Abstract
During mouse B-cell development, Pax5 is an essential transcription factor that acts as an activator of B-cell-specific genes and as a repressor of alternative lineage fates. The repressive function is mediated by the recruitment of members of the Groucho co-repressor family. Using an RNA display approach, we have isolated a transcript, called QD, specifically expressed in human pro-B and pre-B cells, which is derived from the human Groucho TLE4 gene. The QD transcript contains the first four TLE4 exons and an intronic sequence 3′ of exon 4, demonstrating that QD is a splice variant of TLE4. The putative resulting protein of 94 amino acids corresponds to approximately half of an N-terminal tetramerization domain. We also show specific expression of TLE4 transcripts in human B cells and of TLE4 proteins in B-cell nuclei. Moreover, we demonstrate that recombinant QD protein binds to the TLE4 Q domain and is able to abolish the TLE4/Pax5 interaction. Thus, QD could act as a negative regulator of TLE4 function, in early B-cell differentiation.
Introduction
B lymphocytes mature from haematopoietic stem cells through a series of developmental stages (pro-B, pre-B, immature B and mature B), characterized by certain cell-surface markers and sequential DNA rearrangements of immunoglobulin gene segments.1 Gene-inactivation experiments and studies of natural mutants have shown that B cells must pass several checkpoints in order to progress through these stages.2–4 The first checkpoint concerns B-cell commitment and is completely independent of immunoglobulin rearrangements but dependent on transcription factors expressed at the pro-B-cell stage.5 At the subsequent checkpoints, developing B cells are selected for the presence of functional immunoglobulin chains. Pre-B cells that synthesize an immunoglobulin µ-chain able to associate with the surrogate light chain (SLC), and form a functional pre-B-cell receptor (pre-BCR), will pass the second checkpoint. Then, newly formed immature B cells expressing a functional BCR are subjected to drastic selection against self-reactivity before leaving the bone marrow.6
At the onset of B-cell development, E2A7,8 and EBF9 transcription factors up-regulate the B-cell-specific programme,4 which entails activation of Pax5. Pax5 codes for the transcription factor BSAP, which is essential for brain patterning and B lymphopoiesis (reviewed by Nutt et al., ref. 10). Pax5-deficient pro-B cells are not restricted in their lineage fate and can differentiate into various myeloid and lymphoid cell types in vitro as well as in vivo.11,12 Pax5 seems to exert a dual role, as an activator of B-specific gene expression and as a repressor of alternative lineage fates. Recently, it has been shown that Pax5 may exert its repression function by recruiting members of the Groucho co-repressor family.13
The prototype of this conserved family is the Drosophila Groucho protein, which is expressed throughout development and plays important roles in various processes, such as sex determination, segmentation and neurogenesis.14 Groucho proteins are general co-repressors for a vast number of transcription factors (Hairy, Dorsal, Tcf, NF-κB, etc.).15 The Groucho protein has mammalian homologues: the Grg family in mice and the TLE (transducin-like enhancer of split) family in humans.15 While only one gene has been identified in Drosophila, four members have been described in both mouse and human (reviewed by Chen & Courey, ref. 16). The Groucho proteins form high-order complexes,17 bind to histone H3,18 interact with histone deacetylases19,20 and thus exert their function as part of multiprotein–DNA complexes that locally establish a repressive chromatin structure.
In the course of isolating cDNAs specifically expressed in human early B-cell progenitors we identified a splice variant of the Groucho TLE4 gene. We show that the corresponding recombinant protein binds to TLE4 and is able to impede the TLE4/Pax5 interaction. Therefore, this spliced TLE4 protein could act as a negative regulator of TLE4 function in early B-cell differentiation.
Materials and Methods
Cell lines, tissue culture and RNA isolation
The µ− pro-B (BV173, JEA2, Reh), µ+ pre-B (Nalm6, LAZ), µ+ κ/λ+ mature B (Daudi, Namalwa, JY), T (Hsb2, Jurkat) and monocyte (HL60, THP1, U937) cell lines were used. Total RNA was extracted using TRIzol Reagent (Invitrogen SARL, Cergy-Pontoise, France) and subjected to DNAseI-RNAse-free treatment.
Differential display–polymerase chain reaction
Differential display–polymerase chain reaction (DD–PCR) was performed on DNA-free total RNA from BV173, JEA2, Daudi and Hsb2 cell lines using the Kit Display system (Display system; Tandil SA, France). In brief, nine different cDNAs were produced using two base-anchored oligo-dT primers (13mer) and, for each, reverse transcription (RT)–PCR was performed using the same antisense primers combined with 24 arbitrary sense (10mer) primers, in the presence of [α-33P]dATP. RT–PCR combination products were separated on 6% denaturing polyacrylamide gels, and cDNAs in the 200–600 bp range were selected based on the reproducibility of banding across two separate experiments. Bands corresponding to cDNAs specifically expressed in two pro-B (BV173, JEA2) and absent in the B (Daudi) and the T (Hsb2) cell lines were excised from the gels, eluted and precipitated. Reamplifications were performed using the same primers and PCR conditions. PCR products were purified by using the Qiaquick PCR purification kit (Qiagen SA, Courtaboeuf, France) and cloned using the TA-cloning kit (Invitrogen SARL, Cergy-Pontoise, France). Nucleotide sequences were determined and homology with known sequences in nucleotide databases was checked using blast-n and blast-x programs.
RNA expression
Northern and master blots.
