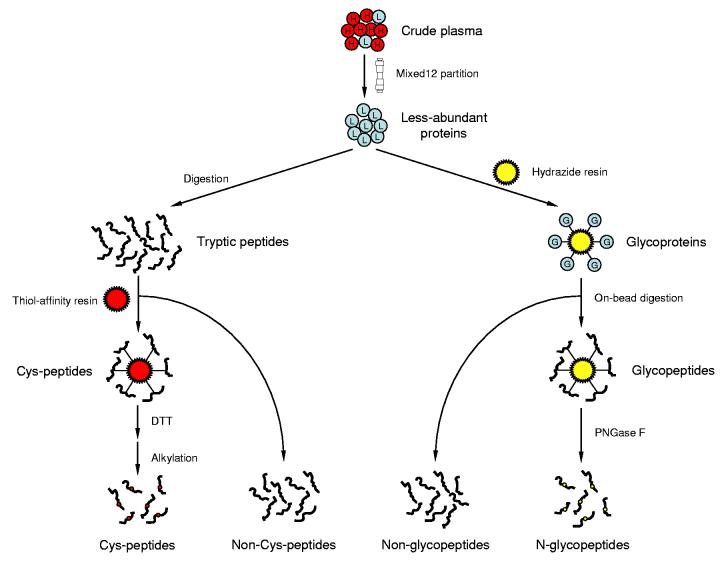
An alternative to plasma protein fractionation is to specifically enrich functional “subproteomes” such as the glycoproteome or the cysteinyl-subproteome by using chemical tagging or capture agents, which significantly reduces overall sample complexity and enhances detection of low abundance proteins. For example, we have recently demonstrated a simple procedure for effectively enriching cysteinyl-peptides from complex proteomes (including human biofluids55), which provides significantly improved proteome coverage when used as a peptide-level fractionation technique27. Additionally, hydrazine chemistry can be applied to specifically enrich N-linked glycopeptides56, 57, and multi-lectin affinity chromatography can be used to isolate and characterize glycoproteins from human plasma and serum samples58. Our laboratory has recently developed a strategy that combines immunoaffinity depletion and subsequent chemical fractionation based on cysteinyl peptide and N-glycoprotein captures with 2D LC-MS/MS for in-depth plasma profiling (Figure 3)59. Application of this “divide-and-conquer” strategy to trauma patient plasma samples resulted in confident identification of ∼1,500 different proteins (with a minimum of two peptides per protein; ∼99.5% confidence level based on reversed database evaluation) and illustrated an overall dynamic range of detection of >107 (low ng/mL concentrations for six identified low abundance proteins were verified by ELISA).
Figure 3.
Schematic representation of a chemical fractionation strategy applied to the plasma proteome characterization. High abundance proteins were firstly removed using immunoaffinity subtraction. The resulting less-abundant proteins were split and subjected to solid-phase cysteinyl peptide and N-glycoprotein captures, independently. Noncysteinyl peptides and non-glycopeptides generated at the same time were also collected. All 4 different peptide populations were then fractionated by SCX chromatography and each fraction was analyzed by capillary LC-MS/MS.
Analysis Throughput
While integration of extensive multi-dimensional fractionation/separations with MS greatly increases the overall proteomic analysis dynamic range and the extent of proteome coverage, this general approach suffers from the limitation of very low throughput. To date, most reports involving extensive fractionation have been limited to small scale studies of one or two pooled clinical samples rather than larger scale quantitative studies. The development of more effective depletion/fractionation strategies and improved LC-MS platforms will most likely reduce the total number of fractions necessary for the detection of low abundance and clinically relevant proteins, and thus provide higher throughput.
Several recent technology developments hold potential for greatly enhancing the overall analysis throughput of clinical samples. The first is the development of very fast LC separations for proteomic analyses. Current automated LC-MS proteomic platforms typically involve LC separations with gradients of 100 min or longer, which limits throughput to ∼10 sample analyses per day per MS instrument. Several reports have explored the use of smaller-particle packed columns or monolithic columns for fast LC separations (10 min or less), as well as multiplex column systems to significantly improve the throughput60, 61. However, it is unclear whether sufficient separation power can be achieved with these fast liquid phase separations since the increase in the solvent gradient speed can degrade the separation peak capacity 60, which in turn reduces the overall dynamic range of detection. Other strategies for achieving robust fast separations include liquid phase chromatographic and electrophoretic separations on a microfluidic chip platform 62-64. Such chip-based separation devices also have the advantage of providing better robustness, reliability, and ease of operation.
Very fast (millisecond scale) gas phase separations based on ion mobility spectrometry (IMS; a separation method that is somewhat analogous to electrophoresis in the gas phase) are another powerful alternative to liquid phase separations for significant improvement in throughput. At its simplest, an IMS stage consists of a drift tube filled with a non-reactive gas (commonly He or N2), and a uniform electric field established along the axis of separation. Mixtures of peptides, proteins, or small molecules are separated by their gas-phase cross-sections (size) in addition to charge, and knowledge of their mobility provides another separation dimension to aid in identification.
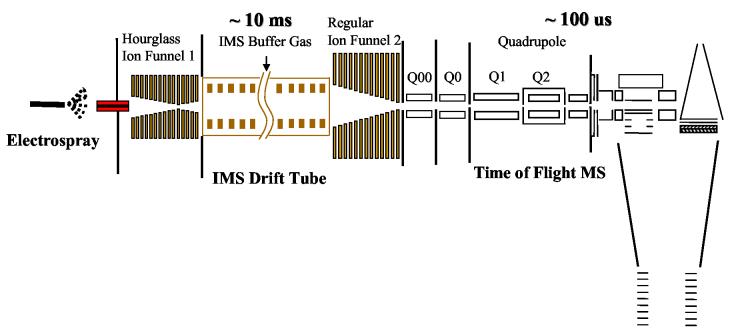
The power of IMS has been advanced by several recent technical developments. IMS coupled with a TOF MS platform and combinatorial libraries 65 has been recently demonstrated for analysis of proteolytic digests 66. Since an IMS separation typically requires 1-100 ms and has a resolving power of 50-200, a single species IMS peak exits the drift tube over a ∼ 0.1-1 ms period. Generation of a typical TOF MS spectrum requires ∼30-100 μs, which allows multiple mass spectra to be obtained during the “elution” of an IMS peak. More recently, LC has been coupled to IMS-TOF MS via an ESI interface, providing 2D separations prior to MS analysis 67. Despite enormous potential for high-throughput analyses of complex samples, the application of IMS-TOF MS has been limited by low sensitivity due to ion losses at the IMS-MS interface; however, the recent implementation of electrodynamic ion funnels at both the ESI-IMS and IMS-TOF MS interfaces has significantly improved the sensitivity of the overall LC-ESI-IMS-TOF MS platform (Figure 4)68 such that the sensitivity is now comparable to that of a commercial ESI-MS. Although still in the development stage, the very fast separation speed and potential high dynamic range of measurements offered by the 2D liquid phase-gas phase separations make the LC-ESI-IMS-TOF MS an attractive and practical platform for high throughput clinical applications.
Figure 4.
Schematic diagram of a prototype ESI-IMS-QTOF instrumentation platform that uses electrodynamic ion funnel interfaces at both ends of the IMS drift tube, and as a result, provides very high sensitivity from high speed analyses.
Confidence of Peptide/Protein Identifications
One of the challenges associated with MS/MS-based proteome profiling is how to assess the confidence levels of peptide and protein identifications that result from automated database searching. It is recognized that a significant portion of the protein identifications in previously published proteomic datasets of human plasma are likely comprised of false positive identifications32, 69-71. For example, four different plasma proteomic datasets that originated from different methodologies were combined into a list that included 1175 non-redundant proteins; however, only 46 of these non-redundant proteins (∼4%) were observed across all four studies.70 This surprisingly low overlap suggests the potential for a very large number of false protein identifications. In a plasma profiling study using nano-scale LC-MS/MS, Shen et al. reported a nearly two-fold difference in the number of identified proteins (ranging from 800 to 1,600) depending on which set of previously published criteria were used to filter the data.69 This criteria dependent difference illustrates the need for more detailed statistical evaluations to ensure high-confidence protein identifications.
