Abstract
While immunoglobulin G (IgG) antibodies to double-stranded (ds)DNA are serological markers of systemic lupus erythematosus (SLE), not all antibodies to DNA (anti-DNA) are able to cause tissue damage to a similar extent. It has been proposed that anti-DNA-induced renal damage could be linked to differences in the fine specificity of the antibodies. In an attempt to gain insight into their fine binding properties, we investigated the cross-reactivity of two human lupus monoclonal IgG anti-dsDNA (B3 and RH14) to a recently described Escherichia coli PolIV (a DNA polymerase). These autoantibodies possess distinct pathogenic properties in severe combined immunodeficient (SCID) mice. Although both antibodies cause proteinuria, only RH14 induces early histological features of lupus nephritis. Both RH14 and B3 bound PolIV; however, they exhibited a marked difference in their reactivity to the PolIV–dsDNA complex. Alhough RH14 exhibited significant activity to the complex, the binding of B3 to PolIV complexed with dsDNA was almost abolished. Furthermore, there was a significant difference in the way the lupus sera recognized naked dsDNA and that presented on PolIV. Although 67% of lupus sera bound naked dsDNA, ≈ 90% of these sera (93% calf thymus DNA; 90% synthetic oligonucleotide) reacted to the complex when dsDNA was presented on PolIV. Thus, the IgG anti-dsDNA likely to exist in lupus patients may be distinguished into those that recognize dsDNA in the context of PolIV and those which do not. This difference in binding ability may help to distinguish those dsDNA antibodies that are more pathogenic.
Keywords: autoantibodies, autoimmunity, human, lupus, PolIV
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune rheumatic disease affecting principally women during their childbearing years. Immunoglobulin G (IgG) antibodies to double-stranded (ds)DNA are serological markers of SLE that often reflect disease activity1, 2 and are closely associated with its pathogenesis.3 These antibodies are considered to be specific for lupus as they are rarely found in other disease conditions.4 However, both clinical and animal model studies clearly show that not all antibodies to DNA (anti-DNA) are equally able to cause tissue damage in SLE. Anti-DNA-induced renal damaging capacity could be linked to differences in their fine specificities (reviewed in ref. 5). By using two prototypic human monoclonal antibodies (mAbs) (B3 and RH14) available in our laboratory, we investigated the properties that make an anti-DNA pathogenic. These antibodies were derived from patients with lupus, who had characteristic disease manifestations6, 7 and possessed distinct and diverse pathogenic properties. In severe combined immunodeficient (SCID) mice, although both types of antibodies cause proteinuria, only RH14 induces early histological features of lupus nephritis detectable by electron microscopy.
In an attempt to gain insight into their fine binding properties, we investigated the cross-reactivity of RH14 and B3 to 7 professional DNA-binding proteins (DNA polymerases and DNA-repair enzymes), including three non-specific (not requiring a specific sequence or length of DNA for binding) and four specific DNA-binders. All three of the nonspecific DNA-binders, but none of the specific DNA-binders, exhibited binding to the synthetic oligonucleotide and, in turn, comparable binding to autoantibodies, in the solid phase (S. Kumar et al., unpublished observations). One of these molecules presently described –Escherichia coli PolIV – is representative of the enzymes studied, and is a member of the newly classified Y-family of DNA polymerases.8 These polymerases display poor processivity, low fidelity, lack any detectable proofreading activity and are involved in the bypass of a variety of DNA lesions that stall the main replicative polymerase.9 PolIV has been shown to extend misaligned primer–template termini.10 PolIV is a well-characterized protein8–12 with its homologues representing key enzymes of a variety of pathogenic genera, including Bacillus, Staphylococcus, Enterococcus, Streptococcus, Mycobacterium, Bordetella, Neisseria, Escherichia, Klebsiella, Legionella, Pasteurella, Pseudomonas, Salmonella, Yersinia, Treponema, Plasmodium and Trypanosoma.13 Recent advances include the discovery that PolIV is a special error-prone DNA polymerase required for adaptive point mutation.11 Humans possess three Y-family polymerases, including a DinB homologue, now designated PolKappa.12 As bacterial infection has been implicated previously as one of the causative agents for autoimmune disease (reviewed in ref. 14), PolIV is of particular interest as, in addition to being of bacterial origin, PolIV homologues have been proposed to contribute to adaptive strategies of pathogens, including antigenic variation.13 Furthermore, PolIV can be overexpressed in E. coli, purified to homogeneity and, most importantly, is able to retain its characteristic DNA-binding ability when bound in a solid phase. Antibodies to DNA-binding proteins associated with DNA metabolism have been shown to occur in lupus sera.15 It is known that antibodies directed to such autoantigens recognize conformation-dependent epitopes that represent active sites and functional domain regions.16 We have recently purified PolIV to homogeneity and have optimized the conditions that allow the enzyme to achieve its correct conformation when bound in a solid phase. The successful adaptation of these conditions in enzyme-linked immunosorbent assays (ELISAs) allowed us to carry out the present investigations.
We now demonstrate that two distinct populations of IgG anti-DNA exist in lupus patients – the population that recognizes dsDNA in the context of PolIV is highly prevalent in lupus sera. We also demonstrate that different subsets of lupus autoantibodies recognize qualitatively different DNAs (calf thymus or synthetic oligonucleotide) presented on PolIV, differently.