For Northern blotting, 15 µg of total RNA were electrophoresed under denaturing conditions and transferred to Hybond N nylon membrane (Amersham Biosciences Europe, Orsay, France). Prehybridization, hybridization and washings were performed using conditions previously published.21 The human RNA master blot A (7770-1) containing different human tissues was obtained from Ozyme (Sáint Quentin, France).
Actin, TLE4 and QD probes were generated by RT–PCR, from Nalm6, the EST 1288038 and the EST 140044, respectively, using the following primers: actin, sense 5′-TACCACTGGCATCGTGATGGACT-3′, antisense 5′-TCCTTCTGCATCCTGTCGGCAAT-3′; TLE4, sense 5′-GTCAAGAGGCTGAATGCTATCT-3′, antisense 5′-CTCCTAGAGCACTGGAGAGG-3′; and QD, sense 5′-AATGACAGTAATACTGGGACAC-3′, antisense 5′-CTAATTGCTAGATTTCAAGACCC-3′.
Probes were radiolabelled with [α-32P]dCTP using a random hexamer oligonucleotide.22
Semiquantitative PCR.
DNA-free total RNA (2 µg) from B, T and myeloid cell lines was reverse transcribed using the reverse transcriptase Superscript II (Gibco-BRL, Cergy-Pontoise, France), 1 µg of random hexamer (dN6), 1 mm dNTPs and the supplied buffer. For semiquantitative RT–PCR, analysis of mRNA expression was carried out during the exponential phase of the amplification, determined by titration of cycle numbers and by quantities of total RNA. RT–PCR was carried out with 400 ng (TLE4 and QD) or 2 ng (actin) of cDNAs in the presence of [α-33P]dATP for 30 cycles (actin, TLE4) or 34 cycles (QD) of 30 seconds at 94°, 1 min at 66° (actin) or 58° (TLE4, QD) and 1 min at 72° with a final 10-min extension at 72°, using Taq DNA polymerase (Gibco-BRL). For actin measurements, the same primers as for probe amplification were used. For TLE4, sense 5′-CAGCGGCATTATGTCATG-3′, antisense 5′-CATGTCCATGTGATAAATGCTG-3′ and for QD, sense 5′-CAGCGGCATTATGTCATG-3′, antisense 5′-CTTTTGTAGCTCCACAGATTTC-3′ primers were used. RT–PCR products were analysed on 2% agarose gels. Dried gels were exposed with a Fuji screen and analysed by Phosphoimager apparatus (Amersham Biosciences, Sunnyvale, CA). Quantification was performed using MacBas software.
Immunostaining and microscopy
Cells were suspended in culture medium at 104–105 cells/ml and aliquots (0·5 ml) were centrifuged (5 g for 5 min) onto polylysine-coated slides in a cytospin 3 (Shandon, Cergy-Pontoise, France). Nuclei preparations were fixed at room temperature for 10 min in 4% (wt/vol) paraformaldehyde/phosphate-buffered saline (PBS), pH 7·2. After washing in PBS, permeabilization was performed with 0·1% Triton-X-100 in PBS for 40 min at room temperature, followed by 15 min in blocking buffer (PBS, 0·05% Triton-X-100, 5% goat serum, 5% fetal calf serum).
Preparations were subjected to indirect immunofluorescence using a rabbit polyclonal antibody (1 : 200 dilution) against a mouse Grg4 recombinant protein (Santa Cruz Biotechnology Inc., Heidelberg, Germany) in PBS, 0·05% Triton-X-100, for 2 hr at room temperature. After washings in PBS, the primary antibody was detected using a 1 : 300-diluted Cyanine 3-conjugated affinity-purified goat anti-rabbit immunoglobulin G (IgG) (Tebu, Le Persay en Yvelines, France), in the same buffer, for 1 hr at room temperature. Preparations were fixed with 4% (wt/vol) paraformaldehyde in PBS for 5 min and counterstained with DAPI (100 ng/ml) diluted in Vectashield (Vector Laboratories, Burlingame, CA). Controls without the primary antibody were performed for each cell line. Preparations were observed using an Axioplan-2 Zeiss fluorescent microscope and the images captured with a CCD camera (Sensys 1400; Photometrics, Tuscon, AZ). Information was collected and merged using IPLabs Spectrum software (Vysis, Voisins Le Bretonneux, France).
Expression, purification and labelling of recombinant proteins
DNA constructs.
The human His-QD coding sequence was amplified by PCR using the EST 140044 cDNA and the sense 5′-ATGGATCCAGTACCCGCAGACCAGACA-3′ and antisense 5′-ATAAGCTTTAGTGTCCCAGTATTACTGTC-3′ primers. The purified PCR product was digested with BamHI and HindIII and ligated into the pQE32 vector (Qiagen). For the GST-TLE4 Q (amino acids 2–132) and GST-TLE4-ΔW (amino acids 2–432) fusion proteins, the human TLE4 cDNA (EST 1288038) was amplified using the sense 5′-ATGAATTCTACCCGCAGACCAGACAC-3′ and antisense 5′-ATGCGGCCGCATGTCCATGTGATAAATGCTG-3′ or 5′-ATGCGGCCGCCAGGTTTGGAGGTATTGCTG-3′ primers, respectively. Purified PCR products were digested with EcoRI and NotI, and ligated into pGEX-4T-1 vector (Amersham Biosciences Europe, Orsay, France).