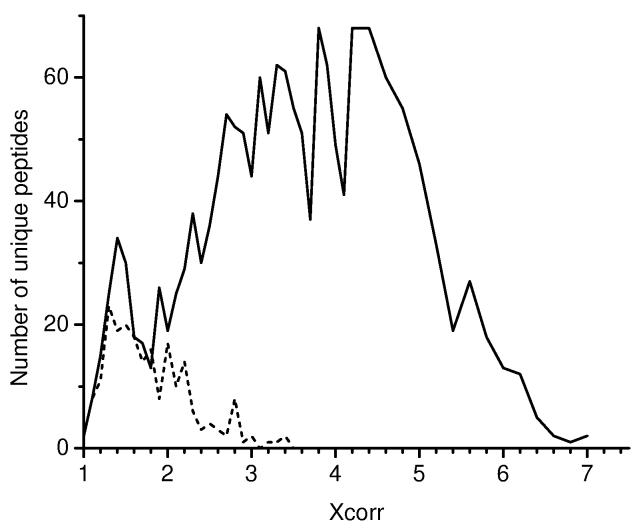
To address the issue of false peptide identifications, we recently performed a probability-based evaluation of peptide identifications derived from LC-MS/MS and SEQUEST analysis in which selected human proteomes, including human plasma were searched against a sequence-reversed human protein database32, similar to a previous report applying the reversed database strategy to the yeast proteome72. The reversed protein database was created by reversing the order of amino acid sequences for each protein (the carboxyl terminus becomes the amino terminus, and vice versa) in the original human protein database. This approach assumes that the numbers of false positives that arise from “random” hits should be the same for both the normal database and the reversed database since the reversed database is identical in number of protein entries, protein size, and distribution of amino acids as the normal database. Figure 5 shows a histogram of Xcorr distribution for unique peptides (charge state 2+; fully tryptic) from a human plasma sample identified by searching the normal (solid line) and reversed (dashed line) databases. The Xcorr distribution allows an estimated confidence level for any given Xcorr bin, as well as the overall false positive rate for a given Xcorr cutoff to be calculated by dividing the area beneath the dashed line (reversed database hits) by the area beneath the solid line (normal database hits) for a given Xcorr range. This study also revealed the high false positive rates for plasma/serum peptide/protein identifications in several previously published studies10, 69, 70, 73, 74. For example, ∼30% false positives were observed when the often-cited Washburn et al. filtering criteria75 were applied to human plasma. Thus, filtering criteria that provided overall >95% confidence at the unique peptide level for both human cell lines and human plasma were proposed. When identical filtering criteria were used, the observed false positive rates of peptide identifications for human plasma were significantly higher than those for the human cell lines, suggesting that the false positive rates are significantly dependent upon sample characteristics, particularly the number of proteins found within the detectable dynamic range for different samples. Additionally, Xie et al. reported the increased potential for false positive identifications for the 2D linear ion trap (LTQ) when compared to a traditional 3D ion trap (LCQ) instrument, and more stringent filtering criteria are required for LTQ compared to LCQ to minimize false positive identifications76. These results suggest that peptide/protein identification confidence levels not only depend on sample characteristics, but also on components of the LCMS platform.
Figure 5.
Relative frequency of different peptides identified from the normal human protein database (solid line) and the reversed human protein database (dashed line) at different Xcorr values. Data shown are for the 2+ charge state fully tryptic peptides identified from human plasma and filtered with ΔCn ≥ 0.1.
Table 3 illustrates differences in filtering criteria stringency by comparing peptide/protein identification results from the same plasma MS/MS dataset (obtained from a recent profiling study using trauma patient plasma samples59) that was filtered using three different sets of criteria77, 78. As shown, the reversed database filtering criteria generated the smallest number of peptide and protein identifications, consistent with the significantly lower percentage of false positive identifications (∼4%), while the human proteome organization (HUPO) plasma proteome project recommended criteria77 and the criteria recently reported by Hood et al.78 generated nearly ∼25% and ∼66% false positives at the peptide level, respectively. The comparison shows that the number of peptide/protein identifications from an individual protein profiling study could be easily inflated if a statistical evaluation of false positives was not performed.
Table 3.
Comparison of peptide and protein identifications from a plasma proteome profiling dataset analyzed using different criteria.
| Filtering criteria | Difference in stringency | Peptides identified | Proteins identifieda | Multi-peptide proteins | Avg. peptides per protein | Estimated false positive rateb |
|---|---|---|---|---|---|---|
| Reversed database32 | >95% confidence at the unique peptide level based on statistical evaluation. | 22267 | 3654 | 1494 (40.9%) | 6.1 | ∼4% |
| Only fully and partially tryptic peptides are considered. | ||||||
| HUPO Plasma Proteome Project77 | Inclusion of partially tryptic peptides with relatively low cutoffs. | 30524 | 7928 | 2850 (35.9%) | 3.9 | ∼25% |
| Hood et al.78 | Inclusion of partially tryptic and other enzymatic cleaved peptides, as well as peptides without protease constraints with relatively low cutoffs. | 66839 | 18958 | 11653 (61.5%) | 3.5 | ∼66% |
Non-redundant protein identifications generated by Protein Prophet80.
False positive rate for each filtering criteria was calculated at unique peptide level based on reversed database evaluation32. The reversed protein database was created by reversing the order of amino acid sequences for each protein (the carboxyl terminus becomes the amino terminus, and vice versa) in the original protein database.
A similar observation was recently reported for proteins identified from data acquired on different instruments from 18 laboratories as part of the large scale HUPO plasma proteome collaborative study77. Application of a rigorous statistical approach that employed multiple hypothesis-testing techniques and took into account the length of coding regions in genes reduced the initial list of 9,504 proteins (of which 3,020 identified with two or more peptides) to 889 proteins (containing both multi-peptide and single-peptide protein identifications) identified with a confidence level of at least 95% 71. Interestingly, this length-dependent statistical approach was applied to re-analyze one of our previously published datasets69 and resulted in 1,073 proteins using the HUPO criteria and 433 proteins using the >95% confidence length-dependent statistics71. Similarly, a ∼two-fold difference in protein identifications between the reversed database filtering results and the HUPO criteria (Table 3) was observed, which suggests similar performance between the length-dependent statistical approach and reversed database filtering with >95% confidence.
PeptideProphet provides another independent statistical model for evaluating potential false positive peptide identifications. The model utilizes the expectation maximum algorithm to derive a mixture of correct and incorrect peptide assignments from the data79. This approach has been directly compared with the reversed database approach for analyzing the same dataset derived from human plasma59. Following filtering with reversed database criteria, 6,279 unique peptides were identified from this dataset with >95% confidence, while 6,341 unique peptides were identified by PeptideProphet using a minimum computed probability of 0.95. Approximately 95% of peptides were common between the two datasets, suggesting comparable results from these two statistical approaches. The use of ProteinProphet, another statistical model that computes the probability of the presence of proteins, addresses the issue of whether peptides are present in more than one entry in the protein database (protein redundancy problem) 80. The list of identified peptides from both the PeptideProphet and the reversed database filtering approaches can serve as input for ProteinProphet to generate a list of non-redundant protein identifications. Several other statistical methods have been recently described for evaluating peptide assignments from MS/MS spectra81-83. Ideally, universal acceptance of a statistical model that optimizes both sensitivity and specificity for confident peptide identifications from MS/MS spectra will allow cross-comparison of protein profiling results from different laboratories, which currently remains as an unresolved challenge.
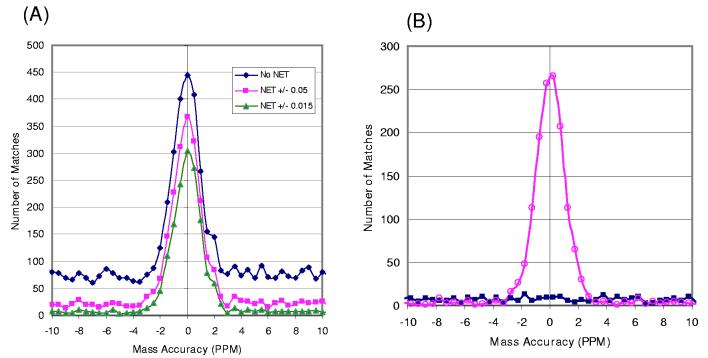
Similar challenges exist for evaluating false positive identifications from MS-only approaches that utilize accurate mass measurements for peptide/protein identifications. The utility of accurate mass measurements initially was demonstrated in the “peptide mass fingerprinting” approach for protein identification, in which a set of peptide fragments unique to each protein are created by digestion and the mass of these peptide fragments used as a “fingerprint” to identify the original protein84-86. Thus far, this approach has been limited to simple protein mixtures or single proteins. The more recently reported AMT tag approach utilizes accurate LC retention time measurements in addition to accurate mass measurements to identify peptides and has been successfully applied to global proteome profiling, including the human plasma proteome31, 87. With the AMT tag approach, peptides are identified by matching LC-MS observed masses and normalized elution time (NET) features to AMT tags in the preestablished reference database (“look-up table” of peptides) with a given mass error and NET error tolerances (typically 1-5 ppm for mass and 1-3% for NET). The potential false positive identifications resulting from random matching of features to the reference database are indicated on histograms of mass error (the difference between observed mass and calculated mass for the matched peptide in the database), exemplified in Figure 6A for a human plasma dataset analyzed by LC-FTICR. Note that the use of the NET constraint significantly reduces the level of random matches as indicated by the background level for each histogram. Similar to the reversed database approach for MS/MS, we have recently applied a shifted database approach for evaluating the false positive rate in the AMT tag process (Petyuk, et al., manuscript submitted). As shown in Figure 6B, ∼3% false positive rate for this human plasma dataset was estimated as the ratio of the area beneath the blue curve that represents matches to the shifted database and the area beneath the magenta curve that represents matches to the normal database within a +/− 2 ppm window. In addition to being used for direct identification in the MS-only approach, the accurate mass information also has been utilized for improving the confidence of peptide identifications by MS/MS through application of the new generation of LTQ-FT and LTQ-Orbitrap mass spectrometers88, 89.
Figure 6.
(A) Mass error histograms of features detected from a single LC-FTICR dataset of a human plasma sample that matched to a human plasma AMT tag database using different levels of normalized elution time (NET) constraints. The LC separation time is normalized to a 0-1 scale in NET. (B) Mass error histograms for features from the same dataset matching to a normal AMT tag database (magenta curve) and to a shifted AMT tag database (blue curve). Blue squares represent random matches to the 11 Da shifted AMT tag database.