Materials and methods
Preparation of PolIV
E. coli PolIV was polymerase chain reaction (PCR) amplified by using the genomic DNA prepared from JM109. The gene, carried on an adapted version of pET28, was N-terminally His6-tagged and expressed in E. coli B834 DE3 cells. Freshly transformed cells, streaked on Luria–Bertani (LB) plates containing 50 µg/ml of kanamycin, were grown overnight at 37° and then used to inoculate 1 L of LB broth, also containing 50 µg/ml of kanamycin. The cells were grown at 37° with agitation until the optical density at 600 nm (OD600) reached 0·4. The temperature was reduced to 20° and the cultures were induced with isopropyl thio-β-d-galactoside (IPTG) (final concentration 1 mm) for 1 hr, prior to harvesting by centrifugation. Cells were resuspended in 20 mm Tris, pH 8·0, containing 1 m NaCl, prior to disruption using an EmulsiFlex-C5 high pressure homogeniser (Avestin, Ottawa, Canada). Cell debris was removed by centrifugation and the supernatant was applied to Talon metal affinity resin (Clontech, Palo Alto, CA) equilibrated in 20 mm Tris, pH 8·0, containing 0·5 m NaCl and 5 mm imidazole. The resin was washed in the same buffer and the protein was eluted in buffer containing 300 mm imidazole. Buffer exchange into 50 mm HEPES, pH 7·0, containing 2 mm MgCl2 and 100 mm NaCl, was performed by using 5-ml HiTrap desalting columns (Pharmacia, Dublin, VA). PolIV was concentrated, via loading, onto a 1-ml SP-sepharose HiTrap column (Pharmacia) equilibrated in the same buffer, followed by elution in buffer containing 50 mm HEPES, pH 7·0, containing 2 mm MgCl2 and 1 m NaCl. To determine the purity of the protein, 6xHis-PolIV (7·5 µg per lane) was separated by sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) on 12% acrylamide gels.
Electrophoretic mobility shift assay
Two synthetic oligonucleotides were designed to form a single primer-template terminus. The primers (5′-CGGTGTCGC-3′ and 5′-GTUGTTGGGCGACACCG-3′) were mixed in a equimolar ratio and heated at 95° for 10 min, incubated at 65° for 10 min, then cooled at 1° per min until a temperature of 15° was obtained. The substrate was mixed in a relatively higher molar excess (1·2: 1) with purified 6xHis-PolIV in 50 mm HEPES, pH 7·0, containing 2 mm MgCl2 and 200 mm NaCl. Each lane contained 6 µg of protein or equivalent amounts of oligonucleotide. Reactions were incubated on ice for 30 min prior to mixing with 6× DNA sample buffer (40% glycerol, 0·25% bromophenol blue). Samples were then run in duplicate (at 200 V and 4°) on an 8% non-denaturing gel in 1 × Tris-Borate-EDTA (TBE). Half of the gel was stained with Coomassie Blue and the other half with ethidium bromide in order to visualize protein and DNA, respectively.
Preparation of IgG
Culture supernatants containing B3 and RH14 IgG were prepared from the hybridomas, as described previously.6 The antibodies were purified via loading onto a protein G column (protein G–Sepharose 4 Fast Flow; Amersham Pharmacia Biotech, Bucks., UK) and elution in 100 mm glycine–HCl buffer, pH 2·7.
Sera from SLE patients and healthy individuals
Thirty sera were studied from the patients attending our SLE clinic. Each patient met four or more of the revised American College of Rheumatology criteria for the disease.17 Of these 30 patients, 15 had biopsy-proven evidence of nephritis, whereas the other 15 did not have renal involvement. Ten sera from normal healthy individuals (controls), were analysed for the presence of reactivity against PolIV and DNA presented on PolIV. The female: male ratios in these three groups were as follows: 13: 2 (with lupus nephritis), 15: 0 (without lupus nephritis) and 10: 0 (controls). The ages [mean + standard deviation (years)] of the patients in the three groups were as follows: 34·7 + 10·9 (with lupus nephritis), 38·3 + 14·0 (without lupus nephritis) and 41·4 + 9·0 (controls).
Assessment of the functional activity (DNA binding) of PolIV in a solid phase
The prerequisite for the solid-phase binding assay was the ability to retain the DNA-binding activity of the enzyme PolIV when bound to polystyrene plates. We have recently optimized the conditions which allow this protein to achieve its correct conformation and to exhibit its characteristic DNA-binding features when bound to polystyrene plates (data not shown). The functional activity of PolIV, as a measure of its achievement of correct conformation in a solid phase, was evaluated by its ability to bind a previously described biotinylated 35mer synthetic oligonucleotide in its single-stranded (ss) or double-stranded (ds) form.18 The biotinylated strand (5′-biotin-CCGAATCAGTTCACTTCCAGCCCAGGTATTTAGCC-3′) and the non-biotinylated complementary strand were annealed (pH 7, 1 hr, 37°) by using a hybridization buffer (1 m sodium chloride, 20 mm monosodium phosphate, 0·1 mm EDTA), as recommended by Amersham Pharmacia Biotech Biosensor AB (Application Note 306, 1995), to achieve a final dsDNA concentration of 4 mg/ml.
Polystyrene plates were coated with the purified protein (20 µg/ml, 250 µl/well, overnight at 4°) prepared in a modified Pfu buffer (20 mm Trizma base, pH 8·8, containing 10 mM potassium chloride, 10 mm ammonium sulphate, 2 mm magnesium sulfate and 2 mm magnesium chloride). The plates were washed (× 3) and blocked [in the modified Pfu buffer containing 2% bovine serum albumin (BSA), at 37° for 2 hr]. One-third of the wells were treated with ss calf thymus DNA and one-third of the wells with ds calf thymus DNA (20 µg/ml, prepared in the modified Pfu buffer) at 37° for 30 min. The ss or ds biotinylated oligonucleotide (20 µg/ml, prepared in the modified Pfu buffer, 37°, 30 min) was then applied to DNA treated and untreated wells. The plates were washed (six times) and binding was detected by incubation with alkaline phosphatase-conjugated streptavidin (1 µg/ml, prepared in the modified Pfu buffer containing 100 µg/ml of BSA) at 37° for 1 hr. The modified Pfu buffer containing 100 µg/ml of BSA was used throughout for washing of the plates.
Assessment of the binding of RH14 and B3 to PolIV and to calf thymus DNA presented on PolIV in a solid phase
Polystyrene plates were coated with the recombinant PolIV, and then two-thirds of the wells were treated with calf thymus ssDNA and dsDNA, washed three times and blocked as described above. RH14 and B3 (5 µg/ml, diluted in the modified Pfu buffer) were applied (37°, 1 hr) to the wells treated or untreated with DNA. The plates were washed six times. The modified Pfu buffer containing 100 µg/ml of BSA, was used throughout for washing of the plates.