For TA cloning into pGEMT-Easy vector (Promega SARL, Charbonnières, France), the QD coding sequence was amplified with the sense 5′-ATGTACCCGCAGACCAGAC-3′ and the antisense 5′-ATGCGGCCGCTAGTGTCCCAGTATTACTGTC-3′ primers, and the TLE4-ΔW sequence with the sense 5′-ATGTACCCGCAGACCAGAC-3′ and the antisense 5′-CAGGTTTGGAGGTATTGCTG-3′ primers. The human Pax5 coding sequence was amplified using LAZ cDNA and the sense 5′-ATAGATCTATGGATTTAGAGAAAAATTATCC-3′ and the antisense 5′-ATTCTAGACTAGTGACGGTCATAGGCAGT-3′ primers.
Production and purification of GST and His fusion proteins.
The pGEX-TLE4 Q and pGEX-TLE4-ΔW plasmids were transfected into DH5α and BL21-RP bacterial strains (Stratagene, Amsterdam, Netherlands), respectively. Transformed bacteria were grown at 37° to reach an optical density at 600 nm (OD600) of 0·6 in 2xYT medium containing the appropriate antibiotics, induced with 1 mm IPTG, and allowed to continue growing for an additional 2 hr. Pelleted cells were resuspended in buffer B1 [Tris–HCl 50 mm, pH 8, KCl 100 mm, EDTA 1 mm, glycerol 5%, dihiothreitol (DTT) 1 mm, phenylmethylsulphonyl fluoride (PMSF) 1 mm], lysed by two runs at 1700 Bars using a French Press (Constant Systems Ltd, Warwick, UK) and bacterial extracts were ultracentrifuged at 350 000 g for 20 min at 4°. Supernatants were dialysed against buffer B2 (HEPES 10 mm, pH 7·0, KCl 10 mm, NaCl 150 mm, MgCl2 1 mm, ZnSO4 1 µm, DTT 1 mm, PMSF 1 mm), clarified by centrifugation, and GST fusion proteins were purified on glutathione-agarose beads, according to the manufacturer's instructions (Pharmacia Biotech). GST-TLE4-ΔW proteins of 70 000 molecular weight (MW) were further purified by electrophoresis on a 10% sodium dodecyl sulphate (SDS)–polyacrylamide preparative gel under reducing conditions.
M15-transformed bacteria containing the pQE32-His-QD plasmid were grown in 2xYT medium, containing the appropriate antibiotics, to reach an OD600 of 1, induced with 0·5 mm IPTG and allowed to grow for an additional 6 hr. Pelleted cells were resuspended in buffer B3 (K2HPO4/HCl 50 mm, pH 7·5, PMSF 1 mm), lysed using a French Press, as described above, and His-QD proteins were purified on Talon Superflow Metal Affinity Resin (Clontech) according to the manufacturer's instructions.
In vitro transcription/translation of pGEMT cloned sequences.
The TLE4-ΔW, QD and hPax5 proteins were synthesized and in vitro labelled using rabbit reticulocyte lysate and 35S-methionine, using the TNT Quick Coupled Transcription/Translation Systems, according to the manufacturer's instructions (Promega).
GST pull-down assay
Purified GST–fusion proteins (4 µg), immobilized on glutathione-sepharose beads (5 µl), were incubated for 2 hr at room temperature with in vitro-synthesized 35S-labelled protein (2 µl of the transcription/translation assay) in 400 µl of buffer B4 (buffer B2 supplemented with 1% bovine serum albumin). Beads were washed in sucrose gradient buffers, as previously described.23 Labelled proteins were revealed by autoradiography after separation on a 17·5% SDS–polyacrylamide gel.
For the Pax5 inhibition experiment, 1 µg of purified GST-TLE4-ΔW protein was incubated with increasing amounts of His-QD (0·5, 1 and 3 µg) for 2 hr at room temperature in 400 µl (final volume) of B4 buffer. Glutathione-agarose beads were added for 1 hr, washed twice with buffer B4 and incubated with 35S-labelled hPax5 (4 µl of the transcription/translation assay) for 2 hr at room temperature. Labelled proteins, separated on a 17·5% SDS-polyacrylamide gel, were analysed by Phosphoimager apparatus (Molecular Dynamics). Quantification was performed using MacBas software.
Protein structure analysis
The QD sequence was analysed for predicted coiled-coil regions using the Network Protein Sequence analysis program through the Pôle Bio-Informatique Lyonnais at pbil.ibcp.fr (see ref. 24).
Results
Identification of human QD and TLE4 cDNAs
Isolation of cDNAs expressed during early B-cell differentiation was undertaken using a differential display screening25 between two pro-B-cell lines (BV173, JEA2), one B-cell line (Daudi) and one T-cell line (Hsb2). After screening through 216 combinations of primers, representing 90% of the genes expressed in a cell,26 a total of 162 ‘pro-B’ cDNAs was identified. Each cDNA was cloned and sequenced and a second expression screening was performed by semiquantitative RT–PCR using a panel of 13 different cell lines, including pro-B, pre-B, B, monocyte and T cells (see below in Fig. 3). We confirmed an early B-cell expression profile for only 18 cDNAs, with 11 and seven cDNAs that presented a pro-B-, and a pro-B plus a pre-B-restricted phenotype, respectively.
