Abstract
This review attempts to illuminate the glycolipid antigen presentation properties of CD1d, how CD1d controls the function of natural T (iNKT) cells and how CD1d and iNKT cells interact to jump-start the immune system. It is postulated that the CD1d-iNKT cell system functions as a sensor, sensing alterations in cellular lipid content by virtue of its affinity for such ligands. The presentation of a neo-self glycolipid, presumably by infectious assault of antigen-presenting cells, activates iNKT cells, which promptly release pro-inflammatory and anti-inflammatory cytokines and jump-start the immune system.
Introduction
Recognition of foreign antigens by the adaptive immune system represents the fundamental underpinning of host defence, which is required for the integrity of a mammalian organism. The evolutionary roots of the adaptive immune system lie within the far older components of the innate immune system whose importance has been appreciated only recently. In fact, the adaptive immune system cannot function to adequately protect the organism without the extensive architecture provided by the innate immune system. Amongst the multiple cellular components of the innate immune system such as the natural killer (NK) cells, dendritic cells (DCs), macrophages and tissues themselves are the innate lymphocytes. Natural T (NKT) cells represent a subset of innate lymphocytes.1 Functions of NKT cells have been intensively investigated, and findings point to their fundamental participation at the earliest stages of the immune response to many pathogens2 and tumours.3 Furthermore, current evidence suggests that NKT cells also function to suppress autoimmunity,4–6 maintain immune privilege7,8 and support engraftment of transplanted tissues.9,10 Thus, it might be inferred that NKT cell function may have evolved not only to provide an impetus during challenge to organismal integrity but also to limit the consequent damage an overly avid immune reaction might inflict on the surrounding healthy tissue.
The CD1d antigen presentation system and the NKT cell antigen recognition system are extraordinarily well conserved from mouse to human, and amongst most other mammalian species. CD1 represents a family of MHC class I-like lipid antigen-presenting molecules. They are comprised of two groups of molecules based on sequence similarities; group I (CD1a, CD1b and CD1c) and group II (CD1d). Most mammalian species studied express group II CD1 molecules, whereas in some species, such as the guinea pig11 and miniature swine,12 only group I CD1 molecules have been discovered so far. Mice carry only group II CD1, CD1d1 and CD1d2; CD1d2 is a pseudogene and, hence, targeted disruption of CD1d1 is sufficient to render mice CD1 deficient.13 Group I CD1 molecules present self and foreign (e.g. mycobacterial) lipid antigens to T cells, while current evidence indicates that the group II CD1d molecules present a self lipid antigen(s) to NKT cells.14 This review focuses on antigen presentation by CD1d, the CD1 molecule that controls NKT cell function.
NKT cells can be broadly divided based on the kind of T-cell receptors (TCR) they express, namely, αβ vs. γδ. The αβ NKT cells predominate in this innate lymphocyte subset. Amongst these are two kinds: (1) the vast majority which are CD1d-restricted and (2) the less frequent whose restriction element remains unknown. Amongst the CD1d-restricted NKT cells are those that express the invariant Va14Ja18 TCR α-chain (iNKT), which represent the majority, and those that express diverse TCR α-chains. The focus here is on iNKT cells because much of our current understanding comes from studies of this lymphocyte subset.
Here, we review recent developments in molecular and cellular aspects of CD1d structure and function as well as iNKT cell ontogeny essential for understanding the physiology of these innate lymphocytes. The literature preceding this review is excellently summarized elsewhere.1–3,6,14–16
CD1d, a self lipid antigen-presenting molecule
Tissue distribution
Expression of CD1 molecules is tissue restricted and closely resembles the expression of MHC class II molecules. Thus, CD1d molecules are expressed by specialized antigen-presenting cells (APCs) such as splenic DCs and marginal zone B lymphocytes. Strikingly, marginal zone B cells as well as a novel subset of follicular B cells express high level of CD1d. Unlike MHC class II, CD1d is also expressed by CD4+8+ (DP) thymocytes, hepatocytes and, arguably, by intestinal epithelium. Whereas group I CD1 molecules are up-regulated by cytokine signals such as granulocyte-monocyte colony-stimulating factor (CSF-2) and interleukin (IL)-4, CD1d expression is constitutive and unresponsive to these cytokines. Whether other factors induce CD1d expression is currently unknown. Strikingly, they are not expressed by thymic cortical epithelium, a site where MHC class I and class II expression is essential for positive and negative selection of conventional T lymphocytes, suggesting that alternative selection mechanisms are operational during iNKT cell development (reviewed in 15).
CD1d structure dictates function
Critical to understanding the mechanism(s) by which CD1 binds and presents lipid antigens to T cells is an appreciation of the biochemical and structural features of CD1–lipid interaction. Solution of the crystal structures of both a group I CD1, human CD1b18 (hCD1b), and a group II CD1, mouse CD1d119 (mCD1d1), has provided invaluable clues to CD1 biology. Topologically, both structures resemble the classical peptide antigen-presenting MHC molecules. Akin to MHC class I molecules, CD1 consists of a heavy chain non-covalently associated with β2-microglobulin (β2m). The heavy chain folds in such a manner that the two membrane distal α1 and α2 domains form a groove made up of two α-helices supported at the bottom by two β-sheets. Both solved CD1 structures show similar features, which include a large, exclusively non-polar antigen-binding groove.18,19 However, the details of the antigen-binding grooves differ between hCD1b and mCD1d1. The structure of mCD1d1 was solved with, possibly, a conglomerate of lipids bound to its antigen-binding groove.19 Therefore, the details of CD1–lipid interaction could not be gathered from the solved structure. In contrast, the structures of hCD1b complexed with phosphatidylinositol (PI) and a sphingolipid GM2 were determined.18 The two structures point to key details of CD1–lipid interaction. Human CD1b has four non-polar hydrophobic channels termed A′, F′, C′ and T′,18 whereas mCD1d1 has only A′ and F′ channels.19 In the hCD1b structure, the T′ channel connects the A′ and F′ channels, allowing very long lipid alkyl chain to occupy its antigen-binding groove.18,20 In comparison to hCD1b, mCD1d1 contains critical amino acid substitutions in the region that makes channels T′ and C′. Hence, these channels are occluded in mCD1d1.18,19
Interestingly, we discovered that PI associates with mCD1d1 during biosynthetic assembly. Curiously, the eluted PI is phospholipase A2 resistant. This would suggest that the configuration of sn2-acyl group of PI bound to mCD1d1 is distinct from the PI bound in the crystal structure of hCD1b.18,21 Given that mCD1d1 may not contain channel C′, we would predict that PI binds mCD1d1 in a manner distinct from that observed in the hCD1b-PI structure.
CD1d assembly and intracellular trafficking
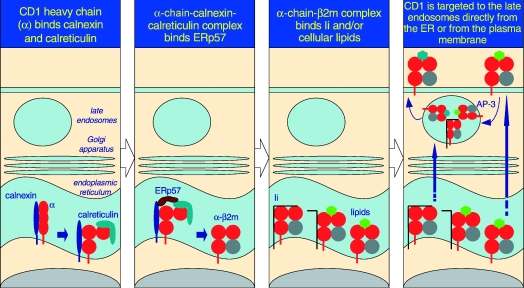
Being a type I integral membrane protein, CD1d folds and assembles within the endoplasmic reticulum (ER) lumen, and thence negotiates the secretory pathway to the plasma membrane. From the plasma membrane, it cycles numerous times through the late endosomes.22 In an effort to understand how CD1 molecules assemble in vivo, the ER protein chaperones that associate with hCD1d have been studied. Akin to hCD1b,23,24 hCD1d also binds calnexin.25 This association between CD1 and calnexin occurs in the ER because, in the absence of β2m, hCD1b was shown to be retained in this compartment by calnexin.23 Human CD1d also associates with calreticulin,25 another ER chaperone that assists protein folding and assembly in cells. Interestingly, the association of hCD1d with calreticulin occurs prior to β2m binding to the CD1 heavy chain.25 This finding is in contrast with that for the MHC class I–calreticulin association, which occurs only after the heavy and light chains have assembled together.26,27 The hCD1d heavy chain–calnexin–calreticulin complex facilitates association with a third ER chaperone, ERp57,25 an oxido-reductase that assists in disulphide bond formation critical for the integrity of CD1 structure (see Fig. 1).
Figure 1.
The predicted pathway for CD1d assembly and intracellular trafficking. As it is a type I integral membrane glycoprotein, the folding and assembly of CD1d occur in the rough ER. Here, calnexin, calreticulin and ERp57 assist the folding of the CD1d α-chain. β2m associates with the folded α-chain, unlike MHC class I molecules, which bind calreticulin only after association with the light chain. The α-chain+β2m+ complex forms a structure receptive to Ii and/or resident lipids and glycolipids. Upon stable association, the CD1d (α-chain and β2m)–glycolipid complexes egress from the ER and negotiate the secretory pathway to the plasma membrane. Because of the Yxxφ internalization sequence within the cytosolic tail of CD1d, it is rapidly internalized into the late endosomes in an AP-3-dependent manner and recycled back to the plasma membrane. In contrast, Ii-associated CD1d may be directly targeted to the late endosomes, and thence egress to the cell surface. During its time in the late endosomes, lipids bound to CD1d in the ER are exchanged for another, presumably cellular, glycolipid that is presented to iNKT cells.
Mouse CD1d1 biosynthetically associates with MHC class II-associated invariant chain (Ii; discussed below in detail).22 In Ii-negative cells, mouse CD1d1 assembles with PI and PI-glycans21,28 and possibly other phospholipids indigenous to the ER. PI and other phospholipids activate a small subset of iNKT hybridomas.29 Mouse CD1d1 assembled in the presence of Ii activates mCD1-restricted non-Va14 T cells22 which are also thought to recognize lipid antigens. Therefore, it is possible that a significant fraction of newly synthesized CD1d molecules assemble with cellular lipids. It also remains possible that Ii does not occlude the antigen-binding site of CD1d, thus permitting biosynthetic assembly of CD1d-Ii complexes with ER lipids. Therefore, we postulate that cellular lipids indigenous to the ER may serve as a chaperone to protect the large, exclusively polar and hydrophobic antigen-binding groove of CD1 from collapse in the aqueous ER lumen. Thus, CD1d utilizes both protein and possibly lipid chaperones for biosynthetic assembly (see Fig. 1).