Assessment of the binding of dsDNA-treated RH14 and B3 to the PolIV–dsDNA complex in a solid phase
RH14 and B3 (5 µg/ml) were incubated (at 37° for 1 hr) with the modified Pfu buffer alone (control) or with the modified Pfu buffer containing calf thymus ssDNA or dsDNA (20 µg/ml, final concentration). Polystyrene plates were coated with the recombinant PolIV, two-thirds of the wells were treated with calf thymus ssDNA and dsDNA, and then washed (three times) and blocked, as described above. The antibody samples, prepared as decribed above, were applied (at 37° for 1 hr) to the wells treated or untreated with DNA. The plates were washed (six times). The modified Pfu buffer containing 100 µg/ml of BSA was used throughout for washing of the plates.
Assessment of the binding of RH14 and B3 antibodies to poly l-lysine and to calf thymus dsDNA presented on poly l-lysine in a solid phase
Polystyrene plates were coated with 50 µg/ml of poly l-lysine (at 37° for 1 hr) prepared in deionized water (dH2O) and washed three times in phosphate-buffered saline (PBS). Half of the wells were treated with calf thymus dsDNA (20 µg/ml, prepared in dH2O) at 37° for 1 hr, the plates were washed three times in PBS and then blocked at 37° for 1 hr with PBS containing 2% casein. RH14 and B3 (5 µg/ml, diluted in PBS) were applied at 37° for 1 hr. The plates were subsequently washed four times in PBS-Tween.
Binding of the antibodies to PolIV or to poly l-lysine alone, or to these proteins complexed with dsDNA, was detected by incubation at 37° for 1 hr, with alkaline phosphatase-conjugated anti-human lambda (1: 1000, prepared either in the modified Pfu buffer containing 100 µg/ml of BSA for PolIV, or in PBS-Tween for poly l-lysine).
By using the following equation, the Pol IV-specific binding of the mAbs was determined after subtracting the background absorbance (A) values from those of the wells containing PolIV:
| (1) |
Assessment of the binding of serum antibodies to calf thymus dsDNA
The sera were tested in a standard anti-DNA ELISA, as described previously, 19 with minor modifications. In brief, polystyrene plates were coated (4°, overnight) with calf thymus dsDNA (previously purified by filtration on a 0·45-µm filter) and blocked for 1 hr at 37° with PBS containing 2% BSA and 2% casein. The serum samples were then applied (1: 300, in PBS) and incubated for 1 hr at 37°. The plates were washed and the binding of serum immunoglobulins to dsDNA was assessed as described above.
The binding of antibodies to dsDNA was determined following the subtraction of background A values from those of the sera tested. British Isles Lupus Assessment Group (BILAG) activity scores for the lupus patients were recorded (Table 1).20 The activity index divides disease activity into eight organs/systems. It is based on the physician's intention-to-treat principle, and the activity of each organ/system is scored from A (the most active) to E (no activity now or previously). BILAG can be converted into a global score by using the following scoring system: A = 9, B = 3, C = 1 and D/E = 0.
Table 1.
Summary of the patients with lupus, their renal British Isles Lupus Assessment Group (BILAG) scores and a comparison of serum antibody reactivity to naked and complexed double-stranded (ds)DNA in an enzyme-linked immunosorbent assay (ELISA)
| Comparative assessment | ||||||||
|---|---|---|---|---|---|---|---|---|
| BILAG score | PolIV-DNA | |||||||
| Serum no. | Age | Gender | Renal | Global | Naked DNA | Calf thymus | Oligo | Major immunosuppressive drug |
| Test sera | ||||||||
| 1 | 33 | F | C | 4 | +++ | ++++ | ++++ | – |
| 2 | 53 | M | B | 7 | ++ | ++ | ++ | Azathiopine 200 mg/day |
| 3 | 28 | F | B | 8 | +++ | +++ | + | – |
| 4 | 35 | F | B | 6 | ++ | + | + | – |
| 5 | 57 | F | C | 11 | – | ++++ | ++++ | – |
| 6 | 40 | F | C | 3 | ++ | ++ | + | Cellsept 2 g/day |
| 7 | 26 | M | B | 4 | – | + | + | Azathiopine 150 mg/day |
| 8 | 37 | F | B | 6 | +++ | ++++ | ++++ | Cyclophosphamide 750 i.v. |
| 9 | 24 | F | B | 6 | +++ | +++ | ++++ | – |
| 10 | 44 | F | B | 5 | – | + | – | Azathiopine 150 mg/day |
| 11 | 40 | F | D | 3 | – | – | – | Azathiopine 150 mg/day |
| 12 | 28 | F | B | 7 | +++ | +++ | + | Cyclophosphamide 750 i.v. |
| 13 | 20 | F | D | 3 | – | +++ | ++ | Cyclophosphamide 750 i.v. |
| 14 | 21 | F | B | 9 | – | – | – | Azathiopine 125 mg/day |
| 15 | 34 | F | C | 8 | ++ | ++ | ++ | – |
| 16 | 62 | F | E | 11 | + | +++ | +++ | – |
| 17 | 32 | F | E | 12 | – | – | + | – |
| 18 | 33 | F | E | 1 | – | ++++ | ++ | – |
| 19 | 57 | F | E | 8 | +++ | +++ | + | – |
| 20 | 46 | F | E | 6 | ++ | ++ | – | Azathiopine 150 mg/day |
| 21 | 48 | F | E | 3 | ++ | ++ | ++ | – |
| 22 | 37 | F | E | 4 | ++ | ++ | + | – |
| 23 | 24 | F | E | 4 | – | +++ | + | – |
| 24 | 25 | F | E | 5 | + | +++ | + | – |
| 25 | 31 | F | E | 7 | ++ | ++ | + | – |
| 26 | 24 | F | E | 6 | – | ++ | + | Azathiopine 50 mg/day |
| 27 | 24 | F | E | 5 | ++ | ++ | + | Azathiopine 50 mg/day |
| 28 | 62 | F | E | 5 | ++ | +++ | + | Azathiopine 50 mg/day |
| 29 | 44 | F | E | 6 | + | + | + | Azathiopine 50 mg/day |
| 30 | 25 | F | E | 9 | +++ | +++ | + | – |
| Control sera | ||||||||
| 31 | 37 | F | ||||||
| 32 | 43 | F | ||||||
| 33 | 48 | F | ||||||
| 34 | 42 | F | ||||||
| 35 | 32 | F | ||||||
| 36 | 36 | F | ||||||
| 37 | 51 | F | ||||||
| 38 | 43 | F | ||||||
| 39 | 26 | F | ||||||
| 40 | 56 | F | ||||||
No renal disease; insufficient data available to provide a global score.