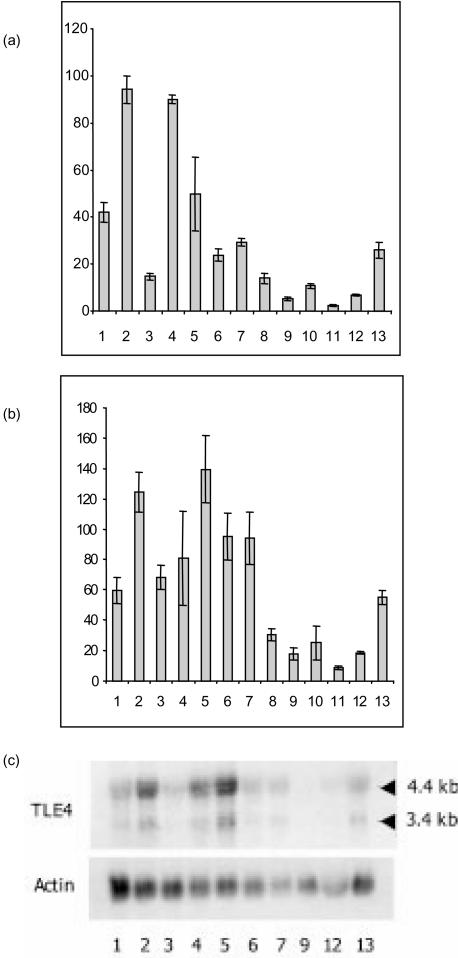
Figure 3.
B-cell-restricted RNA expression of QD and TLE4 transcripts. (a) and (b) Semiquantitative reverse transcription–polymerase chain reaction (RT–PCR) expression of QD (a) and TLE4 (b) transcripts after actin calibration. RT–PCR was performed using a 5′ oligonucleotide in the Q domain common to the TLE4 and QD transcript, and a 3′ oligonucleotide in the 3′-untranslated region of the QD cDNA or in the TLE4 GP domain (see Fig. 1b), leading to PCR products of 515 bp and 238 bp, respectively. The following cell lines were tested: pro-B, lanes 1 (BV173), 2 (JEA2) and 3 (Reh); pre-B, lanes 4 (LAZ) and 5 (Nalm6); B, lanes 6 (Daudi), 7 (Namalwa) and 8 (JY); monocytes, lanes 9 (HL60), 10 (THP1) and 11 (U937); T, lanes 12 (Hsb2) and 13 (Jurkat). Average values of the relative quantity of PCR transcripts for three separate experiments are shown. (c) A Northern blot from the cell lines indicated above was hybridized with the TLE4 (see Fig. 1b) and the actin probes, successively.
Within the latter category, a 250-bp cDNA was isolated and found to be completely identical (Fig. 1a, sequence shown in bold) to the 3′ end sequence of the EST 140044. The complete sequence of this EST was determined and revealed a 92-bp 5′ untranslated sequence, an open reading frame of 282 bp followed by a 578-bp 3′ sequence (Fig. 1a). Sequence comparison revealed that amino acid residues 1–77 are homologous to Groucho family members, especially the rat TLE4, the mouse Grg4 and the Xenopus laevis Grg-4 (data not shown). Search for human homologues was undertaken by comparing the 5′ sequence of the Image clone 140044 with human EST sequences. This allowed characterization of the complete human TLE4 cDNA, the sequence of which was reported as KIAA1261 clone by Nagase et al.27 TLE4 cDNA encompasses 2754 bp and the protein presents an overall conserved ‘Groucho’ structure composed of five domains (Fig. 1b). The structure includes an N-terminal glutamine-rich Q region, a glycine–proline-rich GP domain, a central region with a CcN (nuclear localization) motif, a serine–proline SP domain and a C-terminus region composed of six WD-40 motif repeats.
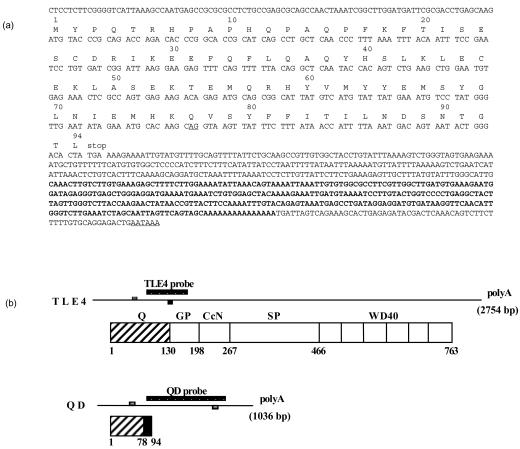
Figure 1.
Characterization of the QD transcript. (a) Sequence of the QD transcript obtained from the Image clone 140044. The cDNA (250 bp) first isolated by the RNA display technique is in bold. The potential polyadenylation site and splicing AG donor site at the end of exon 4 (see Figure 2) are underlined. (b) Schematic representation of TLE4- and QD-spliced mRNA and proteins. The oligonucleotides (small boxes) and the probes used for testing RNA expression of TLE4 and QD transcripts are shown for each mRNA. The different domains of TLE4 and QD proteins are indicated together with the corresponding amino acid positions. Q, glutamine-rich domain; GP, glycine–proline-rich domain; CcN, central region with a nuclear localization motif; SP, serine–proline domain; WD40, WD repeat domain. The C-terminal amino acid sequence (78–94) of the QD protein that differs from the TLE4 protein is indicated in black.
Altogether these data indicate that the early B-specific 140044 cDNA is identical to the 5′ sequence of the human TLE4 cDNA, up to residue 77. By contrast, residues 78–94 are not found in the TLE4 cDNA (Fig. 1b).