Quantitation Strategies
The ability to quantitatively measure relative protein abundance differences between different clinical samples is essential for identifying candidate protein biomarkers; however, the vast majority of proteomic work related to biomarker discovery published to date has been qualitative, which highlights the need for more robust quantitative approaches for such applications. Our initial application for comparative proteome analysis of human plasma following lipopolysaccharide (LPS) administration involved a semi-quantitative strategy based on the total number of peptide identifications per protein (peptide hits or spectrum count)74. In this study, standard SCX-LC-MS/MS analysis was performed at 0 h time point (control) and a 9 h time point following LPS administration and peptide hits were used to obtain a relative quantitative measure between the control and 9 h time point. Several known inflammatory response and acute phase proteins were observed to be up-regulated upon LPS administration. Several other studies have shown that this peptide hits approach can be used as a semi-quantitative approach for initial screening when applied with proper controls and with adequate thresholds90-93.
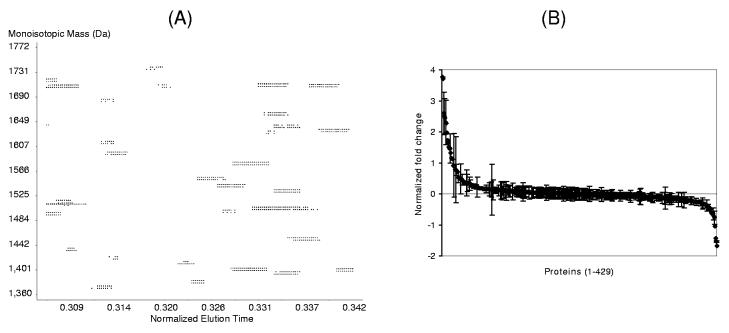
More recently, we have demonstrated 16O/18O labeling combined with the AMT tag strategy as an effective global quantitative approach for quantifying relative protein abundance differences in human plasma31. By incubating tryptic peptides in 18O water55, 94 in the presence of trypsin, the 18O atoms are incorporated into the C-terminus of tryptically cleaved peptides via a post-digestion trypsin-catalyzed oxygen exchange reaction. The 16O/18O labeled peptide pairs provide a 4 Da mass difference (Figure 7A), which allows a high resolution mass spectrometer such as FTICR or TOF to effectively resolve the 16O- and 18O-labeled peptide pairs and accurately measure the relative abundances. The advantage is that all types of samples (e.g., tissues, cells, and biological fluids) can be effectively labeled using this simple and specific enzyme-catalyzed reaction. Figure 7A shows a partial 2D-display of detected peptide pairs in mass vs. time dimensions. The 18O/16O peptides are readily visualized as co-eluting pairs (4 Da apart) and the abundance ratio can be precisely calculated for each 18O/16O pair. In this initial comparative analysis demonstration of two human plasma samples obtained from a healthy individual prior to (control) and following LPS administration, relative abundance differences between the two plasma samples were quantified for a total of 429 plasma proteins. Figure 7B shows the normalized fold changes in 429 quantified proteins and demonstrates the significant changes in abundance for a set of proteins following LPS administration. The combined 16O/18O labeling-AMT tag strategy can also be easily coupled with subsequent peptide-level fractionation approaches such as cysteinyl-peptide enrichment55 and SCX fractionation.
Figure 7.
(A) A partial 2D display of the detected 18O/16O labeled peptide pairs from a LC-FTICR analysis. The elution time is shown as a normalized scale between 0 and 1. Observed peaks (represented by spots) correspond to various eluting peptides. The heavy and light isotope-labeled pairs are easily visualized with a 4 Da mass difference. (B) Normalized fold changes for the 429 quantified proteins following LPS administration. Abundance ratio for each protein shown was normalized to zero (R – 1).118 For ratios smaller than 1, normalized inverted ratios were calculated as [1 – (1/R)]. Error bar for each protein indicates the standard deviation for the abundance ratios from multiple peptides. Proteins without error bars were identified with single peptides.
Other stable isotope labeling methods based on relative peptide/protein abundance measurements include metabolic labeling95-97 and chemical labeling of specific functional groups using reagents such as isotope-coded affinity tags (ICAT)98 and iTRAQ99, 100 have been routinely used for quantitative proteomic analysis. In clinical proteomic applications, these stable isotope labeling techniques are well suited for detecting accurate changes in pair-wise comparisons, provided the samples can be effectively labeled; however, in biomarker discovery applications, it is often challenging to compare across a large number of clinical samples. One alternative to the use of these labeling techniques is the use of a labeled reference sample (often a pooled composite) that is spiked into each normally processed individual clinical sample that allows relative quantitation between each clinical sample and the reference sample, and cross-comparison among the entire set of clinical samples. The 18O labeling strategy is well suited for generating such a labeled reference sample as all other clinical samples can be processed with natural 16O on the C-termini without labeling; 16O/18O peptide pairs are formed after spiking the samples with the 18O-labeled reference.
Alternatively, “label-free” direct quantitation approaches hold interest because of greater flexibility for comparative analyses and simpler sample processing procedures compared to labeling approaches. The isotope labeling and label-free approaches are complementary and each approach has different sources of variations. Several initial studies suggest that the use of normalized LC-MS peak intensities for detected peptides can be used to compare relative abundances between similar complex samples101-103. It has been demonstrated that abundance ratios of separate model proteins may be predicted to within ∼20% in complex proteome digests by using measured peptide ion intensities obtained in LC-MS analyses101. Among the main challenges for label-free quantitation are the multiple issues that affect the usefulness of peptide peak intensities for relative quantitation, such as differences in electrospray ionization efficiencies among different peptides and different samples37, differences in the amount of sample injected in each analysis, and sample preparation reproducibility. These issues are often peptide-dependent, leading to observed disparity among relative abundances of different peptides originating from the same protein. The significant bias and ion suppression effects caused by charge competition (ionization bias) during ESI104 are often considered a major limitation for accurate label-free quantitation. Recent studies have demonstrated substantial advantages for ESI-MS analyses at nano-flow regimes (<100 nL/min) afforded by narrower i.d. capillary columns for separations36, 37. It is well demonstrated that smaller i.d. columns with lower flow rates provide significantly higher sensitivity than larger i.d. columns with higher flow rates34 because of the significant improvements in both ionization and MS sampling efficiencies. Reversed phase packed nanoscale LC and monolithic nanoscale LC separations have been developed and coupled to ESI for improved ionization and quantitation 34, 105. As ionization efficiencies are increased for nano-electrospray, detection biases are decreased since undesired matrix effects and/or ion suppression effects are either reduced or eliminated104-106, which provides the basis for improved quantitation. With further improvements to the robustness of these nano-LC-ESI-MS systems, label-free quantitation may be widely applied in clinical applications.
Another challenge for quantitative clinical proteomic applications is the variability introduced during multiple steps of sample processing. With continued development of clean-up products for more consistent performance and automated sample processing, such reproducibility issues may be minimized, leading to further improvements in quantitation when applying either the stable isotope labeling or label-free approaches.
Implications of Human Heterogeneity in Clinical Proteomic Studies
The ability to identify disease-specific differences by using a proteomic approach relies on multiple factors integral to the overall analysis pipeline, For example, when performing peptide-level measurements, achieving high peptide identification quality is a prerequisite for assuring confidence in all other downstream parameters (i.e., confidence in both protein identification and quantitation), while the ability to quantify differences between any two samples largely depends on the reproducibility of the overall platform. Due to inherent variations that stem from sample preparation and instrument analysis, technical replicates are often performed to evaluate and minimize technical variability arising from the overall analysis pipeline. Technical variability will be minimized as technologies continue to mature and platforms will likely become more robust and reproducible; however, biological variability within the same comparative groups remains as a challenge for identifying real differences between different conditions. Although ideally one would like to either control or minimize such biological variability by utilizing more controlled model systems such as cell cultures, an in vitro model system, or even inbred mouse strains, this is not always possible. Most clinical studies are based on “real world” human clinical samples where inherent human individual heterogeneity makes discovery efforts more difficult. The human heterogeneity challenge in proteomic studies stems from the high probability that two equally “healthy” individuals will have overall significantly different individual protein abundance levels when sampled at any given time. This heterogeneity can be due to individual genetic variability (i.e., gender, race, etc.) and/or to contributing environmental factors such as diet, overall health, detrimental environmental exposures, etc. The complexity of human diseases presents another degree of challenge. For example, in human cancer, each tumor type typically consists of a number of subtypes that differ with regard to their spectrum of genetic alterations107. Therefore, a potential candidate biomarker of disease may be observed to only elevate in a certain percentage of the pool of disease patients.