Following assembly and arrival at the plasma membrane, by virtue of the Yxxφ endosomal/lysosomal-targeting motif within their cytosolic tail, hCD1d and mCD1d1 cycle through the early and late endosomes for lipid sampling30–34 (see Fig. 1). Curiously, however, despite containing the Yxxφ motif, the two CD1 homologues traffic through different endosomal compartments.31,34,35 Thus, amino acid residues within the Yxxφ motif and/or those adjoining it may influence endosomal targeting. Interestingly, clathrin adaptor complex AP-3-deficient Pearl and Mocha mice have defects in iNKT cell development and in lipid antigen presentation to this subset of T lymphocytes (M. Kronenberg, La Jolla Institute for Allergy and Immunology, San Diego, CA, personal communication; Fig. 1). Defective antigen presentation ensues in AP-3-deficient cells because mCD1d1 is unable to negotiate the LAMP-1-positive late endosomes. In contrast, hCD1d, unlike mCD1d1, does not bind AP-3, but traffics to the appropriate endosomal compartment, and hence no defects in iNKT cell development were observed in AP-3-deficient Hermansky–Pudlak syndrome type 2 patients.31 It is noteworthy that the carboxyl terminal residue, proline, of hCD1b, which immediately follows its Yxxφ motif, is critical for AP-3 binding and trafficking to the LAMP-1-positive endosomes.31 Perhaps the inability of hCD1d to bind AP-3 may be attributable to the Y+2 glycine and/or Y+4 leucine within its cytosolic tail.36
In addition to Yxxφ-mediated targeting, CD1 can negotiate the low-pH compartment in association with the Ii22 (Fig. 1) by virtue of the latter's di-leucine/isoleucine motif. Ii-mediated targeting to late endosomes rescues the antigen presentation deficiency of an mCD1d1 mutant incapable of negotiating the low-pH compartment as a result of the deletion of the Yxxφ motif within its cytosolic tail.22 Moreover, Ii-deficient splenic DCs are defective in antigen presentation to iNKT cells.22 These findings underscore the significance of the CD1d–Ii association in cell lines. That notwithstanding, mice with targeted germ-line deletion of the cytosolic tail of mCD1d1 had profound defects in iNKT cell development and in the presentation of endogenous lipid antigens to an iNKT hybridoma.34 The observed defects in the mutant mice may be a result of the fact that only a relatively small fraction (up to 25%) of newly synthesized CD1d1 associates with Ii.22 Ii also has other effects on endosomal compartments37,38 which might provide an alternative explanation for the effect of Ii on intracellular CD1d traffic. Thus, the physiological significance of the CD1–Ii association remains to be established.
Interestingly, hCD1d interacts with Ii-associated HLA-DR in the ER, and this complex negotiates the secretory pathway to the late endosomes and the plasma membrane.35 However, mCD1d1 traffics and functions normally in MHC class II-negative cells. Thus, the functional significance of the hCD1d–class II interaction remains unknown.
In the case of MHC class II-restricted antigen processing, Ii is removed upon arrival at the late endosomes by cathepsin (Cat)-S-mediated proteolysis (reviewed in ref. 39). A similar mechanism may be operable in dissociating Ii from CD1, because Cat-S-deficient mice poorly develop iNKT cells and are unable to present lipid antigens to this subset of T lymphocytes.40 Another study showed that Cat-L, but not Cat-S, is required for iNKT cell development.41 Further, Cat-L-deficient DP thymocytes, but not splenic DCs, have lost their ability to present iNKT cell antigen.41 Biochemical studies indicate that the association between CD1 and Ii is weak,22,40 suggesting that it might easily dissociate in the acidic environment of the late endosomes/lysosomes, and hence may not require proteolysis. Further, the cathepsins may play a role in antigen processing, either directly by trimming lipoproteins or indirectly by activating processing enzymes. In this regard, it is noteworthy that Cat-L is critical for the maturation of arylsulphatase A,42 an enzyme important in sulphatide catabolism.
In summary, intracellular CD1d trafficking occurs by Ii-independent and -dependent mechanisms (Fig. 1). In Ii-negative cells, such as DP thymocytes and hepatocytes, CD1d assembles in the ER, presumably with endogenous lipids, negotiates the secretory pathway and arrives at the plasma membrane (Fig. 1), a path also taken by newly synthesized MHC class I molecules. In contrast, in Ii-positive antigen-presenting cells, Ii-associated CD1d is directly transported to the LAMP-1-positive late endosomes (Fig. 1), akin to MHC class II–Ii complexes. Ii dissociates from CD1d, which then traffics to the plasma membrane. Independent of the mechanism by which CD1d traffics to the plasma membrane, it cycles numerous times through late endosomes (Fig. 1) because of the Yxxφ motif within its cytosolic tail, facilitating antigen presentation to iNKT cells.
Clues to the natural iNKT cell antigen
The diversity and the chemical nature of the natural antigen(s) presented by CD1d and recognized by iNKT cells have not been elucidated. Current evidence suggests that iNKT cells recognize a CD1d-restricted self glycolipid antigen(s). Thus, freshly isolated thymic NKT cells are activated by a variety of CD1d+ cells including thymocytes and tumour lines without the addition of exogenous antigen of any sort. Similarly, iNKT cell-derived hybridomas recognize CD1d+ cells and cell lines.33,43–45 This presentation of self-antigens requires that CD1d traffics through late endosomal vesicles32–34,44 and hence the antigen resides within the late endosomes.
Several approaches have been utilized to determine the nature of the iNKT cell antigen(s). In one approach, the synthetic anti-tumour agent α-d-galactosylceramide (αGalCer), originally extracted from a marine sponge, was identified as a selective and potent activator of all iNKT cells in a CD1d-restricted manner.46–51 The unusual α-anomeric configuration of galactosylceramide has not been described in mammals. Additionally, synthetic α-d-glucosylceramide (αGlcCer), but not α-d-mannosylceramide, confers antigenic activity similar to αGalCer.46,49 Two complementary approaches have been utilized to identify naturally processed iNKT cell antigen(s). In the first approach, as with peptide antigens presented by MHC class I and class II molecules, it was reasoned that iNKT cell antigen(s) would be found in total cellular lipid extracts. Indeed, using an in vitro reconstitution assay cellular phospholipds were identified as antigens of a few iNKT hybridomas.29
We reasoned that, if αGalCer and αGlcCer are synthetic iNKT cell antigens, then cellular βGalCer and/or βGlcCer may be a precursor to the natural antigen. The rationale behind this reasoning is that the biosynthesis of βGalCer and βGlcCer uses UDP-α-galactose or UDP-α-glucose as the sugar donor. Because cells are not known to contain UDP-β-galactose and UDP-β-glucose, βGalCer and βGlcCer may serve as substrates for αGalCer and αGlcCer synthesis, respectively. Therefore, we utilized a genetic approach in which cells deficient in endogenous βGalCer and βGlcCer served as antigen-presenting cells. The data revealed that cellular synthesis of βGlcCer, but not βGalCer, is essential for the presentation of a natural self-antigen to iNKT hybridomas.52 βGlcCer deficiency altered neither the kinetics of intracellular CD1d1 traffic and distribution, nor αGalCer presentation by CD1d1. These data indicate that β-D-GlcCer may be a precursor of a natural iNKT cell antigen or may be involved in an accessory function necessary to generate or load the antigen.52 Taken together, the evidence gathered so far indicates that CD1d molecules have evolved to sample endogenous lipid antigens for presentation to iNKT lymphocytes.
Lipid antigen processing
The original experiments, which demonstrated mycobacterial PI-mannoside (PIM6) specificity of an hCD1b-restricted T lymphocyte clone, suggested that the presentation of some lipid antigens involved processing.53 The lysosomal α-galactosidase has been implicated in generating an active iNKT cell ligand.54 Plate-bound mCD1d1 presents αGalCer, but not the analogue Gal(α1→2)GalCer, to iNKT cells. However, cells from wild-type but not from α-galactosidase-deficient mice efficiently present Gal(α1→2)GalCer to iNKT cells. Further, this presentation is sensitive to an inhibitor of lysosomal α-galactosidase. Moreover, in vitro processing of other αGalCer analogues, GalNAc(α1→3)Glc(α1→2)GalCer with lysosomal α-N-acetylgalactosaminidase or α-glucosidase and Gal(β1→2)GalCer with β-galactosidase, also generates iNKT cell antigens.54 Although the above studies utilized synthetic ligands, which might be related to the natural antigen, the results suggest a role for glycolipid processing in vivo and imply a range of potential precursors for iNKT cell antigen(s).
iNKT cell antigen recognition
The hCD1b-PI and hCD1b-GM2 crystal structures also revealed the TCR interface area on antigen.18 Previous studies suggested that the hydrophobic alkyl/acyl chains bind CD1 exposing the hydrophilic aspect of the lipid to form part of the TCR interface area on the antigen.55 Mutagenesis studies revealed that the TCR interface approximates a diagonal, which includes both α-helices and the glucose residue of the antigen.56 Consistent with the proposed model, both the phosphoinositol of PI and the first glucose residue of GM2 are oriented in a manner that approximates the side chain of the amino acid residue at position 4 of peptides presented by MHC class I molecules.18 However, the glucose residue of GM2 is raised far above the plane of the helices.18 This raised disposition of glucose may be attributable to the β-O-1,1′ linkage of this residue to the ceramide backbone. The α-O-1,1′ linkage of αGalCer and αGlcCer should orient the sugar ring differently so that it may be more closely juxtaposed to the plane of the α-helices of CD1d. Although the molecular definition of the CD1–lipid/TCR interface awaits the solution of the structure of the co-complex, it is clear that more than the hydroxyl groups of the monosaccharide can come in contact with the receptor. Thus, the TCR–lipid antigen interface, in addition to the hydroxyl groups from the sugar residue, includes the phosphate of phospholipids or the amide of sphingolipids.18
Gene transfer and mutagenesis studies have demonstrated that the α and β chains of the TCR are sufficient for CD1–lipid antigen recognition.57,58 Further data suggested that the CDR3 loops of the specific TCR interface the combinatorial epitope formed by the CD1–lipid complex.57 Hence, antigen recognition follows the same structural principles observed in TCR–peptide antigen recognition.59 Interestingly, however, CD1d1-restricted self-antigen recognition did not segregate with the transfer of Va14Ja18/Vb8·2 TCR α and β chains. This result suggests the involvement of an additional molecule(s) along with the Va14Ja18 TCR in the recognition of CD1d-restricted self-antigen(s).57
The binding parameters of CD1–antigen/TCR interaction remain ill-defined. Recent surface plasmon resonance and tetramer-binding studies revealed high-affinity interaction between CD1d1-αGalCer and Va14Ja18 TCR; the relative avidity of this interaction is similar to that of high-affinity MHC class I–peptide/TCR interactions.60 Interestingly, the half-life of the CD1d1-αGalCer/Va14Ja18 TCR interaction was unusually long.60 How these kinetic parameters of lipid antigen–TCR interactions relate to the rapid and robust iNKT cell response remains to be elucidated. In this regard, it is interesting to note that an analogue of αGalCer, OCH, which has a shortened long-chain sphingosine base (C5 vs. C14) and acyl chain (C24 vs. C26), specifically elicits sustained IL-4 with very little interferon (IFN)-γ from iNKT cells.61 The binding parameters of the interactions between the Va14Ja18 TCR and mCD1d1 bound with OCH vs. αGalCer might reveal the mechanism underlying the differential in vivo cytokine response to the two ligands.