The human QD transcript is encoded by the TLE4 gene
The partial identity between QD and TLE4 cDNAs prompted us to look for a common origin between the two transcripts.
A partial genomic sequence of the human TLE4 gene on chromosome 9, determined by the Sanger Centre chromosome 9 Mapping Group (accession numbers: AL 353813 and AL 445252), revealed that the N-terminal part of the TLE4Q domain is encoded by exons 1–4 (Fig. 2). Sequence comparison between the QD cDNA and the TLE4 gene showed that the 3′ end of the QD transcript is encoded by part of the intronic nucleotide sequence located 3′ of exon 4 (Fig. 2, black boxes). This intronic sequence starts at the splicing AG donor site at the end of exon 4 (underlined in Fig. 1a) and encodes amino acids 78–94 (Fig. 1a, Fig. 2). Finally, a genuine polyadenylation site was identified 650 bp after the stop codon (Fig. 1a, Fig. 2).
Figure 2.
TLE4 genomic organization and generation of the QD and TLE4 transcripts. Top: exon/intron organization of the TLE4 gene. Data are from the Sanger Centre chromosome 9 Mapping Group (accession numbers: AL 353813 and AL 445252). The size of the introns is indicated. Bottom: generation of QD and TLE4 transcripts from the TLE4 gene.
B-cell-restricted human QD and TLE4 RNAs
Comparative expression of QD and TLE4 cDNAs was first studied by semiquantitative RT–PCR. A series of lymphoid cell lines, including pro-B, pre-B, mature B, monocyte and T-cell lines were analysed, using a common 5′ oligonucleotide in the Q domain and 3′ specific oligonucleotides either in the 3′ untranslated region of the QD cDNA or in the TLE4 GP domain (see Fig. 1b). Although the Reh pre-B-cell line was not positive for QD expression, results confirm the pro-B- and pre-B-specific expression of the QD transcript (Fig. 3a). These data also indicate that all B-cell lines express TLE4 transcripts (Fig. 3b).
RNA expression was also analysed in lymphoid cell lines by Northern blot, using either a 600-bp QD or a 350-bp TLE4-specific probe (see Fig. 1b). Two main transcripts at 4·4 kb and 3·4 kb were observed using the TLE4 probe (Fig. 3c), but no QD transcripts were detectable. Quantification using the actin probe confirmed the restricted B-cell expression of the TLE4 transcripts. Various human tissues were also analysed using the two probes. The QD probe faintly labelled RNAs present on the dot-blots and, using the TLE4 probe, faint expression was found in approximately all tissues, except the pituitary gland and testis, which strongly express TLE4 (data not shown).
B-cell-restricted humanTLE4 protein
Immunostaining with the rabbit anti-TLE4 antibody revealed a dense punctate pattern to the nuclei of pro-B, pre-B and mature B cells, whereas no labelling was observed in control experiments performed without incubation with the primary antibody (Fig. 4). Moreover, no nuclear signal was detectable on nuclei preparations from myeloblast and lymphocyte T-cell lines, confirming the B-restricted expression of the TLE4 protein in haematopoietic cells (Fig. 4).
Figure 4.
B-cell nuclei-restricted TLE4 protein expression. Representative distribution of the TLE4 protein in a nucleus of pro-B (JEA2), pre-B (Nalm6), B (Namalwa), myeloblast (HL60), and T (HsB2) cells, obtained by immunostaining with the anti-TLE4 antibody. ‘mAb control’ represents a B-lymphocyte nucleus from a preparation incubated without the primary anti-TLE4 antibody.
QD protein interacts with TLE4 and inhibits TLE4/Pax5 interaction
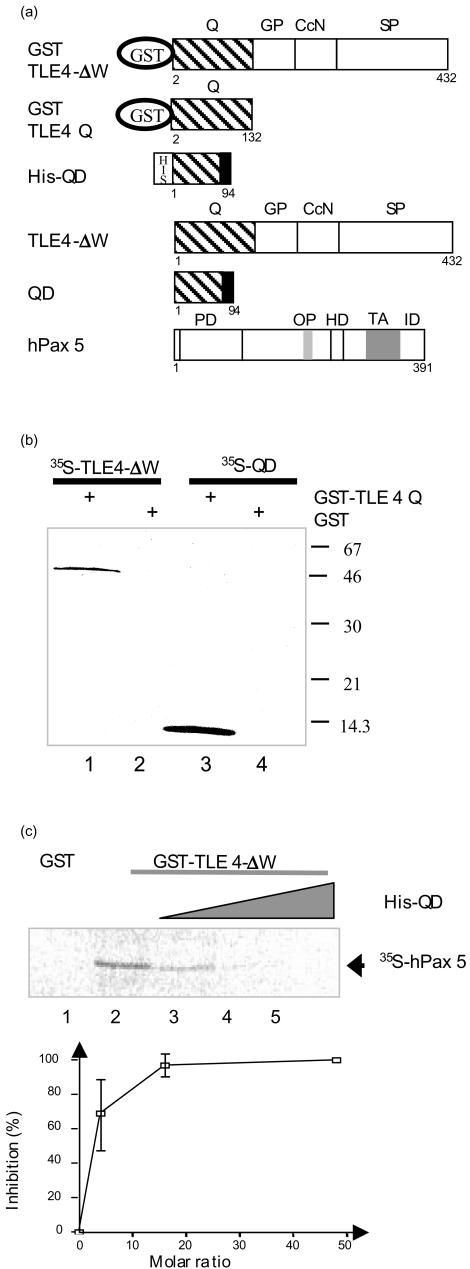
As previous studies suggested that the N-terminal Q domain of Groucho proteins is involved in the oligomerization of the molecule,17 we investigated the capacity of QD protein to interact with the native TLE4 Q domain using an in vitro protein–protein interaction assay.