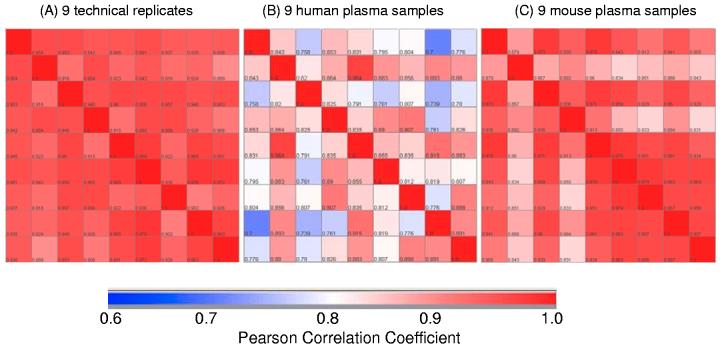
The implications of human heterogeneity in the context of LC-MS based proteomic experiments centers mostly on the measured quantitative values for peptide/protein identifications. Figure 8 shows an initial evaluation of the technical variation and biological variations of human and mouse plasma samples based on the Pearson correlation of the identified peptide intensities between any two individual samples. The technical replicate results (Figure 8A; nine individually processed samples from one pooled reference plasma) show overall good correlation (0.94 ± 0.02), which suggests relatively good reproducibility of the overall analytical platform. The increased variation among human subjects (Figure 8B) appears obvious on the basis of significantly reduced average correlation coefficients (0.85 ± 0.06) compared to the technical replicate results; whereas mouse plasma samples (Figure 8C) show only slightly reduced correlation (0.92 ± 0.05), which suggests relatively small biological variation in these inbred mouse models. Such large variations observed among different healthy control subjects present a challenge for identifying disease-specific differences. To address these challenges and increase the confidence of discovery results, it is essential for the discovery platform to be able to analyze a relatively large number of clinical samples in a high throughput manner to obtain sufficient statistical power.
Figure 8.
Pearson correlation plot comparing peptide intensities of LC-FTICR analyses of plasma samples. (A) Nine technical replicates for a pooled reference plasma sample from multiple healthy subjects. (B) Nine human plasma samples from individual healthy subjects with ages range from 18-26. (C) Nine mouse plasma samples isolated from individual C57BL6 mice. Each sample including the technical replicate was separately processed by ProteomeLab™ IgY-12 (for human) or IgY-R7 (for mouse) depletion and the flow-through portions were digested with trypsin prior to LC-MS analyses.
Other proteomic studies have also described the effects of human heterogeneity in specific model systems. Hu et al. performed a limited study that compared both intra- and inter-individual variability of human cerebrospinal fluid samples obtained from six individuals15. Specific proteins were observed to fluctuate over time with the same individual, but overall there was a higher concordance of inter-individual results than across individuals. Interestingly, results from measuring intra-individual protein levels suggested that certain proteins tended to fluctuate more than others, calling into question the effectiveness of using these proteins as potential disease markers. Other studies include a report by Zhan et al. which showed the heterogeneity in 2DE human pituitary proteome analysis108 and an interesting review by Mann et al. that overviewed the effects of genotypic and phenotypic variations in evaluations of the hemostatic proteome109. They reported that “normal” pro- and anti-coagulant concentrations were observed to vary significantly and influence downstream responses, which demonstrated how heterogeneity in individual phenotypes should influence diagnosis and therapy for hemostatic-based diseases.
Designing experiments to minimize biological variability is imperative for clinical studies. One example is to analyze a serial sample set, i.e., plasma or biopsy tissue samples, from the same individual over a time course or disease progression, which in theory will alleviate a majority of heterogeneity effects, but such samples are traditionally more difficult to obtain in addition to the fact that most patients do not have a “control” blood or tissue sample in storage for comparison against a possible disease diagnosis. For most studies that use cross-sectional approaches, it is desirable to match the patients and controls in terms of age, sex, race, weight, and even diet, if possible. A recent study reported the potential utility of pooling for reducing the effects of biological variation in microarray studies, while retaining the accuracy of identifying differentially expressed genes when biological replicates are retained in the study design and providing the additional benefit of a great reduction in the total number of samples to be analyzed110. Such a strategy might be explored and extended to clinical proteomic studies.
A further implication in heterogeneity is the presence of protein isoforms, splice variants, specific amino acid mutations, proteolytic products, and other post-translational modifications that are likely present in individual samples, but are most often not explicitly included as sequences in the searchable protein database. This exclusion makes it challenging for traditional LC-MS/MS based bottom-up approaches to identify such modified proteins and is possibly one of the main reasons that a large percentage of MS/MS spectra in clinical analyses remain unidentified. The identification of amino acid specific post-translational modifications (e.g., phosphorylation, glycosylation, glycation, nitration, oxidation, and deamination) challenge MS/MS-based approaches due to the vast variety of possible modifications and the potential high false positive rate that originate from database searching. Since it is recognized that many protein biomarkers may be specific protein isoforms or modified proteins, further technical developments for more effective identification and quantitation of protein isoforms and modifications would be greatly desirable.
As an alternative to identifying protein isoforms and modifications, intact protein-level separations can be used to separate different protein isoforms on the basis of their different masses or other properties. The ability to use 2DE for resolving different isoforms and monitoring their abundance changes has been well documented111. The recently developed multi-dimensional intact protein analysis system (IPAS) separates intact proteins on the basis of charge, hydrophobicity, and molecular mass; quantitation is achieved by protein tagging with fluorophores43. The potential for revealing different protein isoforms and specific protein cleavage products in human plasma/serum also has been demonstrated49. The advantages offered by intact protein analysis complements the “bottom-up” proteomic approaches, and better integration of these two approaches may lead to more effective biomarker discovery.
Targeted Proteomic Approaches
The majority of proteomic applications in the search for candidate biomarkers to date have been focused on global proteome characterization focused on identifying multiple protein differences (candidate biomarkers) that correlate with specific human diseases; however, as discussed previously, there are many challenges associated with applying such a strategy to the discovery of low abundance candidate marker proteins. An alternative strategy for biomarker discovery that complements global profiling is the targeted proteomic approach that involves quantitative MS to measure a hypothesis-generated list of candidates112. The targeted proteomic strategy often provides greater sensitivity and allows for detection of low abundance candidate proteins. Anderson et al. recently demonstrated the use of peptide multiple reaction monitoring (MRM) for quantitative assaying of major plasma proteins113. Such MRM assays provide great specificity for peptide/protein identifications and relatively good precision for quantitation. Additionally, MRM can provide a rapid and specific platform for biomarker validation, particularly when coupled with specific enrichment techniques such as the recently published SISCAPA method for enriching target peptides using anti-peptide antibodies114. Activity-based protein profiling is another strategy that uses chemical probes for tagging, enriching, and isolating a specific subset of physiologically important proteins on the basis of enzymatic activity115, 116. Coupling such strategies with LC-MS holds potential for eliminating many issues related to the dynamic range of protein abundance.
A continuing issue for current LC-MS based profiling approaches is that many of the detected species or features from LC-MS and LC-MS/MS analyses remain unidentified. Based on our experience, ∼80% of MS/MS spectra on average are not confidently identified via database searching, and more than 50% of LC-FTICR detected features remain unidentified by the AMT tag approach. Present informatics tools and statistical algorithms have been able to utilize intensity information of these unidentified features to identify “interesting” features as potential biomarkers for specific diseases; effectively targeting these “interesting” features using data-directed or targeted MS/MS approaches is of current interest. One of the informatics challenges associated with identifying these features concerns different post-translational modifications. Current commercial mass spectrometers such as the LTQ offer a targeted MS/MS capability based on the selection of a list of m/z values. Developing an advanced targeted MS/MS approach117 that incorporates “smart selection” of the targets and different, but complementary fragmentation techniques will be an integral component for an effective LC-MS profiling platform suitable for clinical applications.
Conclusions and Perspectives
The amount of effort placed into developments and applications for effective proteomic profiling of serum/plasma and other clinical samples has increased tremendously over the last several years. With the emergence of more effective LC-MS technologies and the variety of fractionation approaches, the number of proteins detectable in human plasma by global profiling has been greatly expanded (e.g., 889 proteins with >95% confidence reported in the recent HUPO study and 1494 proteins with >99% confidence, including confident identification of many low ng/mL level plasma proteins, in our recent study 59. Although this level of detection is still falls short of the 10 orders of magnitude in dynamic range that encompasses plasma protein abundances, it still offers significant potential for the discovery of novel candidate biomarkers from clinical plasma/serum samples.
Currently, there is no single platform that represents the “best” technology for such discovery applications, and integration of multiple technologies is often required for detection and quantitation of low abundance proteins. The need for improved reproducibility, throughput, dynamic range, and quantitation will continue to drive technology development and improvement efforts. Importantly, several new technological developments such as fast LC separations, gas phase IMS separations, and high efficiency nanoESI interfaces look promising for future discovery platforms and applications. With improvements in quantitation accuracy, throughput, and robustness, the LC-MS protein profiling platform may eventually become a powerful tool for clinical diagnostic testing that provides simultaneous measurements of a large number of clinically relevant analytes.
An important component of any integrated profiling platform not previously discussed is the informatics and statistical analysis. The development of more effective software packages will be essential for processing the large number of LC-MS datasets, which may include peak (or feature) detection, run-to-run feature alignment, intensity normalization, feature matching to the database, and statistical analysis to generate a list of high confidence potential candidates.
Finally, due to the complexity of large scale clinical proteomic studies, collaborative efforts from multiple laboratories with different platforms may be required for benchmarking and better cross-validation of the discovery results and eliminating potential biases introduced into any given platform. This implies that a common set of standards is needed so that platform performance in different laboratories may be readily compared and large scale proteomic datasets can be effectively exchanged and shared.