iNKT cell function
NKT cells are innate lymphocytes that express cell-surface markers characteristic of T cells as well as NK cells (reviewed in 1). Their identification has relied primarily upon co-expression of TCR αβ and NK1·1 (CD161). About 80% of mouse NK1·1+TCR+ cells have the invariant Va14-Ja18 rearrangement. Recent development of CD1d tetramer loaded with the synthetic antigen αGalCer allows antigen receptor-specific identification of iNKT cells independent of other cell-surface markers, and hence permits more detailed study of their development and distribution.50,51,62 These studies, in addition to those previously reported based on NK1·1+TCR+ cells, revealed that iNKT cells are CD4+8− or CD4−8− (DN). Their phenotype also reflected characteristics of effector/memory T lymphocytes (CD44, CD122, Ly6C and CD62Llow).63,64 In mice, iNKT cells constitute 10–20% of mature thymic, 20–30% of hepatic and bone-marrow and 2–3% of splenic T cells as well as 0·1–0·5% of lymph node and peripheral blood mononuclear cells (PBMCs).63,65–67 While expression of the Ly49 receptor family members can be used to distinguish thymic and hepatic iNKT cells, the difference in function of these subsets, if any, is currently unknown.65,68–70 Thus, iNKT cells maintain a distinct tissue distribution that may be critical to their function.
iNKT subsets
Recent studies with mouse CD1d tetramer-based identification of human Va24Vb11 iNKT cells isolated from PBMCs revealed the existence of two functionally distinct subsets. The functional distinctions have been found to correlate with CD4 co-receptor expression.71,72 In humans, as in mice, iNKT cells are CD4+8− or DN. Activated DN iNKT cells preferentially secrete Th1 cytokines such as IFN-γ and tumour necrosis factor (TNF)-α, whereas the CD4+8− cells secrete both Th1-type and a plethora of Th2-type cytokines (IL-4, IL-5, IL-10 and IL-13).71,72 Furthermore, DN but not CD4+8− iNKT cells produce intracellular perforin following IL-2, phorbolmyristylacetate and ionomycin, or αGalCer stimulation.72 Interestingly, exposure to IL-2 or IL-12 alone is sufficient to induce production of intracellular perforin, but not IFN-γ, indicating that complete activation of iNKT cells requires stimulation through their TCR.72 Maturation of mouse NK1·1-negative iNKT cells has been correlated with reduced expression of the CD4 co-receptor.73–75 Th1 bias of human DN iNKT cells is in complete agreement with a similar bias observed with mature NK1·1+ mouse iNKT cells.73–75 These observations suggest that human iNKT cells may also undergo a similar developmental regulation of CD4 expression. That notwithstanding, whether CD4 is a more reliable indicator of iNKT cell maturity in humans than in mice remains to be determined.
CD4+8− and DN subsets of human iNKT cells have distinct distributions of chemokine receptor expression, suggesting subset-specific trafficking. Human iNKT cells express high levels of CCR5.71,76 Such iNKT cells are highly susceptible to HIV-1 infection.76,77 In fact, TCR-stimulated iNKT cells are several-fold more susceptible to HIV infection than either memory or naïve TCR-stimulated CD4 T cells.76 In support of these findings, HIV-infected individuals have diminished numbers of CD4+ iNKT cells.76,78,79 Thus, CD4 plays a role in iNKT cell pathology in HIV and is correlated with distinct functional subsets of human iNKT cells.
Rapid and robust iNKT cell cytokine response
Historically, NKT cells were discovered as the source of early IL-4 secretion following in vivo stimulation with antibodies to CD3ε.13,80–83 To this day, the early (i.e. 60–90 min following αGalCer or anti-CD3ε injection) and robust secretion of cytokines remains the most notable functional feature of iNKT cells. Following stimulation through their TCRs, iNKT cells have been reported to secrete IL-2, IL-4, IL-5, IL-10, IL-13, IFN-γ, TNF-α, CSF-2, lymphotactin, macrophage inflammatory protein (MIP)-1α, MIP-1β and RANTES (reviewed in 17). The diversity of cytokines and other soluble messengers synthesized suggests that iNKT cells have a T-helper (Th) phenotype (Fig. 2). It is noteworthy that iNKT cells express cytokines in a developmental and age-dependent manner.73–75,84,85 Immature mouse NK1·1-negative iNKT cells73–75 and neonatal human cord blood iNKT cells preferentially secrete IL-4.85 However, mature iNKT cells73–75,84 and those from older mice84 preferentially secrete IFN-γ. The precise molecular mechanisms and physiologic consequences of the developmental regulation of iNKT cell cytokine secretion remain unknown. However, dysregulation of iNKT cell cytokine secretion and iNKT cell overactivation in SOCS-1-deficient mice result in severe hepatic injury.86 Furthermore, administration of αGalCer to mice results in significant liver injury and death in old mice, an αGalCer effect that could be inhibited by antibodies to IFN-γ, but not IL-4.84 Also, αGalCer activated iNKT cells from older, but not younger, mice exhibit Fas ligand (FasL) up-regulation.84 Antibody blockade of FasL partially ameliorates the in vitro cytotoxicity of activated iNKT cells.84 Thus, inappropriate iNKT cell activation can result in serious hepatotoxicity and perhaps death.
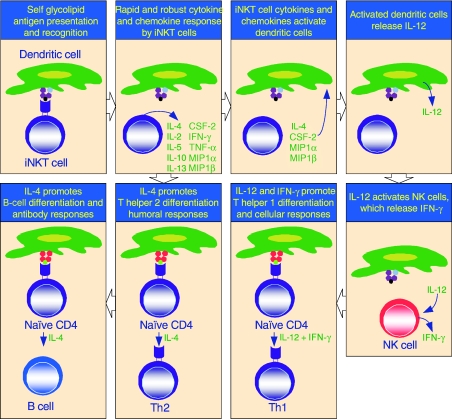
Figure 2.
The immunological functions of iNKT cells. iNKT cells participate in cross talk between members of the innate and the adaptive immune system by deploying cytokine/chemokine messengers. Upon activation in vivo, iNKT cells rapidly secrete several cytokines/chemokines. Of these cytokines/chemokines, IL-4, CSF-2, MIP1α and MIP1β facilitate the recruitment, activation and differentiation of macrophages and dendritic cells resulting in the production of IL-12 and possibly other factors. IL-12, in turn, stimulates NK cells to secrete IFN-γ. Along with IL-12, IFN-γ can polarise the differentiation of antigen-activated CD4+ T cells towards a Th1 phenotype. IL-4 can skew the differentiation of CD4+ T cells towards a Th2 phenotype. IL-4 can also activate B cells in vivo resulting in their differentiation to antibody secreting-plasma cells. Thus iNKT cells have the potential for jump-starting an immune response.
Activation of iNKT cells requires signalling through their TCR and through costimulatory receptors. Thus, a blockade of CD28, CTLA-4, CD80, CD86, CD40 and CD40 ligand (CD40L) prevents iNKT cell activation.46,80 The physiological role of killer inhibitory receptors (KIR) in iNKT cell activation is not fully understood. However, antibody-mediated cross-linking of NK1·1 has been demonstrated to activate NKT cells.15,87,88 Contrarily, activation of iNKT cells by αGalCer is inversely correlated with Ly49 expression.89 Because H2Db, a KIR ligand, interacts with Ly49, a KIR, its expression by immature DCs inhibits the activation of sorted NK1·1+TCR+ cells.90 Thus, iNKT cell activation is tightly regulated, as would be expected for a lymphocyte population exhibiting self-antigen recognition that might ordinarily damage tissues wherein they reside.
The activation of iNKT cells in vivo results in the prompt secretion of both IL-4 and IFN-γ.91,92 A recent study demonstrated that αGalCer-pulsed DC activates iNKT cells in vivo and stimulates IFN-γ secretion.93 However, a careful examination of the cell type that secretes IFN-γ in response to αGalCer in vivo revealed NK cells as the major source of this cytokine.92,94αGalCer-activated iNKT cells stimulate IL-12 production by DCs in a CD40L-dependent manner. Further, antibody inhibition of IL-12 suppressed the IFN-γ levels following αGalCer administration.95–97 Thus, iNKT cell activation mediated by αGalCer presented by DCs in turn elicits IL-12 from DCs and consequent activation and IFN-γ secretion by NK cells (Fig. 2).