Assessment of the binding of serum antibodies to dsDNA presented on PolIV
Polystyrene plates were coated with the recombinant PolIV protein, washed three times and blocked, as described above. One-third of the wells were treated with calf thymus dsDNA and one-third of the wells with synthetic oligonucleotide (non-biotinylated) dsDNA (20 µg/ml, prepared in the modified Pfu buffer) at 37° for 1 hr. The plates were washed six times and the serum samples (1: 300, diluted in the modified Pfu buffer) were then applied and incubated at 37° for 1 hr. The plates were washed six times and the binding of the serum antibodies to PolIV, or to PolIV + dsDNA, was detected by incubation at 37° for 1 hr with a monoclonal anti-human IgG (Fc specific) alkaline phosphatase conjugate (1: 1000, prepared in the modified Pfu buffer containing 100 µg/ml of BSA). The modified Pfu buffer containing 100 µg/ml of BSA was used throughout for washing of the plates.
Binding of the antibodies to the PolIV + dsDNA (calf thymus or synthetic oligonucleotide) complex was determined by using the following equation:
| (2) |
The percentage increase in binding of the antibodies to PolIV following its complexing with calf thymus or synthetic oligonucleotide dsDNA, was evaluated by using the following equation:
| (3) |
Comparative assessment of the binding of serum antibodies to naked and to the PolIV complexed dsDNA
The difference in binding of the serum autoantibodies to naked and to PolIV-complexed calf thymus dsDNA was determined by using the following equation:
| (4) |
The difference in percentage increase in binding of the serum autoantibodies to calf thymus or to synthetic oligonucleotide dsDNA complexed with PolIV, was determined by using the following equation:
 |
(5) |
The Pol IV-specific binding of the serum antibodies (data not shown) was determined following the subtraction of background A values from those of the wells containing PolIV, by using the following equation:
| (6) |
Different antigens, including ‘naked’ DNA, PolIV, PolIV + dsDNA and PolIV + synthetic DNA, were coated on the same plate for reliable comparisons of autoantibody reactivities. The presentation of data as the percentage increase in A (autoantibody binding) following complexing of PolIV with DNA, eliminates possible plate-to-plate variability. For comparisons, raw A values are also presented. The binding assays were repeated at least three times and the results obtained were consistent. Two expressions are used to denote background values: ‘plate background’ refers to A values noted for the non-specific binding of antibodies to the plate, whereas ‘background’ refers to the A values exhibited by normal human sera.
The results obtained on the comparative assessment of binding of lupus sera to naked and to PolIV-complexed dsDNA have been summarized in Table 1. The following arbitrary scales were used to assess the sensitivity levels. Calf thymus ‘naked’ dsDNA: A > background to < 1, +; A = 1–1·5, ++; A = 1·5–2, +++; A > 2, ++++. Pol IV–calf thymus dsDNA: A > background to < 0·5, + A = 0·5–1, ++; A = 1–1·5, +++; A > 1·5, ++++. PolIV–oligonucleotide: A > background to < 0·25, +; A = 0·25–0·5, ++; A = 0·5–0·75, +++; A > 0·75, ++++.
The study was approved by University College London Hospitals (UCLH) ethics committee. The participants had given their informed consent. All chemicals were purchased from Sigma Immunochemicals (St Louis, MO), unless mentioned otherwise. Polystyrene plates (Maxisorp, Rochester, NY) were purchased from Nunc (Roskilde, Denmark). Moα-HL was from Immunostics (London, UK).
Results
Loading of 7·5 µg of 6xHis-PolIV onto SDS–PAGE gels yielded a single band (Fig. 1a), confirming that the protein was purified to homogeneity. An electrophoretic mobility shift assay, in a non-denaturing PAGE system, confirmed the binding of purified PolIV to a primer-template terminus in solution. Relative positions of the unbound oligonucleotide and protein controls compared to the DNA–protein complex, are indicated in Fig. 1(b). In the absence of the oligonucleotide, the positive charge on PolIV (theoretical pI of 9·28) prevents migration of the protein into the gel (Fig. 1b, i). Migration of the small, negatively charged oligonucleotide was severely retarded in the presence of PolIV, as a result of the positive charge on the protein (Fig. 1b, ii). The presence of both DNA and protein in the complex are confirmed by duplicate staining in Coomassie Blue (Fig. 1b, i) and ethidium bromide (Fig. 1b, ii).
Figure 1.
(a) Sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS–PAGE) gel profile of the purified PolIV. The gel was stained with Coomassie Blue. PolIV resolves at 40 500 molecular weight (MW) – the correct size of the molecule. Lane 1, MW markers; and lane 2, purified PolIV (7·5 µg). (b)Electrophoretic mobility shift assay in a non-denaturing PAGE system, demonstrating binding of purified PolIV to a primer-template terminus. Relative positions of the unbound oligonucleotide and of the protein controls are indicated, compared to the DNA–protein complex. Migration of the small, negatively charged oligonucleotide is severely retarded in the presence of PolIV, owing to the positive charge on the protein (theoretical pI of 9·28). In the absence of oligonucleotide, the positive charge on PolIV prevents migration of the protein into the gel. The presence of both DNA and protein in the complex is confirmed by duplicate staining in Coomassie Blue (i) and ethidium bromide (ii). Lane 1, oligonucleotide alone (control, amount equivalent to oligonucelotide present in lane 2); lane 2, PolIV–oligonucleotide complex (6 µg of protein); and lane 3, PolIV alone (control, 6 µg of protein).