ACKNOWLEDGMENTS
The contributions of Marina Gritsenko, Hongliang Jiang, Matt Monroe, Ron Moore, Tom Metz, Angela Norbeck, Sam Purvine, and Yufeng Shen to the work reviewed here are gratefully acknowledged. We thank the U.S. Department of Energy (DOE) Office of Biological and Environmental Research, the National Institutes of Health, through the National Center for Research Resources (RR018522), NIGMS Large Scale Collaborative Research Grant (U54 GM-62119-02), NIDDK grant R21 DK070146, NIDA grant 1P30DA01562501, the Entertainment Industry Foundation (EIF) and the EIF Women's Cancer Research Fund, and the Laboratory Directed Research Development program at Pacific Northwest National Laboratory for support of portions of the reviewed research. Our laboratories are located in the Environmental Molecular Sciences Laboratory, a national scientific user facility sponsored by the DOE and located at Pacific Northwest National Laboratory, which is operated by Battelle Memorial Institute for the DOE under Contract DE-AC05-76RL0 1830.
Abbreviations
- SCX
strong cation exchange chromatography
- NET
normalized elution time
- AMT
accurate mass and time
- TIF
tumor interstitial fluid
- FDA
Food and Drug Administration
- ESI
electrospray ionization
- FTICR
Fourier transform ion cyclotron resonance
- IMS
ion mobility spectrometry
- IEF
isoelectric focusing
References
- 1.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Hanash S. Disease proteomics. Nature. 2003;422:226–232. doi: 10.1038/nature01514. [DOI] [PubMed] [Google Scholar]
- 3.Etzioni R, Urban N, Ramsey S, McIntosh M, Schwartz S, Reid B, Radich J, Anderson G, Hartwell L. The case for early detection. Nat Rev Cancer. 2003;3:243–252. doi: 10.1038/nrc1041. [DOI] [PubMed] [Google Scholar]
- 4.Ludwig JA, Weinstein JN. Biomarkers in cancer staging, prognosis and treatment selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 5.Anderson NL, Anderson NG. The human plasma proteome: history, character, and diagnostic prospects. Mol. Cell. Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 6.Zhou G, Li H, DeCamp D, Chen S, Shu H, Gong Y, Flaig M, Gillespie JW, Hu N, Taylor PR, Emmert-Buck MR, Liotta LA, Petricoin EF, 3rd, Zhao Y. 2D differential in-gel electrophoresis for the identification of esophageal scans cell cancer-specific protein markers. Mol Cell Proteomics. 2002;1:117–124. doi: 10.1074/mcp.m100015-mcp200. [DOI] [PubMed] [Google Scholar]
- 7.Zangar RC, Varnum SM, Bollinger N. Studying cellular processes and detecting disease with protein microarrays. Drug Metab Rev. 2005;37:473–487. doi: 10.1080/03602530500205309. [DOI] [PubMed] [Google Scholar]
- 8.Janzi M, Odling J, Pan-Hammarstrom Q, Sundberg M, Lundeberg J, Uhlen M, Hammarstrom L, Nilsson P. Serum microarrays for large scale screening of protein levels. Mol Cell Proteomics. 2005;4:1942–1947. doi: 10.1074/mcp.M500213-MCP200. [DOI] [PubMed] [Google Scholar]
- 9.Uhlen M, Bjorling E, Agaton C, Szigyarto CA, Amini B, Andersen E, Andersson AC, Angelidou P, Asplund A, Asplund C, Berglund L, Bergstrom K, Brumer H, Cerjan D, Ekstrom M, Elobeid A, Eriksson C, Fagerberg L, Falk R, Fall J, Forsberg M, Bjorklund MG, Gumbel K, Halimi A, Hallin I, Hamsten C, Hansson M, Hedhammar M, Hercules G, Kampf C, Larsson K, Lindskog M, Lodewyckx W, Lund J, Lundeberg J, Magnusson K, Malm E, Nilsson P, Odling J, Oksvold P, Olsson I, Oster E, Ottosson J, Paavilainen L, Persson A, Rimini R, Rockberg J, Runeson M, Sivertsson A, Skollermo A, Steen J, Stenvall M, Sterky F, Stromberg S, Sundberg M, Tegel H, Tourle S, Wahlund E, Walden A, Wan J, Wernerus H, Westberg J, Wester K, Wrethagen U, Xu LL, Hober S, Ponten F. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005;4:1920–1932. doi: 10.1074/mcp.M500279-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Adkins JN, Varnum SM, Auberry KJ, Moore RJ, Angell NH, Smith RD, Springer DL, Pounds JG. Toward a human blood serum proteome: analysis by multidimensional separation coupled with mass spectrometry. Mol. Cell Proteomics. 2002;1:947–955. doi: 10.1074/mcp.m200066-mcp200. [DOI] [PubMed] [Google Scholar]
- 11.Jacobs JM, Adkins JN, Qian WJ, Liu T, Shen Y, Camp DG, 2nd, Smith RD. Utilizing human blood plasma for proteomic biomarker discovery. J Proteome Res. 2005;4:1073–1085. doi: 10.1021/pr0500657. [DOI] [PubMed] [Google Scholar]
- 12.Veenstra TD, Conrads TP, Hood BL, Avellino AM, Ellenbogen RG, Morrison RS. Biomarkers: mining the biofluid proteome. Mol Cell Proteomics. 2005;4:409–418. doi: 10.1074/mcp.M500006-MCP200. [DOI] [PubMed] [Google Scholar]
- 13.Lee HJ, Lee EY, Kwon MS, Paik YK. Biomarker discovery from the plasma proteome using multidimensional fractionation proteomics. Curr Opin Chem Biol. 2006;10:42–49. doi: 10.1016/j.cbpa.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 14.Wright ME, Han DK, Aebersold R. Mass spectrometry-based expression profiling of clinical prostate cancer. Mol Cell Proteomics. 2005;4:545–554. doi: 10.1074/mcp.R500008-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Hu Y, Malone JP, Fagan AM, Townsend RR, Holtzman DM. Comparative proteomic analysis of intra- and interindividual variation in human cerebrospinal fluid. Mol. Cell. Proteomics. 2005;4:2000–2009. doi: 10.1074/mcp.M500207-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Wattiez R, Falmagne P. Proteomics of bronchoalveolar lavage fluid. J Chromatogr B Analyt Technol Biomed Life Sci. 2005;815:169–178. doi: 10.1016/j.jchromb.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 17.Liao H, Wu J, Kuhn E, Chin W, Chang B, Jones MD, O'Neil S, Clauser KR, Karl J, Hasler F, Roubenoff R, Zolg W, Guild BC. Use of mass spectrometry to identify protein biomarkers of disease severity in the synovial fluid and serum of patients with rheumatoid arthritis. Arthritis Rheum. 2004;50:3792–3803. doi: 10.1002/art.20720. [DOI] [PubMed] [Google Scholar]
- 18.Varnum SM, Covington CC, Woodbury RL, Petritis K, Kangas LJ, Abdullah MS, Pounds JG, Smith RD, Zangar RC. Proteomic Characterization of Nipple Aspirate Fluid: Identification of Potential Biomarkers of Breast Cancer. Breast Cancer Research and Treatment. 2003;80:87–97. doi: 10.1023/A:1024479106887. [DOI] [PubMed] [Google Scholar]
- 19.Xie H, Rhodus NL, Griffin RJ, Carlis JV, Griffin TJ. A catalogue of human saliva proteins identified by free flow electrophoresis-based peptide separation and tandem mass spectrometry. Mol Cell Proteomics. 2005;4:1826–1830. doi: 10.1074/mcp.D500008-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Theodorescu D, Wittke S, Ross MM, Walden M, Conaway M, Just I, Mischak H, Frierson HF. Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol. 2006;7:230–240. doi: 10.1016/S1470-2045(06)70584-8. [DOI] [PubMed] [Google Scholar]
- 21.Celis JE, Gromov P, Cabezon T, Moreira JM, Ambartsumian N, Sandelin K, Rank F, Gromova I. Proteomic characterization of the interstitial fluid perfusing the breast tumor microenvironment: a novel resource for biomarker and therapeutic target discovery. Mol Cell Proteomics. 2004;3:327–344. doi: 10.1074/mcp.M400009-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Yates JR, III, Eng JK, McCormack AL. Mining genomes: correlating tandem mass spectra of modified and unmodified peptides to sequences in nucleotide databases. Anal. Chem. 1995;67:3202–3210. doi: 10.1021/ac00114a016. [DOI] [PubMed] [Google Scholar]
- 23.Perkins D, Pappin D, Creasy D, London U. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 25.Mayya V, Rezaul K, Cong YS, Han D. Systematic comparison of a two-dimensional ion trap and a three-dimensional ion trap mass spectrometer in proteomics. Mol Cell Proteomics. 2005;4:214–223. doi: 10.1074/mcp.T400015-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolters DA, Washburn MP, Yates JR. An Automated Multidimensional Protein Identification Technology for Shotgun Proteomics. Anal. Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 27.Wang H, Qian WJ, Chin MH, Petyuk VA, Barry RC, Liu T, Gritsenko MA, Mottaz HM, Moore RJ, Camp Ii DG, Khan AH, Smith DJ, Smith RD. Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. J Proteome Res. 2006;5:361–369. doi: 10.1021/pr0503681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tabb DL, MacCoss MJ, Wu CC, Anderson SD, Yates JR. Similarity among tandem mass spectra from proteomic experiments: detection, significance, and utility. Anal. Chem. 2003;75:2470–2477. doi: 10.1021/ac026424o. [DOI] [PubMed] [Google Scholar]