Immune consequences of iNKT cell cytokines
The prompt IL-4 secretion upon stimulation of iNKT cells by αGalCer in vivo also has functional consequences. IL-4 activates B cells, as inferred from the expression of the early activation marker CD69.92,98 Expression of CD69 is induced within 6 hr of αGalCer treatment.92 Activated B cells are also induced to express CD86, the ligand for the T-cell co-stimulatory molecule CD28.91 Within 5 to 6 days, αGalCer administration also induces IgE secretion,4,91,99 a hallmark of the Th2 immune response. Additionally, the stimulation of iNKT cells by αGalCer at the time of immunization with nominal antigen results in antigen-specific IgE response.4,91 Thus, IL-4 secreted by activated iNKT cells has a Th2 polarizing effect in response to antigen in vivo (Fig. 2).
αGalCer-activated iNKT cells cross-talk with CD8+ T lymphocytes.100,101 Such communication between NKT cells and CD8+ T lymphocytes is absent in CD1d1- and Ja18-deficient mice. Activation of CD8+ cells requires CD1d1 for the presentation of αGalCer as well as CD40–CD40L interactions between iNKT cells and DCs.100,101 CD8+ lymphocytes thus activated possess the ability to secrete IFN-γ as well as mediate cytotoxicity. In the presence of a tumour, specific CD8+ T lymphocytes were elicited rapidly.100 Thus, iNKT cells activated by αGalCer in vivo can communicate with CD8+ T lymphocytes as well. Recent evidence suggests that DC maturation induced by activated iNKT cells facilitates communications with CD8+ T lymphocytes.101
iNKT cells function to enhance immunity in vivo through the adjuvent-like activity of αGalCer.2,101,102 In contrast, they are also known to suppress immunity, as seen in the maintenance of immune privilege7,8 and the down-modulation of autoimmune diseases.4,5,103–107 Mechanistically, how do iNKT cells mediate such opposing effects on the adaptive immune response? iNKT cell activation by DCs to produce IFN-γ may have important functional consequences.93 It is possible that activation of iNKT cells by DCs at the site of injury causes sustained IFN-γ secretion. Alternatively, iNKT cells in the area immediately surrounding the site of injury may preferentially secrete IL-4 and/or promote anergy.108 Also, iNKT cells lose NK1·1 expression upon activation,109 which has been correlated with preferential IL-4 secretion.73–75 IL-4 and CSF-2 secreted by iNKT cells could influence DC maturation which then promotes Th2 responses.104 Thus, iNKT cells during the first few days of an infection may promote the elimination of the invading pathogen, while suppressing the damage of healthy tissues surrounding the site of the infection. This mechanism may explain the duality of iNKT function.
Activation of iNKT cells in vivo results in their cell death, as evidenced by significant binding of annexin-V by 12–24 hr post-activation.110 However, DCs presenting αGalCer stimulate sustained production of IFN-γ from activated iNKT cells over several days.93 Activated iNKT cells also down-regulate the expression of NK1·1,109 precluding their detection. Thus, in contrast to previous assumptions, it is possible that a subset of originally activated iNKT cells survive αGalCer treatment and mediate sustained effector function(s).
iNKT cell ontogeny
Central aspects of iNKT cell development
The developmental origin of iNKT cells has been an issue of much debate, with both thymic111–113 and extrathymic114–116 sites suggested. More recent studies employing CD1d-αGalCer tetramer and real-time RT-PCR for the Va14-Ja18α-chain gene rearrangement in athymic nu/nu mice failed to provide evidence for the development of iNKT cells in the absence of a functional thymus.74,117 Although previous studies described the development of NK1·1+TCR+ lymphocytes in the fetal liver as early as embryonic day 9,114,116 more recent studies failed to demonstrate CD1d1-αGalCer tetramer positive cells within this site.74,75 iNKT cells develop in fetal thymic organ cultures (FTOCs), and, consistent with their postnatal ontogeny in mice, they appear after conventional CD4+8− and CD4−8+ single-positive cells.63,74,118 Additionally, DP thymocytes of CD1d-deficient mice contain precursors of iNKT cells, which develop in CD1d-competent but Ja18-deficient mice following intrathymic, but not intravenous, injection.117 Further elucidation of the developmental pathway revealed that early, immature iNKT cells express their TCR prior to the acquisition of NK1·173–75 and DX5 markers.75 Immature, NK1·1-negative iNKT cells proliferate strongly.73–75,117 Intrathymic injections of fluorescin, and tracking of recent thymic emigrants (RTE) revealed that an unusually large proportion of total thymic efflux consists of iNKT cells in young mice.73,74,119 These studies, taken together, provide strong evidence for late fate specification of iNKT cells and the thymus as the predominant if not the only site of early iNKT cell development.
iNKT cell development critically depends on the interaction of the putative NKT cell precursors with CD1d-positive thymic haematopoietic cells.66,111–113,120,121 This interaction is unusual, in that positive selection of conventional T lymphocytes depends on interactions with MHC molecules of the thymic cortical epithelium (Fig. 3). What part this unconventional interaction plays in iNKT cell fate specification remains unclear.
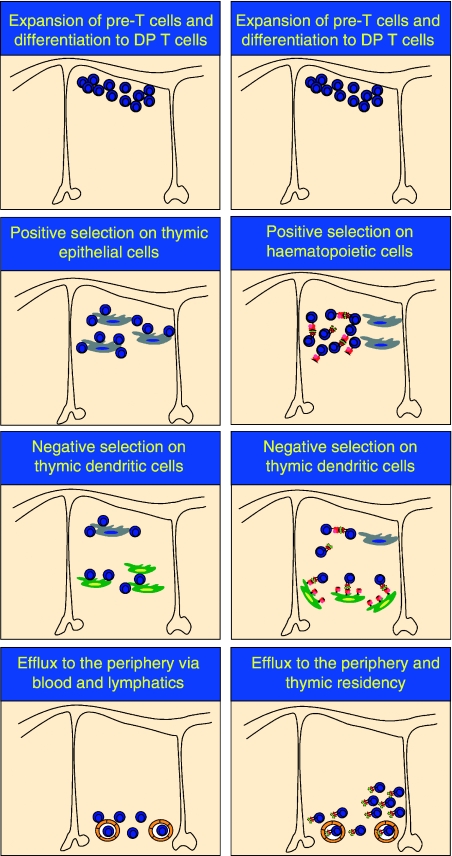
Figure 3.
A schematic of cell-to-cell interactions during positive and negative selection in the thymus. Conventional T and iNKT cells develop in the thymus, and share DN T-cell precursors. However, positive selection of iNKT cells proceeds via interactions with haematopietic cells, while conventional T cells require thymic epithelium for the positive developmental signal. Negative selection of both conventional T and iNKT cells is mediated by thymic dendritic cells. Both conventional T and iNKT cells are exported to the periphery following successful thymic education; however, a subset of iNKT cells remain thymic residents. Thus, conventional T and iNKT cells have distinct cellular requirements during positive and negative selection.
iNKT cells, akin to conventional T lymphocytes, undergo negative selection by a mechanism which appears to depend on affinity and avidity.118 Thus, overexpression of CD1d1 under the control of the H2Kb promoter caused significant deletion of CD1d1-αGalCer tetramer-reactive iNKT cells.118 Furthermore, addition of αGalCer, a high-avidity ligand, to FTOC also resulted in the deletion of developing iNKT cells.118 However, in agreement with previous observations, once iNKT cells develop, they are insensitive to deletion by αGalCer.15,118 Interestingly, addition of αGalCer-loaded DCs, but not thymic epithelial cells, to FTOC mediates negative selection.118 Thus, iNKT cells undergo both positive and negative selection under the control of CD1d-presenting endogenous glycolipid antigens (Fig. 3).
The molecular and developmental bases for the acquisition of an unconventional phenotype by iNKT cells are poorly understood. However, current evidence suggests a developmental relatedness of iNKT cells to NK cells. iNKT cell development is impaired in mice deficient in Ets1,122 myeloid elf–l–like factor (Mef),123 interferon-regulatory factor (Irf)-1124 and lymphotoxin (Lt)αβ125 at distinct stages (see Fig. 4). Ets1- and Fyn-deficient mice are severely impaired in iNKT cell development and function.122,126,127 Interestingly, Ets1 is expressed in both conventional T lymphocytes and NK cells.128,129 Ets1- and Fyn-deficient T cells display defects in TCR signal transduction.128,130 Conventional T cells develop in Ets1- and Fyn-deficient mice.128,130 These results taken together suggest that the quality of TCR signalling during iNKT cell commitment is critical for their development.
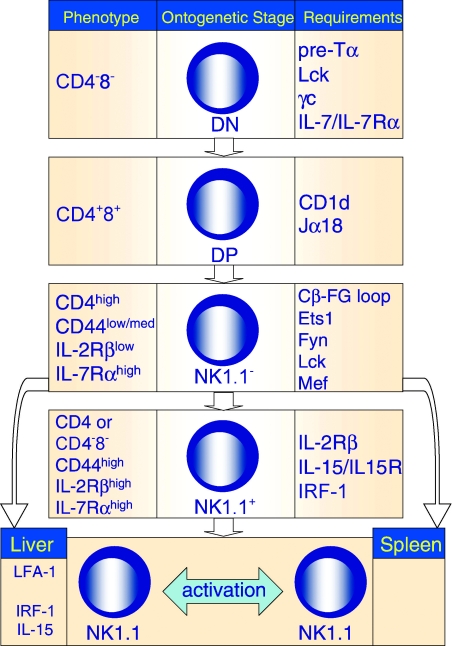
Figure 4.
Molecular control of iNKT cell development. The development of a functional repertoire of iNKT cells requires progression through several developmental intermediates in the thymus, export to the periphery and homeostatic proliferation in resident organs. In the column on the right are molecular factors required for iNKT cell progression past the stage shown in the middle column. The phenotype of the developmental intermediate is shown in the left column.