The enzyme bound both ssDNA and dsDNA in a binding assay adopted in ELISA format (Fig. 2a). The binding of ssDNA was higher than that of dsDNA. The binding of the oligonucleotides was almost completely abolished following pretreatment of PolIV with calf thymus ssDNA or dsDNA. The results obtained suggest that the enzyme was functionally active, at least to the extent that it bound DNA. Adapting these conditions for use in an ELISA allowed us to measure reliably the antibody activity to conformation-dependent PolIV epitopes and, in particular, to the PolIV–DNA complex. Both RH14 and B3 exhibited significant reactivity to PolIV (Fig. 2b). RH14 exhibited a significant increase in the binding, whereas B3 binding was almost abolished when DNA was presented on PolIV, indicating that this complex is not recognized by all anti-dsDNA. In contrast, the binding of both of the antibodies was significantly increased when dsDNA was presented on a non-specific binder protein, poly l-lysine (Fig. 2c). The results suggest that the observed inhibition in the binding of B3 to PolIV–dsDNA was not caused by the presentation of dsDNA as a complex with any protein, and that the PolIV–dsDNA complex could identify differences in the reactivity of human anti-dsDNA. To investigate whether the increased binding of RH14 to the PolIV–dsDNA complex was caused by its reactivity to dsDNA and/or to a novel conformation-dependent epitope that may have appeared on PolIV following its binding to dsDNA, the binding of dsDNA-preblocked RH14 and B3 (used as a control), to the PolIV–dsDNA complex, was evaluated. The binding of the preblocked RH14 to the PolIV–dsDNA complex was almost completely abolished (Fig. 2d), suggesting that the binding of RH14 was exclusively directed towards the dsDNA bound to PolIV.
Figure 2.
(a) Binding of the 35mer biotinylated oligonucleotides in their single-stranded (ss) or double-stranded (ds) forms, to PolIV alone or to PolIV-preblocked calf thymus dsDNA, in a solid phase. (b) Binding of the monoclonal human dsDNA antibodies RH14 and B3 to PolIV or PolIV + ds/ss DNA or (c) to poly-l-lysine or poly-l-lysine + dsDNA, in a solid phase. (d) Binding of calf thymus ds/ssDNA-preblocked RH14 and B3 to PolIV or PolIV + ds/ss DNA, in a solid phase.
The upper limit of the reactivity (mean A405 + 2 standard deviations) of the normal sera in the anti-dsDNA assay was set at 0·871 for naked dsDNA, at 0·227 for the calf thymus dsDNA–PolIV complex, or at 0·108 for the oligonucleotide–PolIV complex. A significant proportion of lupus sera had high levels of antibodies to naked dsDNA (67%, Fig. 3) or to that presented on PolIV (≈ 90%, Fig. 4a,4c), compared to the normal sera. Sera from two healthy controls (numbers 38 and 39; Fig. 3) exhibited high normal binding to the naked dsDNA, resulting in the high background in the anti-DNA assay. Importantly, however, the binding of these sera to the DNA presented on PolIV was substantially reduced. Notably, 93% (calf thymus, Fig. 4a, 4b) and 90% (synthetic oligonucleotide, Fig. 4c,4d) of the lupus sera showed a significant percentage increase in their reactivity to the PolIV–dsDNA complex over their reactivity to PolIV alone, compared to the control sera. However, the reactivity of many sera to the PolIV–oligonucleotide dsDNA was significantly reduced compared with that to PolIV–calf thymus dsDNA. Although most of the normal healthy sera had significant binding to naked dsDNA (Fig. 3), their reactivity to the PolIV–dsDNA complex was significantly reduced (Fig. 4a).
Figure 3.
Binding of 30 sera from the patients with lupus (numbers 1–15 with nephritis; 16–30, without nephritis) and of 10 sera from normal healthy individuals (numbers 31–40) to naked calf thymus double-stranded (ds)DNA in a traditional enzyme-linked immunosorbent assay (ELISA). The highest absorbance (A) at 405 nm (0·763) recorded for the normal sera, was used as the background and the lower limit to determine the anti-naked dsDNA activity of lupus sera.
Figure 4.
Binding of 30 sera from the patients with lupus (numbers 1–15 with or numbers 16–30 without lupus nephritis) and of 10 sera from normal healthy individuals (numbers 31–40) to a PolIV–calf thymus double-stranded (ds)DNA complex (a) or to a PolIV–oligonucleotide dsDNA complex (c), and the percentage increase in their binding to these complexes, in relation to their binding to PolIV alone, following the presentation of calf thymus (b)or oligonucleotide (d)dsDNA on PolIV.
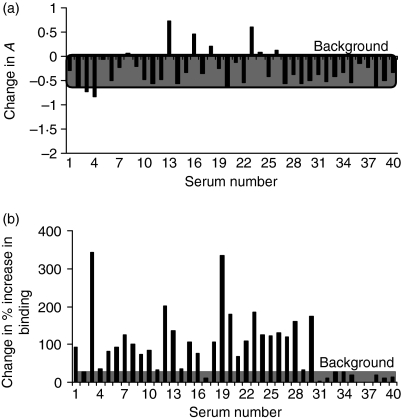
Seven of the 30 sera (23%) from patients with SLE showed increased (Fig. 5a) binding to the PolIV–dsDNA complex (Fig. 4a) compared with binding to naked dsDNA (Fig. 3). There was a marked difference in the serum reactivity to PolIV complexed with calf thymus dsDNA or synthetic oligonucleotide dsDNA (Fig. 5b); 28 of the 30 sera had significantly higher binding to the former. Six out of nine patients who exhibited higher than ++ levels of activity against PolIV–oligonucleotide dsDNA belonged to the B or C category of the renal BILAG score, with nearly half of the patients with a B or a C score possessing higher than ++ levels of activity to this complex (Table 1). A description of the patients and a comparison of the serum autoantibody reactivity to naked or to PolIV-complexed dsDNA (ELISA), is summarized in Table 1.