TLE4-ΔW and QD proteins (Fig. 5a) were synthesized and labelled with 35S-methionine by in vitro transcription/translation, and incubated with either GST-TLE4 Q fusion protein or GST alone (as a control), immobilized on glutathione-agarose beads. 35S-bound proteins were revealed by autoradiography after gel electrophoresis. Under such conditions, TLE4-ΔW and QD proteins were detected at 50 000 and 11 000 MW, respectively (Fig. 5b, lanes 1 and 3), demonstrating that both proteins could interact with the native Q domain. This result suggests that the QD protein could modify the oligomerization status of the TLE4 protein. As it has recently been reported that the mouse Pax5 can interact with Grg413 (the TLE4 mouse equivalent), we performed experiments to test the potential role of the QD protein on such an interaction. We first checked that GST-TLE4-ΔW on glutathione-agarose beads could bind to 35S-hPax5 protein (Fig. 5c, lane 2). Then, by increasing the amount of recombinant His-tag QD protein that could interact with GST-TLE4-ΔW before adding the 35S-hPax5 protein, we demonstrated that QD was able to inhibit the TLE4–hPax5 interaction (Fig. 5c, lanes 3–5). This inhibition is dose dependent and 50% inhibition (IC50) is obtained with a QD/TLE4 molar ratio of 3·0 (Fig. 5c, lower panel). These results indicate that the binding between QD and TLE4 proteins alters TLE4–Pax5 interactions.
Figure 5.
Analysis of QD, TLE4 and Pax5 protein interactions. (a) Schematic diagram of TLE4, QD and Pax5 fusion proteins. The different domains of each protein are indicated (PD, paired domain; OP, octapeptide; HD, partial homeodomain; TA, transactivation region; ID, inhibitory domain; also see Fig. 1 legend). (b) The QD recombinant protein interacts with the Q domain of TLE4. GST pull-down assays were used to analyse the interactions between in vitro-translated 35S-labelled TLE4-ΔW (lanes 1 and 2) and 35S-labelled QD proteins (lanes 3 and 4) and GST or GST-TLE4Q proteins bound to glutathione-sepharose. (c) The QD recombinant protein inhibits the TLE4/Pax5 interactions. Upper panel: GST pull-down assays were performed with 35S-labelled hPax5 protein and GST (lane 1), GST-TLE4-ΔW (lanes 2–5) bound to glutathione-sepharose. The GST-TLE4-ΔW proteins were incubated with an increasing amount of His-QD proteins (0·5, 1 and 3 µg, lanes 3, 4 and 5, respectively). Lower panel: for three independent experiments the percentage of inhibition was calculated for each QD/TLE4 molar ratio.
Altogether, our data indicate that QD was able to interfere with the normal function of the TLE4 protein and thus may be considered a dominant negative form of TLE4.
Discussion
The mRNA differential display technique was applied to haematopoietic cell lines to identify genes differentially expressed in early B cells compared to B, monocyte and T cells. We isolated and characterized the QD cDNA that encodes part of the N-terminal Q domain of the TLE4 Groucho protein. This cDNA corresponds to the normal splice of the first four TLE4 exons followed by part of the intronic sequence 3′ of exon 4 that is converted to a new exon, with a polyadenylation site 650 bp downstream (Fig. 2). Interestingly, the QD and TLE4 genomic organization resembles that of the immunoglobulin µ-secreted and immunoglobulin µ-membrane forms of the immunoglobulin H-chain.28 It has been reported previously that the switch from membrane-bound to secreted immunoglobulin µ form, which occurs during the differentiation of B lymphocytes, involves regulated processing of the heavy chain pre-mRNA.29 In particular, overexpression of one subunit of Cst-64 is sufficient to switch immunoglobulin H-chain expression from the membrane to the secreted form.30 This transcriptional regulation might be specific to B-cell development and may involve a similar molecular mechanism.
Although TLE4 mRNA has been detected in a variety of tissues31,32 its expression is restricted to B cells in the haematopoietic lineage (Fig. 3b, c). This was confirmed at the protein level by using an antibody directed against TLE4 (Fig. 4), which moreover revealed an exclusive nucleus staining, in agreement with the nuclear co-repressive function of Groucho protein members. The number of labelled sites indicates that TLE4 may be involved in the repression of numerous loci. The expression of the QD transcript, although lower than that of TLE4, is also restricted to B cells within the haematopoietic lineage (Fig. 3a). The expression of QD and TLE4 in the same B-restricted cells points to a possible regulatory role of QD on TLE4 function. Other short forms of Groucho family proteins, encoded by specific genes, have been described that could modulate the activity of the long forms.16 For example, the Xenopus Grg5 protein could act as an anti-repressor of endogenous long Groucho proteins.33
It has been demonstrated that Pax5 functions as a positive regulator of B-specific genes; however, B-cell commitment also depends on the repression of lineage-inappropriate genes.11 By identifying Grg4 as a Pax5 partner, Eberhard et al.13 have elucidated a molecular mechanism by which Pax5 can be converted from a transcriptional activator to a repressor of gene transcription, in mice. Grg4 possesses two interacting domains with Pax5, the Q and the SP domains, which act in cooperation for the binding to Pax5 protein.13 Similarly, our results show that QD protein binds with TLE4-ΔW protein (Fig. 5b) but does not interact directly with Pax5 (data not shown). By contrast, preincubation of TLE4 with QD proteins is sufficient to abolish the interaction between the TLE4 and the Pax5 proteins. Thus, QD could act as a negative regulator of the repressive activity of Pax5 mediated by TLE4.