Another interesting similarity between iNKT cells and NK cells is the ontogenetic expression of KIR such as Ly49 family members. NK cells during development interact with stromal MHC class I molecules. Through the mediation of Src homology region 2 domain-containing phosphatese (Shp)-1 phosphatase, the interaction of KIR with self MHC inhibits NK cell activation and hence the killing of self.131,132 Expression of KIR by iNKT cells correlates with prolifation arrest and concurrent maturation.73–75 Thus, subsequent interactions with self through the TCR may be regulated by Shp-1.70
In adult mice, iNKT cells express CD4 or neither CD4 nor CD8 co-receptors. Further, transgenic overexpression of CD8 inhibits the development of iNKT cells.63 An interpretation of this finding is that CD8 enhances the already putatively strong interaction of the Va14Ja18 TCR and the selecting self-antigen and causes negative selection of developing iNKT cells. If true, this would preclude a DP precursor iNKT cell. An alternative possibility is that forced overexpression of CD8 might alter the delicate balance of Src kinases associated with co-receptor chains on developing iNKT cells. This is especially pertinent considering the following points. (1) iNKT cell development is very sensitive to alterations in the levels of cellular Src kinase. Both Lck0/0 and Fyn0/0 animals completely lack iNKT cells.126,127 In contrast, a significant degree of Lck-Fyn cross-compensation is observed in conventional T-cell development.133 (2) The cytoplasmic chain of CD8 interacts with Lck, albeit less avidly than with CD4134. Endogenous CD8 is often alternatively spliced in thymocytes to remove the Lck-interacting site.135 Thus, one would hypothesize that transgenically overexpressed CD8136 might bind and remove a significant amount of limiting Lck from the iNKT cell synapse during ontogeny. This quenching of Lck activity might impact iNKT cell development far more severely than it does conventional T-cell ontogeny. This alternative explanation fully supports a model for iNKT cell development via a DP precursor.
Cytokine control of iNKT cell development
Akin to conventional T-cell development, cytokines provide important signals for iNKT cell ontogeny. For example, mice deficient in IL-7Rα or IL-7 develop very few iNKT cells and conventional T cells.88,137 In the thymus, both populations are equivalently affected by deficient IL-7 signalling.88 In contrast, peripheral iNKT cells are significantly impacted by IL-7Rα deficiency compared with conventional T cells.88 Thus, signalling via receptors utilizing IL-7Rα not only impairs the early DN stage of lymphopoiesis138 but also plays a preferential role in providing survival signal to exported iNKT cells.88,137
Explanted thymocytes incubated with IL-7 for 5 days contain an enriched population of iNKT cells, which have undergone several cell divisions as demonstrated by [5–(6–) carboxyfluoresceindiacetate succinimidyl ester (CFSE) dye dilution experiments.75 IL-7Rα levels on immature (NK1·1−) and mature (NK1·1+) iNKT cells are roughly equal.75,88 Mature iNKT cells preferentially express IL-2/15Rβ, whereas neither mature nor immature iNKT cells express IL-2Rα.65 Preferential expression of IL-2/15Rβ but not IL-2Rα on mature iNKT cells suggests that IL-15 may play a significant role in their homeostasis. Indeed, mice deficient in IL-1588,139 or its receptor140,141 have diminished numbers of thymic iNKT cells (Fig. 4). There is a specific reduction in the proportion of mature but not immature thymic iNKT cells; about a two-fold reduction in the splenic and a much greater reduction in the hepatic iNKT cells.88,139 The importance of IL-15 to liver iNKT cells will be discussed below.
Deficiency of both IL-7 and IL-15 results in a complete absence of iNKT cells.88 Both IL-7 and IL-15 have the common γ chain (γc) as a component of their cognate receptor complexes. It is therefore surprising that mice deficient in γc have been reported to have a significant population of iNKT cells.142 These cells are NK1·1-negative, Ly49C-negative, and IL-2/15Rβlow; they express a biased Vβ repertoire, produce IL-4 upon in vitro stimulation, and have mRNA encoding rearranged Va14-Ja18 gene segments.142 This finding was interpreted to mean that these cells are immature iNKT cell precursors,142 consistent with more recent work.73–75 Interestingly, poor survival of these iNKT cells in vitro was significantly reconstituted by the addition of thymic stromal lymphopoietin (TSLP), a cytokine which shares the IL-7Rα subunit with the IL-7 receptor,143 suggesting that TSLP may function as a survival factor for iNKT cells.142 However, the physiological role of TSLP in vivo has not yet been investigated. While the recent study of the function of IL-7 and IL-15, cytokines utilizing γc as a part of their cognate receptor complex, was carried out with older (6–15 weeks old) mice,88 the earlier study used younger (4–10 weeks old) γc-deficient animals.142 As iNKT cells develop postnatally,63,117 the role of cytokines utilizing γc may be more important in older mice.
The importance of IL-15 in the maturation and/or survival of iNKT cells is underscored in mice deficient in Irf-1.124 They show a similar iNKT cell phenotype to IL-15-deficient mice. Irf-1-deficient mice have diminished IL-15 mRNA expression following stimulation of bone marrow-derived cells. In vitro supplementation with IL-15 results in increased numbers of NK and NK1·1+TCR+ cells.124 Thus, dependence on signalling through the IL-15 receptor complex is a common feature of iNKT and NK cells.
Molecular and cellular features of peripheral iNKT cells
Molecular and cellular features of peripheral iNKT cells are exported from the thymus as both NK1·1− immature and NK1·1+ mature lymphocytes. Interestingly, iNKT cells form a large proportion (about 4%) of RTE.73,74 The majority of RTE iNKT cells are NK1·1-negative and incorporate bromodeoxyuridine (BrdU).73,74 Other experiments have demonstrated that splenic iNKT cells show significant labelling with annexin-V at steady state, which is highly up-regulated following stimulation with αGalCer, anti-CD3ε or IL-12.50,110,144,145 These results strongly suggest that iNKT cells are in a dynamic flux between cell proliferation and cell death. Somewhat surprisingly, splenic iNKT cells express high levels of Bcl-2, and are highly radio-resistant compared with conventional T cells.146,147 Thus, the dynamics of Bcl-2 family member expression and the mechanisms of cell survival and cell death of iNKT cells at steady state and following various types of stimulation warrant further study.
The size of the iNKT cell pool in the periphery is dependent on thymic eflux as well as the balance of proliferation, survival and death. Surprisingly, and in contrast to the requirement for trafficking-competent CD1d during iNKT cell development, CD1d expression appears not to be important for iNKT cell survival and proliferation in the peripheral organs.88 However, cytokines such as IL-15, and to a lesser extent IL-7, provide a critical impetus for their proliferation and survival in the periphery.88 Interestingly, iNKT cells do not seem to have a separate niche, as the extent of proliferation upon cell transfer into wild-type and iNKT cell-deficient Ja180/0 mice is roughly equal.88 However, NK and CD8 memory T cells strongly compete with iNKT cells for IL-15.88 Furthermore, depletion of NK cells found in RAG10/0 by anti-asialo-GM1 or irradiation strongly enhanced the homeostatic proliferation of iNKT cells in vivo.88 Transgenic overexpression of IL-7 in IL-15-deficient animals did not increase the thymic complement of iNKT cells, and only partially rescued the liver complement.88 In addition, the absence of IL-7 in recipient animals did not impact homeostatic proliferation of iNKT cells, arguing against a preferential role of this cytokine in peripheral NKT cell survival.88 As noted earlier, IL-7 deficient mice develop few iNKT cells; in the thymus their deficit is proportional to conventional T lymphocytes but in the periphery iNKT cells are impacted more than conventional T cells.88 It is likely that this disparity may have to do with iNKT cell efflux to the periphery under conditions of IL-7 deficiency. Thus, iNKT cells share ‘space’ with NK cells and to a lesser extent with memory CD8 lymphocytes.88
The development of a full complement of hepatic iNKT cells has additional requirements compared with the splenic population. In addition to IL-15 and Irf-1 deficiencies, which mostly affect hepatic iNKT cells, leukocyte function-associated antigen (LFA)-1 deficiency also diminishes their number in the liver.68,148 iNKT cells express LFA-1, as well as its ligands intercellular adhesion molecule (ICAM)-1 and ICAM-2.149 Interestingly, while the expression of LFA-1 on iNKT cells was not essential for their migration to the liver, bone-marrow reconstitution experiments demonstrated that LFA-1 on hematopoietic cells was essential for the development of liver iNKT cells.68 A resolution of this conundrum was offered by the finding that a full NK cell complement in the liver was essential for the homing of iNKT cells to that organ.149 These results also help explain the preferential requirement for IL-15 in the development of a full liver iNKT cell compartment. As IL-15-deficient animals have an almost complete loss of NK cells,139 iNKT cells may be unable to home to the liver as a consequence of the NK cell deficit. Thus, the evidence suggests that LFA-1 on NK cells interacts with its ligands on iNKT cells during the latter's circulation and causes their accumulation in the liver.149
As alluded to earlier, iNKT cells respond to a CD1d-restricted self-antigen(s). This property of iNKT cells prompts an important question: How does an innate self-recognizing lymphocyte develop and maintain tolerance, but acquire the capability to respond to self-antigen upon breach of tissue integrity? Although this question remains unanswered, the solution of the puzzle might lie within ontogenetic programming of iNKT cells. Thus, further studies of factors that influence iNKT cell ontogeny are warranted.
Epilogue: A hypothetical model for how CD1d controls iNKT cell function
The control of iNKT cell response by CD1d depends on its function as a lipid antigen-presenting molecule. The recognition of CD1d-bound lipid by iNKT cells depends on the nature of its antigen-specific receptor. The identity of the naturally processed antigen(s) recognized by iNKT cells remains elusive. On the basis of our current knowledge of CD1d-iNKT cell biology, we can surmise the following regarding the features of the naturally processed iNKT cell antigen(s). (1) Despite the fact that PI and PI-glycans are natural CD1d ligands,21,28 they are rare iNKT cell antigens.29,43 (2) An α-anomeric glycolipid may be an elusive natural iNKT cell antigen.52 (3) α-anomeric glycolipids are rare in the animal kingdom, and have not been detected in mammals.150 We predict that the recognition of infectious agents by antigen-presenting cells induces the synthesis of an α-anomeric glycolipid transiently, which when presented by CD1d stimulates iNKT cells.