- 29.Smith RD, Anderson GA, Lipton MS, Pasa-Tolic L, Shen Y, Conrads TP, Veenstra TD, Udseth HR. An Accurate Mass Tag Strategy for Quantitative and High Throughput Proteome Measurements. Proteomics. 2002;2:513–523. doi: 10.1002/1615-9861(200205)2:5<513::AID-PROT513>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 30.Qian WJ, Camp DG, Smith RD. High Throughput Proteomics Using Fourier Transform Ion Cyclotron Resonance (FTICR) Mass Spectrometry. Expert Review of Proteomics. 2004;1(1):89–97. doi: 10.1586/14789450.1.1.87. [DOI] [PubMed] [Google Scholar]
- 31.Qian WJ, Monroe ME, Liu T, Jacobs JM, Anderson GA, Shen Y, Moore RJ, Anderson DJ, Zhang R, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Camp DG, Smith RD. Quantitative Proteome Analysis of Human Plasma following in Vivo Lipopolysaccharide Administration Using 16O/18O Labeling and the Accurate Mass and Time Tag Approach. Mol. Cell. Proteomics. 2005;4:700–709. doi: 10.1074/mcp.M500045-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, Smith RD. Probability-Based Evaluation of Peptide and Protein Identifications from Tandem Mass Spectrometry and SEQUEST Analysis: The Human Proteome. J. Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- 33.Tolley L, Jorgenson JW, Moseley MA. Very high pressure gradient LC/MS/MS. Anal Chem. 2001;73:2985–2991. doi: 10.1021/ac0010835. [DOI] [PubMed] [Google Scholar]
- 34.Shen Y, Zhao R, Berger SJ, Anderson GA, Rodriguez N, Smith RD. High-Efficiency Nanoscale Liquid Chromatography Coupled On-line with Mass Spectrometry using Nanoelectrospray Ionization for Proteomics. Anal. Chem. 2002;74:4235–4249. doi: 10.1021/ac0202280. [DOI] [PubMed] [Google Scholar]
- 35.Shen Y, Zhang R, Moore RJ, Kim J, Metz TO, Hixson KK, Zhao R, Livesay EA, Udseth HR, Smith RD. Automated 20 kpsi RPLC-MS and MS/MS with chromatographic peak capacities of 1000-1500 and capabilities in proteomics and metabolomics. Anal Chem. 2005;77:3090–3100. doi: 10.1021/ac0483062. [DOI] [PubMed] [Google Scholar]
- 36.Wilm MS, Mann M. Electrospray and Taylor-Cone theory, Dole's beam of macromolecules at last? International Journal of Mass Spectrometry and Ion Processes. 1994;136:167–180. [Google Scholar]
- 37.Smith RD, Shen Y, Tang K. Ultrasensitive and Quantitative Analyses from Combined Separations-Mass Spectrometry for the Characterization of Proteomes. Accounts of Chemical Research. 2004;37:269–278. doi: 10.1021/ar0301330. [DOI] [PubMed] [Google Scholar]
- 38.Zolotarjova N, Martosella J, Nicol G, Bailey J, Boyes BE, Barrett WC. Differences among techniques for high-abundant protein depletion. Proteomics. 2005;5:3304–3313. doi: 10.1002/pmic.200402021. [DOI] [PubMed] [Google Scholar]
- 39.Huang L, Harvie G, Feitelson JS, Gramatikoff K, Herold DA, Allen DL, Amunngama R, Hagler RA, Pisano MR, Zhang WW, Fang X. Immunoaffinity separation of plasma proteins by IgY microbeads: meeting the needs of proteomic sample preparation and analysis. Proteomics. 2005;5:3314–3328. doi: 10.1002/pmic.200401277. [DOI] [PubMed] [Google Scholar]
- 40.Echan LA, Tang HY, Ali-Khan N, Lee K, Speicher DW. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005;5:3292–3303. doi: 10.1002/pmic.200401228. [DOI] [PubMed] [Google Scholar]
- 41.Cho SY, Lee EY, Lee JS, Kim HY, Park JM, Kwon MS, Park YK, Lee HJ, Kang MJ, Kim JY, Yoo JS, Park SJ, Cho JW, Kim HS, Paik YK. Efficient prefractionation of low-abundance proteins in human plasma and construction of a two-dimensional map. Proteomics. 2005;5:3386–3396. doi: 10.1002/pmic.200401310. [DOI] [PubMed] [Google Scholar]
- 42.Liu T, Qian WJ, Mottaz HM, Gritsenko MA, Norbeck AD, Moore RJ, Purvine SO, Camp DG, 2nd, Smith RD. Evaluation of multi-protein immunoaffinity subtraction for plasma proteomics and candidate biomarker discovery using mass spectrometry. Mol Cell Proteomics. 2006 doi: 10.1074/mcp.T600039-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang H, Clouthier SG, Galchev V, Misek DE, Duffner U, Min CK, Zhao R, Tra J, Omenn GS, Ferrara JL, Hanash SM. Intact-protein-based high-resolution three-dimensional quantitative analysis system for proteome profiling of biological fluids. Mol Cell Proteomics. 2005;4:618–625. doi: 10.1074/mcp.M400126-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, Hanash S. Intact-protein based sample preparation strategies for proteome analysis in combination with mass spectrometry. Mass Spectrom Rev. 2005;24:413–426. doi: 10.1002/mas.20018. [DOI] [PubMed] [Google Scholar]
- 45.Sheng S, Chen D, Van Eyk JE. Multidimensional liquid chromatography separation of intact proteins by chromatographic focusing and reversed phase of the human serum proteome: optimization and protein database. Mol Cell Proteomics. 2006;5:26–34. doi: 10.1074/mcp.T500019-MCP200. [DOI] [PubMed] [Google Scholar]
- 46.Barnea E, Sorkin R, Ziv T, Beer I, Admon A. Evaluation of prefractionation methods as a preparatory step for multidimensional based chromatography of serum proteins. Proteomics. 2005;5:3367–3375. doi: 10.1002/pmic.200401221. [DOI] [PubMed] [Google Scholar]
- 47.Moritz RL, Clippingdale AB, Kapp EA, Eddes JS, Ji H, Gilbert S, Connolly LM, Simpson RJ. Application of 2-D free-flow electrophoresis/RP-HPLC for proteomic analysis of human plasma depleted of multi high-abundance proteins. Proteomics. 2005;5:3402–3413. doi: 10.1002/pmic.200500096. [DOI] [PubMed] [Google Scholar]
- 48.Heller M, Michel PE, Morier P, Crettaz D, Wenz C, Tissot JD, Reymond F, Rossier JS. Two-stage Off-Gel isoelectric focusing: protein followed by peptide fractionation and application to proteome analysis of human plasma. Electrophoresis. 2005;26:1174–1188. doi: 10.1002/elps.200410106. [DOI] [PubMed] [Google Scholar]
- 49.Misek DE, Kuick R, Wang H, Galchev V, Deng B, Zhao R, Tra J, Pisano MR, Amunugama R, Allen D, Walker AK, Strahler JR, Andrews P, Omenn GS, Hanash SM. A wide range of protein isoforms in serum and plasma uncovered by a quantitative intact protein analysis system. Proteomics. 2005;5:3343–3352. doi: 10.1002/pmic.200500103. [DOI] [PubMed] [Google Scholar]
- 50.Tang HY, Ali-Khan N, Echan LA, Levenkova N, Rux JJ, Speicher DW. A novel four-dimensional strategy combining protein and peptide separation methods enables detection of low-abundance proteins in human plasma and serum proteomes. Proteomics. 2005;5:3329–3342. doi: 10.1002/pmic.200401275. [DOI] [PubMed] [Google Scholar]
- 51.Herbert B, Righetti PG. A Turning Point in Proteome Analysis: Sample Prefractionation via Multicompartment Electrolyzers with Isoelectric Membranes. Electrophoresis. 2000;21:3639–3648. doi: 10.1002/1522-2683(200011)21:17<3639::AID-ELPS3639>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 52.Tu CJ, Dai J, Li SJ, Sheng QH, Deng WJ, Xia QC, Zeng R. High-sensitivity analysis of human plasma proteome by immobilized isoelectric focusing fractionation coupled to mass spectrometry identification. J. Proteome Res. 2005;4:1265–1273. doi: 10.1021/pr0497529. [DOI] [PubMed] [Google Scholar]
- 53.Sheng S, Chen D, Van Eyk JE. Multidimensional liquid chromatography separation of intact proteins by chromatographic focusing and reversed phased of the human serum proteome: optimization and protein database. Mol. Cell. Proteomics. 2005;5:26–34. doi: 10.1074/mcp.T500019-MCP200. [DOI] [PubMed] [Google Scholar]
- 54.Jin WH, Dai J, Li SJ, Xia QC, Zou HF, Zeng R. Human plasma proteome analysis by multidimensional chromatography prefractionation and linear ion trap mass spectrometry identification. J Proteome Res. 2005;4:613–619. doi: 10.1021/pr049761h. [DOI] [PubMed] [Google Scholar]
- 55.Liu T, Qian WJ, Strittmatter EF, Camp DG, Anderson GA, Thrall BD, Smith RD. High throughput comparative proteome analysis using a quantitative cysteinyl-peptide enrichment technology. Anal. Chem. 2004;76:5345–5353. doi: 10.1021/ac049485q. [DOI] [PubMed] [Google Scholar]