Figure 5.
A comparison of the binding of 30 sera from patients with lupus (numbers 1–15 with or numbers 16–30 without lupus nephritis) and of 10 sera from normal healthy individuals (numbers 31–40), to naked double stranded (ds)DNA and to a PolIV–dsDNA complex. (a) The difference in binding of the serum autoantibodies to naked or to PolIV-complexed calf thymus dsDNA was determined by using the following equation: Change in A = (APolIV+dsDNA − APolIV) − (AdsDNA –Aplate background). (b)The difference in percentage increase in the binding of the serum autoantibodies to calf thymus or to synthetic oligonucleotide dsDNA complexed with PolIV, was determined by using the following equation: Change in percentage increase in binding = (% increase in binding to PolIV–calf thymus dsDNA) − (% increase in binding to PolIV-oligonucleotide dsDNA).
Discussion
Both RH14 and B3 bind to PolIV, suggesting that the two antibodies recognize an epitope present on the ‘empty’ molecule (i.e. not complexed to DNA). However, the distinct differential binding of B3 and RH14 to PolIV–dsDNA complexes establishes that by this criterion, the two antibodies probably represent different populations of anti-dsDNA in patients with lupus. Similar results were obtained using two other ‘professional’ DNA-binding proteins: B3 lost its anti-DNA activity, whereas RH14 continued to exhibit significant anti-DNA activity against DNA presented on either a different DNA polymerase or a DNA repair enzyme (data not shown), suggesting that the observed autoantibody reactivity was not unique to the PolIV–DNA complex. The loss of anti-PolIV activity of B3 in the context of the PolIV–dsDNA complex suggests that this antibody recognizes an epitope present only on PolIV itself. This epitope may be lost in the complex (owing to the conformational change) formed by PolIV binding to dsDNA. Given that the formation of the complex may give rise to novel, conformation-dependent epitopes, we investigated whether the observed high reactivity of RH14 to the complex was directed towards the bound dsDNA and/or to a novel conformation-dependent epitope(s). The loss of anti-PolIV dsDNA activity of dsDNA-preblocked RH14 (Fig. 2d) suggests that the observed reactivity of RH14 to the PolIV–dsDNA complex was directed towards the bound dsDNA, and that, like B3, the epitope recognized by RH14 on ‘empty’ PolIV was also conformation-dependent. Whether both the antibodies recognized the same epitope on PolIV is not known. RH14-type anti-dsDNA can thus react to dsDNA presented on a DNA-binding protein, whereas B3-type antibodies are unable to do so. As both RH14 and B3 can bind ‘naked’ dsDNA, these antibodies appear to share an epitope present on dsDNA and ‘empty’ PolIV. Although the shared epitope between the DNA-binding proteins and nucleic acid has not yet been identified, it could arise from the conformational apposition of negative charges in the tertiary structure of such cross-reactive proteins.21 It has been proposed that human S1 protein resembles DNA in its tertiary structure.22 Studies are currently underway to investigate why B3 could no longer recognize dsDNA having complexed with PolIV.
Nearly 67% of the lupus sera bound to ‘naked’ dsDNA (Fig. 3). This observation confirms previous reports that 50–70% of the patients with SLE possess anti-dsDNA.23 However, ≈ 90% (93% calf thymus DNA; 90% synthetic oligonucleotide) of the lupus sera exhibited reactivity to the PolIV–dsDNA complex. Six (serum numbers 5, 7, 13, 18, 23 and 26) of the eight sera (serum numbers 5, 7, 11, 13, 17, 18, 23 and 26) that were negative for their activity to ‘naked’ dsDNA (Fig. 3), exhibited convincing activity to the complex (Fig. 4a–4d). Given that DNA is relatively difficult to adhere to polystyrene, it could be argued that coating ELISA wells with Po1IV increases the total amount of DNA bound in each well, which could have resulted in the observed increase in the signal. However, the fact that some sera, for example serum numbers 2, 3 and 4, bound ‘naked’ DNA better than the DNA presented on Po1IV, argues against this possibility.
Although the binding of lupus autoantibodies to naked oligonucleotide dsDNA was previously found to be better than their binding to naked calf thymus dsDNA, 18, 24 there was a significant decrease in their binding to PolIV-complexed oligonucleotide dsDNA (Fig. 4c,4d) compared with their binding to complexed calf thymus dsDNA (Fig. 4a,4b). PolIV was shown to bind oligonucleotide as well as calf thymus dsDNA (Fig. 2a,2b). Complexed calf thymus dsDNA could present a qualitatively different dsDNA on PolIV, which may well account for the observed difference in the serum reactivities to the two DNAs. Thus, it is a combination of the way in which dsDNA is presented (naked or complexed), and the qualitative nature of the presented DNA, which determines autoantibody reactivity. Although the importance of various aspects (e.g. length, 25–27 secondary structure, 28 base sequences, 29–32 and poly(dT) component33–35) of ‘naked’ DNA in determining the DNA antigenicity and autoantibody reactivity has been reported, no information is available on the autoantibody recognition of DNA presented on a DNA-binding protein.
Although anti-dsDNA possess the characteristics of antibodies arising in an antigen-driven response, such as high affinity for dsDNA, class switching to IgG, and somatic mutations in the variable-region gene segments, 36 the stimulus to their production – either self or foreign – remains uncertain. Unmodified mammalian34 or bacterial37–39 dsDNA does not lead to the production of lupus high-affinity IgG anti-DNA that binds eukaryotic DNA. Given that eukaryotic DNA (hapten) complexed to a DNA-binding protein (carrier) possesses immunogenic potential, 40, 41 it has been hypothesized that the immunogen in lupus is not naked DNA, but consists of DNA complexed to DNA-binding proteins.40 The original trigger for the two antibodies could thus be a PolIV-like dsDNA-binding protein, either ‘empty’ (B3) or complexed with dsDNA (RH14). Although only RH14 causes lupus nephrits-like disease in mice and exhibits the potential to recognize dsDNA complexed to a DNA-binding protein, it remains to be established if the serum antibodies detected by the PolIV–dsDNA complex are only RH14-like autoantibodies and more representative of lupus pathogenic antibodies.