As QD expression is restricted to pro-B and pre-B cells we can speculate that it may regulate genes specifically expressed at these stages. Such early B-specific genes would be negatively regulated by Pax5/TLE4 in late B-cell stages, in the absence of QD. For the non-B-specific genes, because Pax534 and TLE4 (this paper) have the same B-cell-restricted expression pattern, we can speculate that Pax5 associated with its TLE4 co-repressor may exert a long-range transcriptional repression. The fact that TLE4 is associated with a large number of nuclear targets in B cells is consistent with this hypothesis.
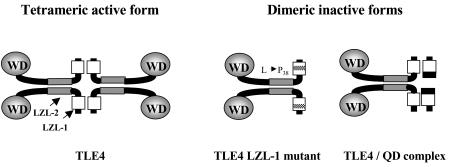
Chen et al.17 have shown that tetramerization of Gro proteins is a prerequisite for efficient repression and is mediated by two putative amphipathic α-helices present in the conserved N-terminal Q domain (Fig. 6). It also has been postulated that Gro protein oligomerization along the template DNA, facilitated by interactions with histones,35 could be necessary for repression and for long-range transcriptional silencing.17 Moreover, Chen et al. demonstrated that point mutations in the leucine zipper-like motifs, Leu to Pro replacement at amino acid 38 (Fig. 6), block the tetramerization and also partially Gro-mediated repression. However, in this case mutant Gro proteins can still form dimers. By analogy, the putative QD protein containing the first N-leucine zipper motif would be able to block the tetramerization of the TLE4 protein and thus its repressive activity (Fig. 6). Further genetic studies will be necessary to confirm this hypothesis.
Figure 6.
Speculative model of TLE4 function inhibition by QD protein. The tetrameric co-repressive TLE4 active form postulated by Chen et al.17 could be disrupted by breaking one of the two leucine-zipper like (LZL) motifs present in the N-terminal Q domain. Potential dimeric co-repressive inactive forms could be generated by a Leu to Pro amino acid substitution at position 38 in the first LZL117, or by interaction with the QD proteins.
Acknowledgments
We thank Danielle DePetris for excellent technical assistance in immunostaining and microscopy, and Maleck Djabali and Pierre Golstein for critical reading of the manuscript. This work was supported by CNRS (Centre National de la Recherche Scientifique) and INSERM (Institut National de la Santé et de la Recherche Médicale).
References
- 1.Alt FW, Yancopoulos GD, Blackwell TK, et al. Ordered rearrangement of immunoglobulin heavy chain variable region segments. EMBO J. 1984;3:1209–19. doi: 10.1002/j.1460-2075.1984.tb01955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix–loop–helix genes, E2A, E2-2, and HEB. Mol Cell Biol. 1996;16:2898–905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nutt SL, Urbanek P, Rolink A, Busslinger M. Essential functions of Pax5 (BSAP) in pro-B cell development: difference between fetal and adult B lymphopoiesis and reduced V-to-DJ recombination at the IgH locus. Genes Dev. 1997;11:476–91. doi: 10.1101/gad.11.4.476. [DOI] [PubMed] [Google Scholar]
- 4.O'Riordan M, Grosschedl R. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity. 1999;11:21–31. doi: 10.1016/s1074-7613(00)80078-3. [DOI] [PubMed] [Google Scholar]
- 5.Bain G, Robanus Maandag EC, te Riele HP, et al. Both E12 and E47 allow commitment to the B cell lineage. Immunity. 1997;6:145–54. doi: 10.1016/s1074-7613(00)80421-5. [DOI] [PubMed] [Google Scholar]
- 6.Goodnow CC, Cyster JG, Hartley SB, et al. Self-tolerance checkpoints in B lymphocyte development. Adv Immunol. 1995;59:279–368. doi: 10.1016/s0065-2776(08)60633-1. [DOI] [PubMed] [Google Scholar]
- 7.Bain G, Maandag EC, Izon DJ, et al. E2A proteins are required for proper B cell development and initiation of immunoglobulin gene rearrangements. Cell. 1994;79:885–92. doi: 10.1016/0092-8674(94)90077-9. [see comments]. [DOI] [PubMed] [Google Scholar]
- 8.Zhuang Y, Soriano P, Weintraub H. The helix–loop–helix gene E2A is required for B cell formation. Cell. 1994;79:875–84. doi: 10.1016/0092-8674(94)90076-0. [DOI] [PubMed] [Google Scholar]
- 9.Lin H, Grosschedl R. Failure of B cell differentiattion in mice lacking the transcription factor EBF. Nature. 1995;376:263–7. doi: 10.1038/376263a0. [DOI] [PubMed] [Google Scholar]
- 10.Nutt SL, Thevenin C, Busslinger M. Essential functions of Pax-5 (BSAP) in pro-B cell development. Immunobiology. 1997;198:227–35. doi: 10.1016/S0171-2985(97)80043-5. [DOI] [PubMed] [Google Scholar]
- 11.Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–62. doi: 10.1038/44076. [see comments]. [DOI] [PubMed] [Google Scholar]
- 12.Rolink AG, Nutt SL, Melchers F, Busslinger M. Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature. 1999;401:603–6. doi: 10.1038/44164. 10.1038/44164. [DOI] [PubMed] [Google Scholar]