- 56.Zhang H, Li X.-j., Martin DB, Aerbersold R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 2003;21:660–665. doi: 10.1038/nbt827. [DOI] [PubMed] [Google Scholar]
- 57.Liu T, Qian WJ, Gritsenko MA, Camp DG, 2nd, Monroe ME, Moore RJ, Smith RD. Human plasma N-glycoproteome analysis by immunoaffinity subtraction, hydrazide chemistry, and mass spectrometry. J Proteome Res. 2005;4:2070–2080. doi: 10.1021/pr0502065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang ZP, Hancock WS, Chew TR, Bonilla L. A study of glycoproteins in human serum and plasma reference standards (HUPO) using multilectin affinity chromatography coupled with RPLC-MS/MS. Proteomics. 2005;5:3353–3366. doi: 10.1002/pmic.200401190. [DOI] [PubMed] [Google Scholar]
- 59.Liu T, Qian WJ, Gritsenko MA, Xiao W, Moldawer LL, Kaushal A, Monroe ME, Varnum SM, Moore RJ, Purvine SO, Maier RV, Davis RW, Tompkins RG, Camp DG, 2nd, Smith RD. High dynamic range characterization of the trauma patient plasma proteome. Mol Cell Proteomics. 2006 doi: 10.1074/mcp.M600068-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shen Y, Smith RD, Unger KK, Kumar D, Lubda D. Ultrahigh-throughput proteomics using fast RPLC separations with ESI-MS/MS. Anal Chem. 2005;77:6692–6701. doi: 10.1021/ac050876u. [DOI] [PubMed] [Google Scholar]
- 61.Chen HS, Rejtar T, Andreev V, Moskovets E, Karger BL. High-speed, high-resolution monolithic capillary LC-MALDI MS using an off-line continuous deposition interface for proteomic analysis. Anal Chem. 2005;77:2323–2331. doi: 10.1021/ac048322z. [DOI] [PubMed] [Google Scholar]
- 62.Xie J, Miao Y, Shih J, Tai YC, Lee TD. Microfluidic platform for liquid chromatography-tandem mass spectrometry analyses of complex peptide mixtures. Anal Chem. 2005;77:6947–6953. doi: 10.1021/ac0510888. [DOI] [PubMed] [Google Scholar]
- 63.He B, Regnier F. Microfabricated liquid chromatography columns based on collocated monolith support structures. J Pharm Biomed Anal. 1998;17:925–932. doi: 10.1016/s0731-7085(98)00060-0. [DOI] [PubMed] [Google Scholar]
- 64.Li J, LeRiche T, Tremblay TL, Wang C, Bonneil E, Harrison DJ, Thibault P. Application of microfluidic devices to proteomics research: identification of trace-level protein digests and affinity capture of target peptides. Mol Cell Proteomics. 2002;1:157–168. doi: 10.1074/mcp.m100022-mcp200. [DOI] [PubMed] [Google Scholar]
- 65.Srebalus CA, Li J, Marshall WS, Clemmer DE. Determining synthetic failures in combinatorial libraries by hybrid gas-phase separation methods. J. Am. Soc. Mass Spectrom. 2000;11:352–355. doi: 10.1016/s1044-0305(00)00099-4. [DOI] [PubMed] [Google Scholar]
- 66.Henderson SC, Valentine SJ, Counterman AE, Clemmer DE. ESI/Ion trap/Ion mobility/Time-of-flight mass spectrometry for rapid and sensitive analysis of biomolecular mixtures. Analytical Chemistry. 1999;71:291–301. doi: 10.1021/ac9809175. [DOI] [PubMed] [Google Scholar]
- 67.Valentine SJ, Kulchania M, Srebalus Barnes CA, Clemmer DE. Multidimensional separations of complex peptide mixtures: a combined high-performance liquid chromatography/ion mobility/time-of-flight mass spectrometry approach. Int. J. Mass Spectrom. 2001;212:97–109. [Google Scholar]
- 68.Tang K, Shvartsburg AA, Lee HN, Prior DC, Buschbach MA, Li F, Tolmachev AV, Anderson GA, Smith RD. High-sensitivity ion mobility spectrometry/mass spectrometry using electrodynamic ion funnel interfaces. Anal Chem. 2005;77:3330–3339. doi: 10.1021/ac048315a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shen Y, Jacobs JM, Camp DG, Fang R, Moore RJ, Smith RD, Xiao W, Davis RW, Tompkins RG. High Efficiency SCXLC/RPLC/MS/MS for High Dynamic Range Characterization of the Human Plasma Proteome. Anal. Chem. 2004;76:1134–1144. doi: 10.1021/ac034869m. [DOI] [PubMed] [Google Scholar]
- 70.Anderson NL, Polanski M, Pieper R, Gatlin T, Tirumalai RS, Conrads TP, Veenstra TD, Adkins JN, Pounds JG, Fagan R, Lobley A. The Human Plasma Proteome: A Nonredundant List Developed by Combination of Four Separate Sources. Mol. Cell Proteomics. 2004;3:311–316. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 71.States DJ, Omenn GS, Blackwell TW, Fermin D, Eng J, Speicher DW, Hanash SM. Challenges in deriving high-confidence protein identifications from data gathered by a HUPO plasma proteome collaborative study. Nat Biotechnol. 2006;24:333–338. doi: 10.1038/nbt1183. [DOI] [PubMed] [Google Scholar]
- 72.Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- 73.Tirumalai RS, Chan KC, Prieto DA, Issaq HJ, Conrads TP, Veenstra TD. Characterization of the low molecular weight human serum proteome. Mol. Cell. Proteomics. 2003;2:1096–1103. doi: 10.1074/mcp.M300031-MCP200. [DOI] [PubMed] [Google Scholar]
- 74.Qian WJ, Jacobs JM, Camp DG, II, Monroe ME, Moore RJ, Gritsenko MA, Calvano SE, Lowry SF, Xiao W, Moldawer LL, Davis RW, Tompkins RG, Smith RD. Comparative proteome analyses of human plasma following in vivo lipopolysaccharide administration using multidimensional separations coupled with tandem mass spectrometry. Proteomics. 2005;5:572–584. doi: 10.1002/pmic.200400942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Washburn MP, Wolters D, Yates JR. Large-scale Analysis of the Yeast Proteome by Multidimensional Protein Identification Technology. Nat. Biotechnol. 2001;19:242–247. doi: 10.1038/85686. [DOI] [PubMed] [Google Scholar]
- 76.Xie H, Griffin TJ. Trade-off between high sensitivity and increased potential for false positive peptide sequence matches using a two-dimensional linear ion trap for tandem mass spectrometry-based proteomics. J Proteome Res. 2006;5:1003–1009. doi: 10.1021/pr050472i. [DOI] [PubMed] [Google Scholar]
- 77.Omenn GS, States DJ, Adamski M, Blackwell TW, Menon R, Hermjakob H, Apweiler R, Haab BB, Simpson RJ, Eddes JS, Kapp EA, Moritz RL, Chan DW, Rai AJ, Admon A, Aebersold R, Eng J, Hancock WS, Hefta SA, Meyer H, Paik YK, Yoo JS, Ping P, Pounds J, Adkins J, Qian X, Wang R, Wasinger V, Wu CY, Zhao X, Zeng R, Archakov A, Tsugita A, Beer I, Pandey A, Pisano M, Andrews P, Tammen H, Speicher DW, Hanash SM. Overview of the HUPO Plasma Proteome Project: results from the pilot phase with 35 collaborating laboratories and multiple analytical groups, generating a core dataset of 3020 proteins and a publicly-available database. Proteomics. 2005;5:3226–3245. doi: 10.1002/pmic.200500358. [DOI] [PubMed] [Google Scholar]
- 78.Hood BL, Zhou M, Chan KC, Lucas DA, Kim GJ, Issaq HJ, Veenstra TD, Conrads TP. Investigation of the mouse serum proteome. J. Proteome Res. 2005 doi: 10.1021/pr050107r. ASAP article. [DOI] [PubMed] [Google Scholar]
- 79.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical Statistical Model To Estimate the Accuracy of Peptide Identifications Made by MS/MS and Database Search. Anal. Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 80.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A Statistical Model for Identifying Proteins by Tandem Mass Spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 81.MacCoss MJ, Wu CC, Yates JR. Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal. Chem. 2002;74:5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- 82.Anderson DC, Li W, Payan DG, Noble WS. A new algorithm for the evaluation of shotgun peptide sequencing in proteomics: support vector machine classification of peptide MS/MS spectra and SEQUEST scores. J. Proteome Res. 2003;2:137–146. doi: 10.1021/pr0255654. [DOI] [PubMed] [Google Scholar]
- 83.Fenyo D, Beavis RC. A method for assessing the statistical significance of mass spectrometry-based protein identifications using general scoring schemes. Analytical Chemistry. 2003;75:768–774. doi: 10.1021/ac0258709. [DOI] [PubMed] [Google Scholar]