In conclusion, at least two types of anti-dsDNA exist in the sera of patients with lupus; antibodies that recognize dsDNA presented on a DNA-binding protein are more lupus specific and may thus be linked to lupus pathology. The study also provides evidence that serum-derived lupus autoantibody reactivity to dsDNA depends not only on its qualitative make-up, but also on the way that it is presented, i.e. naked or in a complex.
Acknowledgments
This work was supported by the Arthritis Research Campaign, UK.
References
- 1.Swaak AJG, Aarden LA, Statius van Eps LW, Feltkamp TE. Anti-dsDNA and complement profiles as prognostic guides in systemic lupus erythematosus. Arthritis Rheum. 1979;22:226–35. doi: 10.1002/art.1780220304. [DOI] [PubMed] [Google Scholar]
- 2.Isenberg DA, Garton M, Reichlin MW, Reichlin M. Long-term follow-up of autoantibody profiles in black female lupus patients and clinical comparison with Caucasian and Asian patients. Br J Rheumatol. 1997;36:229–33. doi: 10.1093/rheumatology/36.2.229. [DOI] [PubMed] [Google Scholar]
- 3.Okamura M, Kanayama Y, Amastu K, Negoro N, Kohda S, Takeda T, Inoue T. Significance of enzyme linked immunosorbant assay (ELISA) for antibodies to double stranded and single stranded DNA in patients with lupus nephritis: correlation with severity of renal histology. Ann Rheum Dis. 1993;52:14–20. doi: 10.1136/ard.52.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Isenberg DA, Shoenfeld Y, Walport M, et al. Detection of cross-reactive anti-DNA antibody idiotypes in the serum of systemic lupus erythematosus patients and of their relatives. Arthritis Rheum. 1985;28:999–1007. doi: 10.1002/art.1780280907. [DOI] [PubMed] [Google Scholar]
- 5.Cabral AR, Alarcon-Segovia D. Autoantibodies in systemic lupus erythematosus. Curr Opin Rheumatol. 1997;9:387–92. doi: 10.1097/00002281-199709000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Ehrenstein M, Longhurst C, Isenberg DA. Production and analysis of IgG monoclonal anti-DNA antibodies from systemic lupus erythematosus (SLE) patients. Clin Exp Immunol. 1993;92:39–45. doi: 10.1111/j.1365-2249.1993.tb05945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravirajan C, Rahman T, Papadaki MA, et al. Genetic, structural and functional properties of an IgG DNA-binding monoclonal antibody from a lupus patient with nephritis. Eur J Immunol. 1998;28:339–50. doi: 10.1002/(SICI)1521-4141(199801)28:01<339::AID-IMMU339>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 8.Ohmori H, Friedberg EC, Fuchs RP, et al. The Y-family of DNA polymerases. Mol Cell. 2001;8:7–8. doi: 10.1016/s1097-2765(01)00278-7. [DOI] [PubMed] [Google Scholar]
- 9.Friedberg EC, Fischhaber PL, Kisker C. Error-prone DNA polymerases. novel structures and the benefits of infidelity. Cell. 2001;107:9–12. doi: 10.1016/s0092-8674(01)00509-8. [DOI] [PubMed] [Google Scholar]
- 10.Wagner J, Gruz P, Kim SR, Yamada M, Matsui K, Fuchs RP, Nohmi T. The dinB gene encodes a novel E. coli DNA polymerase, DNA pol IV, involved in mutagenesis. Mol Cell. 1999;4:281–86. doi: 10.1016/s1097-2765(00)80376-7. [DOI] [PubMed] [Google Scholar]
- 11.McKenzie GJ, Lee PL, Lombardo MJ, Hastings PJ, Rosenberg SM. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol Cell. 2001;7:571–79. doi: 10.1016/s1097-2765(01)00204-0. [DOI] [PubMed] [Google Scholar]
- 12.Johnson RE, Prakash S, Prakash L. The human DINB1 gene encodes the DNA polymerase Poltheta. Proc Natl Acad Sci USA. 2000;97:3838–43. doi: 10.1073/pnas.97.8.3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McKenzie GJ, Rosenberg SM. Adaptive mutations, mutator DNA polymerases and genetic change strategies of pathogens. Curr Opin Microbiol. 2001;4:586–94. doi: 10.1016/s1369-5274(00)00255-1. [DOI] [PubMed] [Google Scholar]
- 14.Ho G., Jr Bacterial arthritis. Curr Opin Rheumatol. 1992;4:509–15. [PubMed] [Google Scholar]
- 15.Astaldi Ricotti GC, Pazzaglia M, Martelli AM, et al. Autoantibodies to purified nuclear proteins related to DNA metabolism during ageing and in SLE patients. Immunology. 1987;61:375–84. [PMC free article] [PubMed] [Google Scholar]
- 16.Tan EM, Muro Y, Pollard KM. Autoantibody-defined epitopes on nuclear antigens are conserved, conformation-dependent and active site regions. Clin Exp Rheumatol. 1994;11:S27–31. [PubMed] [Google Scholar]
- 17.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40:1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 18.Kumar S, Kalsi J, Ravirajan CT, Rahman A, Athwal D, Latchman DS, Isenberg DA, Pearl LH. Molecular cloning and expression of the Fabs of human autoantibodies in Escherichia coli. Determination of the heavy or light chain contribution to the anti-DNA/-cardiolipin activity of the Fab. J Biol Chem. 2000;275:35129–36. doi: 10.1074/jbc.M001976200. [DOI] [PubMed] [Google Scholar]