The activation of iNKT cells in the absence of microbe-derived antigen presentation by CD1d is noteworthy.151 iNKT cells function in immune surveillance against spontaneously arising tumours, which do not express CD1d.46 Additionally, iNKT cells are quiescent in the absence of infectious stimuli.152 For these reasons, we hypothesize that the iNKT cell antigen is inducible. Recently, a role for CD1 in monitoring membrane integrity has been evoked because CD1 molecules bind and present lipids and glycolipids to specific T cells.153 As an extension of this hypothesis, we speculate that infectious assault or glucose starvation induces an unfolded protein response.154 The unfolded protein response increases cytosolic inositol levels, a key step in the induction of membrane biogenesis,154 through its role in regulating phospholipid biosynthesis.155,156 Phospholipids and glycolipids play an important structural role in maintaining membrane integrity. Thus the induction of the unfolded protein response may include the biosynthesis of neo-self lipids that are normally not encountered by iNKT cells. Encounters with the neo-self glycolipid, probably the elusive α-anomeric glycosylceramide, would stimulate iNKT cells. Because the products of the unfolded protein response are similar, irrespective of the cause,154 the induction and presentation of a common antigen are postulated to occur during microbial infections. A virtue of involving the unfolded protein response in the CD1d-iNKT cell antigen presentation-recognition system is the rapidity and short half-life (about 2 min) of the induced products.154 These features may explain the ephemeral nature of the iNKT cell antigen.
Thus, in our model, CD1d functions as a sensor, to detect alterations in cellular lipid content by virtue of its inherent affinity for such ligands. The presentation of a neo-self lipid upon infectious assault of antigen-presenting cells activates iNKT cells, which promptly release pro- and anti-inflammatory cytokines and chemokines to jump-start the immune system (Fig. 2). In this regard, iNKT cells may be akin to an enzyme in that they might function by lowering the ‘energy of activation’ required for the initiation of an immune response. In doing so, the immune system is alerted by the entry of only a few intruders, as occurs in natural infections. Identification of the elusive natural antigen of iNKT cells will shed more light on how the CD1d-iNKT cell system functions in vivo, especially with regard to how the antigen is expressed and presented to this unique subset of T lymphocytes.
References
- 1.Bendelac A, Bonneville M, Kearney JF. Autoreactivity by design: innate B and T lymphocytes. Nature Rev Immunol. 2001;1:177–86. doi: 10.1038/35105052. [DOI] [PubMed] [Google Scholar]
- 2.Bendelac A, Medzhitov R. Adjuvants of immunity: harnessing innate immunity to promote adaptive immunity. J Exp Med. 2002;195:F19–23. doi: 10.1084/jem.20020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smyth MJ, Crowe NY, Hayakawa Y, Takeda K, Yagita H, Godfrey DI. NKT cells – conductors of tumor immunity? Curr Opin Immunol. 2002;14:165–71. doi: 10.1016/s0952-7915(02)00316-3. [DOI] [PubMed] [Google Scholar]
- 4.Hong S, Wilson MT, Serizawa I, et al. The natural killer T-cell ligand α-galactosylceramide prevents autoimmune diabetes in non-obese diabetic mice. Nature Med. 2001;7:1052–6. doi: 10.1038/nm0901-1052. [DOI] [PubMed] [Google Scholar]
- 5.Singh AK, Wilson MT, Hong S, et al. Natural killer T cell activation protects mice against experimental autoimmune encephalomyelitis. J Exp Med. 2001;194:1801–11. doi: 10.1084/jem.194.12.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wilson MT, Singh AK, Van Kaer L. Immunotherapy with ligands of natural killer T cells. Trends Mol Med. 2002;8:225–31. doi: 10.1016/s1471-4914(02)02325-0. [DOI] [PubMed] [Google Scholar]
- 7.Sonoda KH, Exley M, Snapper S, Balk SP, Stein-Streilein J. CD1-reactive natural killer T cells are required for development of systemic tolerance through an immune-privileged site. J Exp Med. 1999;190:1215–26. doi: 10.1084/jem.190.9.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hong S, Van Kaer L. Immune privilege: keeping an eye on natural killer T cells. J Exp Med. 1999;190:1197–1200. doi: 10.1084/jem.190.9.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seino KI, Fukao K, Muramoto K, et al. Requirement for natural killer T (NKT) cells in the induction of allograft tolerance. Proc Natl Acad Sci USA. 2001;98:2577–81. doi: 10.1073/pnas.041608298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zeng D, Lewis D, Dejbakhsh-Jones S, Lan F, Garcia-Ojeda M, Sibley R, Strober S. Bone marrow NK1.1− and NK1.1+ T cells reciprocally regulate acute graft versus host disease. J Exp Med. 1999;189:1073–81. doi: 10.1084/jem.189.7.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dascher CC, Hiromatsu K, Naylor JW, et al. Conservation of a CD1 multigene family in the guinea pig. J Immunol. 1999;163:5478–88. [PubMed] [Google Scholar]
- 12.Chun T, Wang K, Zuckermann FA, Gaskins HR. Molecular cloning and characterization of a novel CD1 gene from the pig. J Immunol. 1999;162:6562–71. [PubMed] [Google Scholar]
- 13.Mendiratta SK, Martin WD, Hong S, Boesteanu A, Joyce S, Van Kaer L. CD1d1 mutant mice are deficient in natural T cells that promptly produce IL-4. Immunity. 1997;6:469–77. doi: 10.1016/s1074-7613(00)80290-3. [DOI] [PubMed] [Google Scholar]
- 14.Porcelli SA, Modlin RL. The CD1 system: antigen-presenting molecules for T cell recognition of lipids and glycolipids. Annu Rev Immunol. 1999;17:297–329. doi: 10.1146/annurev.immunol.17.1.297. [DOI] [PubMed] [Google Scholar]
- 15.Kronenberg M, Gapin L. The unconventional lifestyle of NKT cells. Nature Rev Immunol. 2002;2:557–68. doi: 10.1038/nri854. [DOI] [PubMed] [Google Scholar]
- 16.Taniguchi M, Harada M, Kojo S, Nakayama T, Wakao H. The regulatory role of Vα14 NKT cells in innate and adaptive immune response. Annu Rev Immunol. 2003;21:483–513. doi: 10.1146/annurev.immunol.21.120601.141057. [DOI] [PubMed] [Google Scholar]
- 17.Joyce S. CD1d and natural T cells: how their properties jump start the immune system. Cell Mol Life Sci. 2001;58:442–69. doi: 10.1007/PL00000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gadola SD, Zaccai NR, Harlos K, et al. Structure of human CD1b with bound ligands at 2.3A, a maze for alkyl chains. Nature Immunol. 2002;3:721–26. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 19.Zeng Z, Castano AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: an MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–45. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 20.Moody DB, Briken V, Cheng TY, et al. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nature Immunol. 2002;3:435–42. doi: 10.1038/ni780. [DOI] [PubMed] [Google Scholar]
- 21.De Silva AD, Park J-J, Matsuki N, Stanic AK, Brutkiewicz RR, Medof ME, Joyce S. Lipid protein interactions: the assembly of CD1d1 with cellular phospholipids occurs in the endoplasmic reticulum. J Immunol. 2002;168:723–33. doi: 10.4049/jimmunol.168.2.723. [DOI] [PubMed] [Google Scholar]
- 22.Jayawardena-Wolf J, Benlagha K, Chiu YH, Mehr R, Bendelac A. CD1d endosomal trafficking is independently regulated by an intrinsic CD1d-encoded tyrosine motif and by the invariant chain. Immunity. 2001;15:897–908. doi: 10.1016/s1074-7613(01)00240-0. [DOI] [PubMed] [Google Scholar]
- 23.Sugita M, Porcelli SA, Brenner MB. Assembly and retention of CD1b heavy chains in the endoplasmic reticulum. J Immunol. 1997;159:2358–65. [PubMed] [Google Scholar]
- 24.Huttinger R, Staffler G, Majdic O, Stockinger H. Analysis of the early biogenesis of CD1b: involvement of the chaperones calnexin and calreticulin, the proteasome and beta (2)-microglobulin. Int Immunol. 1999;11:1615–23. doi: 10.1093/intimm/11.10.1615. [DOI] [PubMed] [Google Scholar]
- 25.Kang SJ, Cresswell P. Calnexin, calreticulin, and ERp57 cooperate in disulfide bond formation in human CD1d heavy chain. J Biol Chem. 2002;277:44838–44. doi: 10.1074/jbc.M207831200. [DOI] [PubMed] [Google Scholar]
- 26.Dick TP, Bangia N, Peaper DR, Cresswell P. Disulfide bond isomerization and the assembly of MHC class I-peptide complexes. Immunity. 2002;16:87–98. doi: 10.1016/s1074-7613(02)00263-7. [DOI] [PubMed] [Google Scholar]
- 27.Gao B, Adhikari R, Howarth M, et al. Assembly and antigen-presenting function of MHC class I molecules in cells lacking the ER chaperone calreticulin. Immunity. 2002;16:99–109. doi: 10.1016/s1074-7613(01)00260-6. [DOI] [PubMed] [Google Scholar]
- 28.Joyce S, Woods AS, Yewdell JW, et al. Natural ligand of mouse CD1d1: cellular glycosylphosphatidylinositol. Science. 1998;279:1541–44. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- 29.Gumperz JE, Roy C, Makowska A, Lum D, et al. Murine CD1d-restricted T cell recognition of cellular lipids. Immunity. 2000;12:211–21. doi: 10.1016/s1074-7613(00)80174-0. [DOI] [PubMed] [Google Scholar]