- 84.Henzel WJ, Billeci TM, Stults JT, Wong SC, Grimley C, Watanabe C. Identifying proteins from two-dimensional gels by molecular mass searching of peptide fragments in protein sequence databases. Proc. Natl. Acad.of Sci., USA. 1993;90:5011–5015. doi: 10.1073/pnas.90.11.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pappin DJ, Hojrup P, Bleasby AJ. Rapid Identification of Proteins by Peptide-Mass Fingerprinting. Current Biology. 1993;3:327–332. doi: 10.1016/0960-9822(93)90195-t. [DOI] [PubMed] [Google Scholar]
- 86.Yates JR, Speicher S, Griffin PR, Hunkapiller T. Peptide mass maps: a highly informative approach to protein identification. Analytical Biochemistry. 1993;214:397–408. doi: 10.1006/abio.1993.1514. [DOI] [PubMed] [Google Scholar]
- 87.Zimmer JS, Monroe ME, Qian WJ, Smith RD. Advances in proteomics data analysis and display using an accurate mass and time tag approach. Mass Spectrom Rev. 2006;25:450–482. doi: 10.1002/mas.20071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Olsen JV, Mann M. Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc Natl Acad Sci U S A. 2004;101:13417–13422. doi: 10.1073/pnas.0405549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Dieguez-Acuna FJ, Gerber SA, Kodama S, Elias JE, Beausoleil SA, Faustman D, Gygi SP. Characterization of mouse spleen cells by subtractive proteomics. Mol Cell Proteomics. 2005;4:1459–1470. doi: 10.1074/mcp.M500137-MCP200. [DOI] [PubMed] [Google Scholar]
- 90.Gao J, Opiteck GJ, Friedrichs MS, Dongre AR, Hefta SA. Changes in the Protein Expression of Yeast as a Function of Carbon Source. J. Proteome Res. 2003;2:643–649. doi: 10.1021/pr034038x. [DOI] [PubMed] [Google Scholar]
- 91.Liu H, Sadygov RG, Yates JR. A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 92.Jacobs JM, Diamond DL, Chan EY, Gritsenko MA, Qian WJ, Stastna M, Camp DG, Rice CM, Carithers RL, Katze MG, Smith RD. Proteome Analysis of Huh-7.5 Cells Containing Full-Length Hepatitis C Virus Replicon and Application to HCV Infected Liver Biopsy Samples. J. Virol. 2005;79:7558–7569. doi: 10.1128/JVI.79.12.7558-7569.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zybailov B, Coleman MK, Florens L, Washburn MP. Correlation of Relative Abundance Ratios Derived from Peptide Ion Chromatograms and Spectrum Counting for Quantitative Proteomic Analysis Using Stable Isotope Labeling. Anal. Chem. 2005 doi: 10.1021/ac050846r. ASAP article. [DOI] [PubMed] [Google Scholar]
- 94.Heller M, Mattou H, Menzel C, Yao X. Trypsin catalyzed 16O-to-18O exchange for comparative proteomics: tandem mass spectrometry comparison using MALDI-TOF, ESI-QTOF, and ESI-ion trap mass spectrometers. J. Am. Soc. Mass Spectrom. 2003;14:704–718. doi: 10.1016/S1044-0305(03)00207-1. [DOI] [PubMed] [Google Scholar]
- 95.Pasa-Tolic L, Jensen PK, Anderson GA, Lipton MS, Peden KK, Martinovic S, Tolic N, Bruce JE, Smith RD. High Throughput Proteome-Wide Precision Measurements of Protein Expression using Mass Spectrometry. J. Am. Chem. Soc. 1999;121:7949–7950. [Google Scholar]
- 96.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate Quantitation of Protein Expression and Site-Specific Phosphorylation. Proc. Natl. Acad. Sci. USA. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 98.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative Analysis of Complex Protein Mixtures Using Isotope -Coded Affinity Tags. Nat. Biotechnol. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 99.Zhang Y, Wolf-Yadlin A, Ross PL, Pappin DJ, Rush J, Lauffenburger DA, White FM. Time-resolved mass spectrometry of tyrosine phosphorylation sites in the epidermal growth factor receptor signaling network reveals dynamic modules. Mol Cell Proteomics. 2005;4:1240–1250. doi: 10.1074/mcp.M500089-MCP200. [DOI] [PubMed] [Google Scholar]
- 100.DeSouza L, Diehl G, Rodrigues MJ, Guo J, Romaschin AD, Colgan TJ, Siu KW. Search for cancer markers from endometrial tissues using differentially labeled tags iTRAQ and cICAT with multidimensional liquid chromatography and tandem mass spectrometry. J Proteome Res. 2005;4:377–386. doi: 10.1021/pr049821j. [DOI] [PubMed] [Google Scholar]
- 101.Wang W, Shaler TA, Norton SM, Hill LR, Becker CH. Orlando, FL, USA: Jun 2-6, 2002. p. 1620. [Google Scholar]
- 102.Chelius D, Bondarenko PV. Quantitative Profiling of Proteins in Complex Mixtures Using Liquid Chromatography and Mass Spectrometry. Journal of Proteome Research. 2002;1:317–323. doi: 10.1021/pr025517j. [DOI] [PubMed] [Google Scholar]
- 103.Fang R, Elias DA, Monroe ME, Shen Y, McIntosh M, Wang P, Goddard CD, Callister SJ, Moore RJ, Gorby YA, Adkins JN, Fredrickson JK, Lipton MS, Smith RD. Differential Label-free Quantitative Proteomic Analysis of Shewanella oneidensis Cultured under Aerobic and Suboxic Conditions by Accurate Mass and Time Tag Approach. Mol Cell Proteomics. 2006;5:714–725. doi: 10.1074/mcp.M500301-MCP200. [DOI] [PubMed] [Google Scholar]
- 104.Tang K, Page JS, Smith RD. Charge competition and the linear dynamic range of detection in electrospray ionization mass spectrometry. J Am Soc Mass Spectrom. 2004;15:1416–1423. doi: 10.1016/j.jasms.2004.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Luo Q, Shen Y, Hixson KK, Zhao R, Yang F, Moore RJ, Mottaz HM, Smith RD. Preparation of 20-mum-i.d. Silica-Based Monolithic Columns and Their Performance for Proteomics Analyses. Anal Chem. 2005;77:5028–5035. doi: 10.1021/ac050454k. [DOI] [PubMed] [Google Scholar]
- 106.Juraschek R, Dulcks T, Karas M. Nanoelectrospray--more than just a minimized-flow electrospray ionization source. J Am Soc Mass Spectrom. 1999;10:300–308. doi: 10.1016/S1044-0305(98)00157-3. [DOI] [PubMed] [Google Scholar]
- 107.Alaiya A, Al-Mohanna M, Linder S. Clinical cancer proteomics: promises and pitfalls. J Proteome Res. 2005;4:1213–1222. doi: 10.1021/pr050149f. [DOI] [PubMed] [Google Scholar]
- 108.Zhan X, Desiderio DM. Heterogeneity analysis of the human pituitary proteome. Clin Chem. 2003;49:1740–1751. doi: 10.1373/49.10.1740. [DOI] [PubMed] [Google Scholar]
- 109.Mann KG, Brummel-Ziedins K, Undas A, Butenas S. Does the genotype predict the phenotype? Evaluations of the hemostatic proteome. J Thromb Haemost. 2004;2:1727–1734. doi: 10.1111/j.1538-7836.2004.00958.x. [DOI] [PubMed] [Google Scholar]
- 110.Kendziorski C, Irizarry RA, Chen KS, Haag JD, Gould MN. On the utility of pooling biological samples in microarray experiments. Proc Natl Acad Sci U S A. 2005;102:4252–4257. doi: 10.1073/pnas.0500607102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sickmann A, Marcus K, Schafer H, Butt-Dorje E, Lehr S, Herkner A, Suer S, Bahr I, Meyer HE. Identification of post-translationally modified proteins in proteome studies. Electrophoresis. 2001;22:1669–1676. doi: 10.1002/1522-2683(200105)22:9<1669::AID-ELPS1669>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 112.Anderson L. Candidate-based proteomics in the search for biomarkers of cardiovascular disease. J Physiol. 2005;563:23–60. doi: 10.1113/jphysiol.2004.080473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Anderson L, Hunter CL. Quantitative Mass Spectrometric Multiple Reaction Monitoring Assays for Major Plasma Proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- 114.Anderson NL, Anderson NG, Haines LR, Hardie DB, Olafson RW, Pearson TW. Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA) J Proteome Res. 2004;3:235–244. doi: 10.1021/pr034086h. [DOI] [PubMed] [Google Scholar]
- 115.Berger AB, Vitorino PM, Bogyo M. Activity-based protein profiling: applications to biomarker discovery, in vivo imaging and drug discovery. Am J Pharmacogenomics. 2004;4:371–381. doi: 10.2165/00129785-200404060-00004. [DOI] [PubMed] [Google Scholar]
- 116.Speers AE, Cravatt BF. Chemical strategies for activity-based proteomics. Chembiochem. 2004;5:41–47. doi: 10.1002/cbic.200300721. [DOI] [PubMed] [Google Scholar]
- 117.Masselon C, Pasa-Tolic L, Tolic N, Anderson GA, Bogdanov B, Vilkov AN, Shen Y, Zhao R, Qian WJ, Lipton MS, Camp DG, 2nd, Smith RD. Targeted comparative proteomics by liquid chromatography-tandem fourier ion cyclotron resonance mass spectrometry. Anal Chem. 2005;77:400–406. doi: 10.1021/ac049043e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Andersen JS, Lam YW, Leung AK, Ong SE, Lyon CE, Lamond AI, Mann M. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]