- 19.Putterman P, Diamond B. Immunization with a peptide surrogate for double-stranded DNA (dsDNA) induces autoantibody production and renal immunoglobulin deposition. J Exp Med. 1998;188:29–38. doi: 10.1084/jem.188.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hay EM, Bacon PA, Gordon C, et al. The BILAG index: a reliable and valid instrument for measuring clinical disease activity in systemic lupus erythematosus. Q J Med. 1993;86:447–58. [PubMed] [Google Scholar]
- 21.Khalil M, Spatz L, Diamond B. Anti-DNA antibodies. In: Lahita RG, editor. Systemic Lupus Erythematosus. London, UK: Academic Press; 1999. pp. 197–217. [Google Scholar]
- 22.Tsuzaka K, Leu AK, Frank MB, Movafagh BF, Koscec M, Winkler TH, Kalden JR, Reichlin M. Lupus autoantibodies to double-stranded DNA cross-react with ribosomal protein S1. J Immunol. 1996;156:1668–75. [PubMed] [Google Scholar]
- 23.Cervera R, Khamashta MA, Font J, et al. Systemic lupus erythematosus: clinical and immunologic patterns of disease expression in a cohort of 1,000 patients. The European Working Party on Systemic Lupus Erythematosus. Medicine (Baltimore) 1993;72:113–24. [PubMed] [Google Scholar]
- 24.Kumar S, Kalsi J, Latchman DS, Pearl LH, Isenberg DA. Expression of the Fabs of human auto-antibodies in Escherichia coli: optimization and determination of their fine binding characteristics and cross-reactivity. J Mol Biol. 2001;308:527–39. doi: 10.1006/jmbi.2001.4534. [DOI] [PubMed] [Google Scholar]
- 25.Sano H, Morimoto C. Isolation of DNA from DNA/anti-DNA antibody immune complexes in systemic lupus erythematosus. J Immunol. 1981;126:538–39. [PubMed] [Google Scholar]
- 26.Sano H, Morimoto C. DNA isolated from DNA/anti-DNA antibody immune complexes in systemic lupus erythematosus is rich in guanine-cytosine content. J Immunol. 1982;128:1341–45. [PubMed] [Google Scholar]
- 27.Sano H, Takai O, Harata N, Yoshinaga K, Kodama-Kamada I, Sasaki T. Binding properties of human anti-DNA antibodies to cloned human DNA fragments. Scand J Immunol. 1989;30:51–63. doi: 10.1111/j.1365-3083.1989.tb01188.x. [DOI] [PubMed] [Google Scholar]
- 28.Casperson GF, Voss EW., Jr. Specificity of anti-DNA antibodies in SLE – II. Relative contribution of backbone, secondary structure and nucleotide sequence to DNA binding. Mol Immunol. 1983;20:581–88. doi: 10.1016/0161-5890(83)90002-0. [DOI] [PubMed] [Google Scholar]
- 29.Impraim CC, Conner BJ, Klotz JL, Lee TE, Teplitz RL. A method for binding specificity analysis of anti-DNA autoantibodies in SLE. J Immunol Methods. 1985;78:191–98. doi: 10.1016/0022-1759(85)90075-4. [DOI] [PubMed] [Google Scholar]
- 30.Herrmann M, Winkler TH, Fehr H, Kalden JR. Preferential recognition of specific DNA motifs by anti-dsDNA autoantibodies. Eur J Immunol. 1995;25:1897–1904. doi: 10.1002/eji.1830250716. [DOI] [PubMed] [Google Scholar]
- 31.Radic MZ, Cocca BA, Seal SN. Initiation of systemic autoimmunity and sequence specific anti-DNA autoantibodies. Crit Rev Immunol. 1999;19:117–26. [PubMed] [Google Scholar]
- 32.Stevens SY, Glick GD. Evidence for sequence specific recognition of DNA by anti-ssDNA autoantibodies. Biochemistry. 1999;38:560–69. doi: 10.1021/bi981899o. [DOI] [PubMed] [Google Scholar]
- 33.Lee JS, Dombroski DF, Mosmann TR. Specificity of autoimmune monoclonal Fab fragments binding to single-stranded deoxyribonucleic acid. Biochemistry. 1982;21:4940–45. doi: 10.1021/bi00263a017. [DOI] [PubMed] [Google Scholar]
- 34.Swanson PC, Cooper BC, Glick GD. High resolution epitope mapping of an anti-DNA autoantibody using model DNA ligands. J Immunol. 1994;152:2601–12. [PubMed] [Google Scholar]
- 35.Swanson PC, Ackroyd C, Glick GD. Ligand recognition by anti-DNA autoantibodies. Affinity, specificity, and mode of binding. Biochemistry. 1996;35:1624–33. doi: 10.1021/bi9516788. [DOI] [PubMed] [Google Scholar]
- 36.Diamond B, Katz JB, Paul E, Aranow C, Lustgarten D, Scharff MD. The role of somatic mutation in the pathogenic anti-DNA response. Annu Rev Immunol. 1992;10:731–57. doi: 10.1146/annurev.iy.10.040192.003503. [DOI] [PubMed] [Google Scholar]
- 37.Madaio MP, Hodder S, Schwartz RS, Stollar BD. Responsiveness of autoimmune and normal mice to nucleic acid antigens. J Immunol. 1984;132:872–76. [PubMed] [Google Scholar]
- 38.Pisetsky DS, Grudier JP, Gilkeson GS. a role for immunogenic DNA in the pathogenesis of systemic lupus erythematosus. Arthritis Rheum. 1990;33:153–59. doi: 10.1002/art.1780330202. [DOI] [PubMed] [Google Scholar]
- 39.Gilkeson GS, Pippen AM, Pisetsky DS. Induction of cross-reactive anti-dsDNA antibodies in preautoimmune NZB/NZW mice by immunization with bacterial DNA. J Clin Invest. 1995;95:1398–1402. doi: 10.1172/JCI117793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desai DD, Krishnan MR, Swindle JT, Marion TN. Antigen-specific induction of antibodies against native mammalian DNA in nonautoimmune mice. J Immunol. 1993;151:1614–26. [PubMed] [Google Scholar]
- 41.Rekvig OP, Moens U, Fredrikson K, Traavik T. Human polyomavirus and immunogenicity of mammalian DNA: a conceptual framework. Methods. 1997;11:44–54. doi: 10.1006/meth.1996.0386. [DOI] [PubMed] [Google Scholar]