- 30.Sugita M, Grant EP, van Donselaar E, Hsu VW, Rogers RA, Peters PJ, Brenner MB. Separate pathways for antigen presentation by CD1 molecules. Immunity. 1999;11:743–52. doi: 10.1016/s1074-7613(00)80148-x. [DOI] [PubMed] [Google Scholar]
- 31.Sugita M, Cao X, Watts GF, Rogers RA, Bonifacino JS, Brenner MB. Failure of trafficking and antigen presentation by CD1 in AP-3-deficient cells. Immunity. 2002;16:697–706. doi: 10.1016/s1074-7613(02)00311-4. [DOI] [PubMed] [Google Scholar]
- 32.Roberts TJ, Sriram V, Spence PM, et al. Recycling CD1d1 molecules present endogenous antigens processed in an endocytic compartment to NKT cells. J Immunol. 2002;168:5409–14. doi: 10.4049/jimmunol.168.11.5409. [DOI] [PubMed] [Google Scholar]
- 33.Chiu YH, Jayawardena J, Weiss A, Lee D, Park SH, Dautry-Varsat A, Bendelac A. Distinct subsets of CD1d-restricted T cells recognize self-antigens loaded in different cellular compartments. J Exp Med. 1999;189:103–10. doi: 10.1084/jem.189.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chiu YH, Park SH, Benlagha K, Forestier C, Jayawardena-Wolf J, Savage PB, Teyton L, Bendelac A. Multiple defects in antigen presentation and T cell development by mice expressing cytoplasmic tail-truncated CD1d. Nat Immunol. 2002;3:55–60. doi: 10.1038/ni740. [DOI] [PubMed] [Google Scholar]
- 35.Kang SJ, Cresswell P. Regulation of intracellular trafficking of human CD1d by association with MHC class II molecules. EMBO J. 2002;21:1650–60. doi: 10.1093/emboj/21.7.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bilsland CAG, Milstein C. The identification of the β2-microglobulin binding antigen encoded by the human CD1D gene. Eur J Immunol. 1991;21:71–80. doi: 10.1002/eji.1830210112. [DOI] [PubMed] [Google Scholar]
- 37.Lagaudriere-Gesbert C, Newmyer SL, Gregers TF, Bakke O, Ploegh HL. Uncoating ATPase Hsc70 is recruited by invariant chain and controls the size of endocytic compartments. Proc Natl Acad Sci USA. 2002;99:1515–20. doi: 10.1073/pnas.042688099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nordeng TW, Gregers TF, Kongsvik TL, Meresse S, Gorvel JP, Jourdan F, Motta A, Bakke O. The cytoplasmic tail of invariant chain regulates endosome fusion and morphology. Mol Biol Cell. 2002;13:1846–56. doi: 10.1091/mbc.01-10-0478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Villadangos JA, Ploegh HL. Proteolysis in MHC class II antigen presentation: who's in charge? Immunity. 2000;12:233–9. doi: 10.1016/s1074-7613(00)80176-4. [DOI] [PubMed] [Google Scholar]
- 40.Riese RJ, Shi GP, Villadangos J, et al. Regulation of CD1 function and NK1.1+ T cell selection and maturation by cathepsin S. Immunity. 2001;15:909–19. doi: 10.1016/s1074-7613(01)00247-3. [DOI] [PubMed] [Google Scholar]
- 41.Honey K, Benlagha K, Beers C, Forbush KA, Teyton L, Rudensky A, Bendelac A. Thymocyte expression of cathepsin L is essential for NK T cell development. Nature Immunol. 3:1069–74. doi: 10.1038/ni844. [DOI] [PubMed] [Google Scholar]
- 42.von Bulow R, Schmidt B, Dierks T, Schwabauer N, Schilling K, Weber E, Uson I, von Figura K. Defective oligomerization of arylsulfatase a as a cause of its instability in lysosomes and metachromatic leukodystrophy. J Biol Chem. 2002;277:9455–61. doi: 10.1074/jbc.M111993200. [DOI] [PubMed] [Google Scholar]
- 43.Molano A, Park SH, Chiu YH, Nosseir S, Bendelac A, Tsuji M. The IgG response to the circumsporozoite protein is MHC class II-dependent and CD1d-independent: exploring the role of GPIs in NK T cell activation and antimalarial responses. J Immunol. 2000;164:5005–9. doi: 10.4049/jimmunol.164.10.5005. [DOI] [PubMed] [Google Scholar]
- 44.Brossay L, Tangri S, Bix M, Cardell S, Locksley R, Kronenberg M. Mouse CD1-autoreactive T cells have diverse patterns of reactivity to CD1+ targets. J Immunol. 1998;160:3681–8. [PubMed] [Google Scholar]
- 45.Park S-H, Roark JH, Bendelac A. Tissue-specific recognition of mouse CD1 molecules. J Immunol. 1998;160:3128–34. [PubMed] [Google Scholar]
- 46.Kawano T, Cui J, Koezuka Y, et al. CD1d-restricted and TCR-mediated activation of Vα14 NKT cells by glycosylceramides. Science. 1997;278:1626–9. doi: 10.1126/science.278.5343.1626. [DOI] [PubMed] [Google Scholar]
- 47.Burdin N, Brossay L, Koezuka Y, et al. Selective ability of mouse CD1 to present glycolipids: α-galactosylceramide specifically stimulates Vα14+ NK T lymphocytes. J Immunol. 1998;161:3271–81. [PubMed] [Google Scholar]
- 48.Brossay L, Chioda M, Burdin N, Koezuka Y, Casorati G, Dellabona P, Kronenberg M. CD1d-mediated recognition of an α-galactosylceramide by natural killer T cells is highly conserved through mammalian evolution. J Exp Med. 1998;188:1521–8. doi: 10.1084/jem.188.8.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spada FM, Koezuka Y, Porcelli SA. CD1d-restricted recognition of synthetic glycolipid antigens by human natural killer T cells. J Exp Med. 1998;188:1529–34. doi: 10.1084/jem.188.8.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsuda JL, Naidenko OV, Gapin L, Nakayama T, Taniguchi M, Wang C-R, Koezuka Y, Kronenberg M. Tracking the response of natural killer T cells to a glycolipid antigen using CD1d tetramers. J Exp Med. 2000;192:741–53. doi: 10.1084/jem.192.5.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benlagha K, Weiss A, Beavis A, Teyton L, Bendelac A. In vivo identification of glycolipid antigen-specific T cells using fluorescent CD1d tetramers. J Exp Med. 2000;191:1895–1904. doi: 10.1084/jem.191.11.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanic AK, De Silva AD, Park JJ, et al. Defective presentation of the CD1d1-restricted natural Va14Ja18 NKT lymphocyte antigen caused by β-d-glucosylceramide synthase deficiency. Proc Natl Acad Sci USA. 1995;100:1849–54. doi: 10.1073/pnas.0430327100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sieling PA, Chatterjee D, Porcelli SA, et al. CD1 restricted T cell recognition of microbial lipoglycan antigen. Science. 2003;269:227–30. doi: 10.1126/science.7542404. [DOI] [PubMed] [Google Scholar]
- 54.Prigozy TI, Naidenko O, Qasba P, et al. Glycolipid antigen processing for presentation by CD1d molecules. Science. 2001;291:664–7. doi: 10.1126/science.291.5504.664. [DOI] [PubMed] [Google Scholar]
- 55.Moody DB, Reinhold BB, Guy MREMB, et al. Structural requirements for glycolipid recognition by CD1b-restricted T cells. Science. 1997;278:283–6. doi: 10.1126/science.278.5336.283. [DOI] [PubMed] [Google Scholar]
- 56.Melian A, Watts GFM, Shamshiv A, et al. Molecular recognition of human CD1b antigen complexes: evidence for a common pattern of interaction with αβ TCRs. J Immunol. 2000;165:4494–504. doi: 10.4049/jimmunol.165.8.4494. [DOI] [PubMed] [Google Scholar]
- 57.Gui M, Li J, Wen LJ, Hardy RR, Hayakawa K. TCRβ chain influences but does not solely control autoreactivity of Vα14J281 T cells. J Immunol. 2001;167:6239–46. doi: 10.4049/jimmunol.167.11.6239. [DOI] [PubMed] [Google Scholar]
- 58.Grant EP, Degano M, Rosat JP, Stenger S, Modlin RL, Wilson IA, Porcelli SA, Brenner MB. Molecular recognition of lipid antigens by T cell receptors. J Exp Med. 1999;189:195–205. doi: 10.1084/jem.189.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Garcia KC, Teyton L, Wilson IA. Structural basis for T cell recognition. Annu Rev Immunol. 1999;17:369–98. doi: 10.1146/annurev.immunol.17.1.369. [DOI] [PubMed] [Google Scholar]
- 60.Sidobre S, Naidenko OV, Sim BC, Gascoigne NR, Garcia KC, Kronenberg M. The Vα14 NKT cell TCR exhibits high-affinity binding to a glycolipid/CD1d complex. J Immunol. 2002;169:1340–8. doi: 10.4049/jimmunol.169.3.1340. [DOI] [PubMed] [Google Scholar]
- 61.Miyamoto K, Miyake S, Yamamura T. A synthetic glycolipid prevents autoimmune encephalomyelitis by inducing Th2 bias of natural killer T cells. Nature. 2001;413:531–4. doi: 10.1038/35097097. [DOI] [PubMed] [Google Scholar]
- 62.MacDonald HR. CD1d-glycolipid tetramers: a new tool to monitor natural killer T cells in health and disease. J Exp Med. 2000;192:F15–F19. doi: 10.1084/jem.192.5.f15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bendelac A, Killeen N, Litman DR, Schwartz RH. A subset of CD4+ thymocytes selected by MHC class I molecules. Science. 1994;263:1774–8. doi: 10.1126/science.7907820. [DOI] [PubMed] [Google Scholar]
- 64.Eberl G, Lees R, Smiley ST, Taniguchi M, Grusby MJ, MacDonald HR. Tissue-specific segregation of CD1d-dependent and CD1d-independent NK T cells. J Immunol. 1999;162:6410–9. [PubMed] [Google Scholar]
- 65.Godfrey DI, Hammond KJ, Poulton LD, Smyth MJ, Baxter AG. NKT cells: facts, functions and fallacies. Immunol Today. 2000;21:573–83. doi: 10.1016/s0167-5699(00)01735-7. [DOI] [PubMed] [Google Scholar]
- 66.Ohteki T, MacDonald HR. Major histocompatibility complex class I related molecules control the development of CD4+8− and CD4−8− subsets of natural killer 1.1+ T cell receptor-α/β cells in the liver of mice. J Exp Med. 1994;180:699–704. doi: 10.1084/jem.180.2.699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Arase H, Arase N, Agasawara K, Good RA, Onoe K. An NK1.1+ CD4+8− single positive thymocyte subpopulation that expresses a highly skewed T cell antigen receptor family. Proc Natl Acad Sci USA. 1992;89:6506–10. doi: 10.1073/pnas.89.14.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ohteki T, Maki C, Koyasu S, Mak TW, Ohashi PS. LFA-1 is required for liver NK1.1+TCR αβ+ cell development: evidence that liver NK1.1+TCR αβ+ cells originate from multiple pathways. J Immunol. 1999;162:3753–6. [PubMed] [Google Scholar]