- 13.Eberhard D, Jimenez G, Heavey B, Busslinger M. Transcriptional repression by Pax5 (BSAP) through interaction with corepressors of the Groucho family. EMBO J. 2000;19:2292–303. doi: 10.1093/emboj/19.10.2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paroush Z, Finley RL, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–15. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- 15.Fisher AL, Caudy M. Groucho proteins transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev. 1998;12:1931–40. doi: 10.1101/gad.12.13.1931. [DOI] [PubMed] [Google Scholar]
- 16.Chen G, Courey AJ. Groucho/TLE family proteins and transcriptional repression. Gene. 2000;249:1–16. doi: 10.1016/s0378-1119(00)00161-x. [DOI] [PubMed] [Google Scholar]
- 17.Chen G, Nguyen PH, Courey AJ. A role for Groucho tetramerization in transcriptional repression. Mol Cell Biol. 1998;18:7259–68. doi: 10.1128/mcb.18.12.7259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–10. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- 19.Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–30. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi CY, Kim YH, Kwon HJ, Kim Y. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J Biol Chem. 1999;274:33194–7. doi: 10.1074/jbc.274.47.33194. [DOI] [PubMed] [Google Scholar]
- 21.Schiff C, Milili M, Zucman-Rossi J, Djabali M, Fougereau M. Composite exon structure of an unusual Ig lambda-like gene located at human 22q11 position. Mamm Genome. 1996;7:598–602. doi: 10.1007/s003359900177. [DOI] [PubMed] [Google Scholar]
- 22.Feinberg AP, Vogelstein B. A technique for radiolabelling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 23.Lemmers B, Gauthier L, Guelpa-Fonlupt V, Fougereau M, Schiff C. The human (PsiL+mu-) proB complex: cell surface expression and biochemical structure of a putative transducing receptor. Blood. 1999;93:4336–46. [PubMed] [Google Scholar]
- 24.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–4. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 25.Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992;257:967–71. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 26.Bauer D, Muller H, Reich J, Riedel H, Ahrenkiel V, Warthoe P, Strauss M. Identification of differentially expressed mRNA species by an improved display technique (DDRT–PCR) Nucl Acids Res. 1993;21:4272–80. doi: 10.1093/nar/21.18.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagase T, Ishikawa K, Kikuno R, Hirosawa M, Nomura N, Ohara O. Prediction of the coding sequences of unidentified human genes. XV. The complete sequences of 100 new cDNA clones from brain which code for large proteins in vitro. DNA Res. 1999;6:337–45. doi: 10.1093/dnares/6.5.337. [DOI] [PubMed] [Google Scholar]
- 28.Word CJ, White MB, Kuziel WA, Shen AL, Blattner FR, Tucker PW. The human immunoglobulin C mu-C delta locus: complete nucleotide sequence and structural analysis. Int Immunol. 1989;1:296–309. doi: 10.1093/intimm/1.3.296. [DOI] [PubMed] [Google Scholar]
- 29.Peterson ML, Perry RP. Regulated production of mu m and mu s mRNA requires linkage of the poly(A) addition sites and is dependent on the length of the mu s-mu m intron. Proc Natl Acad Sci USA. 1986;83:8883–7. doi: 10.1073/pnas.83.23.8883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takagaki Y, Manley JL. Levels of polyadenylation factor CstF-64 control IgM heavy chain mRNA accumulation and other events associated with B cell differentiation. Mol Cell. 1998;2:761–71. doi: 10.1016/s1097-2765(00)80291-9. [DOI] [PubMed] [Google Scholar]
- 31.Grbavec D, Lo R, Liu Y, Greenfield A, Stifani S. Groucho/transducin-like enhancer of split (TLE) family members interact with the yeast transcriptional co-repressor SSN6 and mammalian SSN6-related proteins: implications for evolutionary conservation of transcription repression mechanisms. Biochem J. 1999;337:13–7. 10.1042/0264-6021:3370013. [PMC free article] [PubMed] [Google Scholar]
- 32.Brantjes H, Roose J, van De Wetering M, Clevers H. All Tcf HMG box transcription factors interact with Groucho-related co-repressors. Nucl Acids Res. 2001;29:1410–9. doi: 10.1093/nar/29.7.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roose J, Molenaar M, Peterson J, et al. The Xenopus Wnt effector XTcf-3 interacts with Groucho-related transcriptional repressors. Nature. 1998;395:608–12. doi: 10.1038/26989. [DOI] [PubMed] [Google Scholar]
- 34.Kaneko H, Ariyasu T, Inoue R, et al. Expression of Pax5 gene in human haematopoietic cells and tissues: comparison with immunodeficient donors. Clin Exp Immunol. 1998;111:339–44. doi: 10.1046/j.1365-2249.1998.00509.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flores-Saaib RD, Courey AJ. Analysis of Groucho–histone interactions suggests mechanistic similarities between Groucho- and Tup1-mediated repression. Nucl Acids Res. 2000;28:4189–96. doi: 10.1093/nar/28.21.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]