- 69.MacDonald HR, Lees RK, Held W. Developmentally regulated extinction of Ly-49 receptor expression permits maturation and selection of NK1.1+ T cells. J Exp Med. 1998;187:2109–14. doi: 10.1084/jem.187.12.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.MacDonald HR. T before NK. Science. 2002;296:481–2. doi: 10.1126/science.1071492. [DOI] [PubMed] [Google Scholar]
- 71.Lee PT, Benlagha K, Teyton L, Bendelac A. Distinct functional lineages of human Vα24 natural killer T cells. J Exp Med. 2002;195:637–41. doi: 10.1084/jem.20011908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med. 2002;195:625–36. doi: 10.1084/jem.20011786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benlagha K, Kyin T, Beavis A, Teyton L, Bendelac A. A thymic precursor to the NK T cell lineage. Science. 2002;296:553–5. doi: 10.1126/science.1069017. [DOI] [PubMed] [Google Scholar]
- 74.Pellicci DG, Hammond KJ, Uldrich AP, Baxter AG, Smyth MJ, Godfrey DI. A natural killer T (NKT) cell developmental pathway involving a thymus-dependent NK1.1− CD4+ CD1d-dependent precursor stage. J Exp Med. 2002;195:835–44. doi: 10.1084/jem.20011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gadue P, Stein PL. NK T cell precursors exhibit differential cytokine regulation and require Itk for efficient maturation. J Immunol. 2002;169:2397–406. doi: 10.4049/jimmunol.169.5.2397. [DOI] [PubMed] [Google Scholar]
- 76.Motsinger A, Haas DW, Stanic AK, Van Kaer L, Joyce S, Unutmaz D. CD1d-restricted human natural killer T cells are highly susceptible to human immunodeficiency virus 1 infection. J Exp Med. 2002;195:869–79. doi: 10.1084/jem.20011712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fleuridor R, Wilson B, Hou R, Landay A, Kessler H, Al-Harthi L. CD1d-restricted natural killer T cells are potent targets for human immunodeficiency virus infection. Immunology. 2003;108:3–9. doi: 10.1046/j.1365-2567.2003.01560.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sandberg JK, Fast NM, Palacios EH, et al. Selective loss of innate CD4+ Vα24 natural killer T cells in human immunodeficiency virus infection. J Virol. 2002;76:7528–34. doi: 10.1128/JVI.76.15.7528-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.van der Vliet HJ, von Blomberg BM, Hazenberg MD, et al. Selective decrease in circulating Vα24+Vβ11+ NKT cells during HIV type 1 infection. J Immunol. 2002;168:1490–5. doi: 10.4049/jimmunol.168.3.1490. [DOI] [PubMed] [Google Scholar]
- 80.Yoshimoto T, Paul WE. CD4pos NK1pos T cells promptly produce IL-4 in response to in vivo challenge with anti-CD3. J Exp Med. 1994;179:1285–95. doi: 10.1084/jem.179.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zlotnik A, Godfrey DI, Fischer M, Suda I. Cytokine production by mature and immature CD4−8− T cells. αβ-T cell receptor+ CD4−8− T cells produce IL-4. J Immunol. 1992;149:1211–5. [PubMed] [Google Scholar]
- 82.Chen Y-H, Chiu NM, Mandal M, Wang N, Wang C-R. Impaired NK1+ T cell development and early IL-4 production in CD1 deficient mice. Immunity. 1997;6:459–67. doi: 10.1016/s1074-7613(00)80289-7. [DOI] [PubMed] [Google Scholar]
- 83.Smiley ST, Kaplan MH, Grusby MJ. Immunoglobulin E production in the absence of interleukin-4 secreting CD1 dependent cells. Science. 1997;275:977–9. doi: 10.1126/science.275.5302.977. [DOI] [PubMed] [Google Scholar]
- 84.Inui T, Nakagawa R, Ohkura S, et al. Age-associated augmentation of the synthetic ligand-mediated function of mouse NK1.1 Ag+ T cells: their cytokine production and hepatotoxicity in vivo and in vitro. J Immunol. 2002;169:6127–32. doi: 10.4049/jimmunol.169.11.6127. [DOI] [PubMed] [Google Scholar]
- 85.Kadowaki N, Antonenko S, Ho S, Rissoan MC, Soumelis V, Porcelli SA, Lanier LL, Liu YJ. Distinct cytokine profiles of neonatal natural killer T cells after expansion with subsets of dendritic cells. J Exp Med. 2001;193:1221–6. doi: 10.1084/jem.193.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Naka T, Tsutsui H, Fujimoto M, et al. SOCS-1/SSI-1-deficient NKT cells participate in severe hepatitis through dysregulated cross-talk inhibition of IFN-γ and IL-4 signaling in vivo. Immunity. 2001;14:535–45. doi: 10.1016/s1074-7613(01)00132-7. [DOI] [PubMed] [Google Scholar]
- 87.Arase H, Arase N, Saito T. Interferonγ production by natural killer (NK) cells and NK1.1+ T cells upon NKR-P1 cross-linking. J Exp Med. 1996;183:2391–6. doi: 10.1084/jem.183.5.2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matsuda JL, Gapin L, Sidobre S, Kieper WC, Tan JT, Ceredig R, Surh CD, Kronenberg M. Homeostasis of Vα14i NKT cells. Nature Immunol. 2002;3:966–74. doi: 10.1038/ni837. [DOI] [PubMed] [Google Scholar]
- 89.Maeda M, Lohwasser S, Yamamura T, Takei F. Regulation of NKT cells by Ly49: analysis of primary NKT cells and generation of NKT cell line. J Immunol. 2001;167:4180–6. doi: 10.4049/jimmunol.167.8.4180. [DOI] [PubMed] [Google Scholar]
- 90.Ikarashi Y, Mikami R, Bendelac A, et al. Dendritic cell maturation overrules H-2D-mediated natural killer T (NKT) cell inhibition: critical role for B7 in CD1d-dependent NKT cell interferon γ production. J Exp Med. 2001;194:1179–86. doi: 10.1084/jem.194.8.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Singh N, Hong S, Scherer DC, Serizawa I, Burdin N, Kronenberg M, Koezuka Y, Van Kaer L. Activation of NK T cells by CD1d and α-galactosylceramide directs conventional T cells to the acquisition of a Th2 phenotype. J Immunol. 1999;163:2373–7. [PubMed] [Google Scholar]
- 92.Carnaud C, Lee D, Donnars O, Park S-H, Beavis A, Koezuka Y, Bendelac A. Cross-talk between cells of the innate immune system: NKT cells rapidly activate NK cells. J Immunol. 1999;163:4647–50. [PubMed] [Google Scholar]
- 93.Fujii S, Shimizu K, Kronenberg M, Steinman RM. Prolonged IFN-γ-producing NKT response induced with α-galactosylceramide-loaded DCs. Nature Immunol. 2002;3:867–74. doi: 10.1038/ni827. [DOI] [PubMed] [Google Scholar]
- 94.Eberl G, MacDonald HR. Selective induction of NK cell proliferation and cytotoxicity by activated NKT cells. Eur J Immunol. 2000;30:985–92. doi: 10.1002/(SICI)1521-4141(200004)30:4<985::AID-IMMU985>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 95.Kawamura T, Takeda K, Mendiratta SK, Kawamura H, Van Kaer L, Yagata H, Abo T, Okumura K. Critical role of NK1+ T cells in IL-12-induced immune responses in vivo. J Immunol. 1998;160:16–9. [PubMed] [Google Scholar]
- 96.Kitamura H, Iwakabe K, Yahata T, et al. The natural killer T (NKT) cell ligand α-galactosylceramide demonstrates its immunopotentiating effect by inducing interleukin (IL)-12 production by dendritic cells and IL-12 receptor expression on NKT cells. J Exp Med. 1999;189:1121–7. doi: 10.1084/jem.189.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tomura M, Yu WG, Ahn HJ, et al. A novel function of Vα14+CD4+ NKT cells: stimulation of IL-12 production by antigen-presenting cells in the innate immune system. J Immunol. 1999;163:93–10. [PubMed] [Google Scholar]
- 98.Kitamura H, Ohta A, Sekimoto M, et al. α-galactosylceramide induces early B-cell activation through IL-4 production by NKT cells. Cell Immunol. 2000;199:37–42. doi: 10.1006/cimm.1999.1602. [DOI] [PubMed] [Google Scholar]
- 99.Burdin N, Brossay L, Kronenberg M. Immunization with α-galactosylceramide polarizes CD1-reactive NK T cells towards Th2 cytokine synthesis. Eur J Immunol. 1999;29:2014–25. doi: 10.1002/(SICI)1521-4141(199906)29:06<2014::AID-IMMU2014>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 100.Nishimura T, Kitamura H, Iwakabe K, et al. The interface between innate and acquired immunity: glycolipid antigen presentation by CD1d-expressing dendritic cells to NKT cells induces the differentiation of antigen-specific cytotoxic T lymphocytes. Int Immunol. 2000;12:987–94. doi: 10.1093/intimm/12.7.987. [DOI] [PubMed] [Google Scholar]
- 101.Stober D, Jomantaite I, Schirmbeck R, Reimann J. NKT cells provide help for dendritic cell-dependent priming of MHC class I-restricted CD8+ T cells in vivo. J Immunol. 2003;170:2540–8. doi: 10.4049/jimmunol.170.5.2540. [DOI] [PubMed] [Google Scholar]
- 102.Gonzalez-Aseguinolaza G, Van Kaer L, Bergmann CC, et al. Natural killer T cell ligand alpha-galactosylceramide enhances protective immunity induced by malaria vaccines. J Exp Med. 2002;195:617–24. doi: 10.1084/jem.20011889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shi FD, Flodstrom M, Balasa B, Kim SH, Van Gunst K, Strominger JL, Wilson SB, Sarvetnick N. Germ line deletion of the CD1 locus exacerbates diabetes in the NOD mouse. Proc Natl Acad Sci USA. 2001;98:6777–82. doi: 10.1073/pnas.121169698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Naumov YN, Bahjat KS, Gausling R, et al. Activation of CD1d-restricted T cells protects NOD mice from developing diabetes by regulating dendritic cell subsets. Proc Natl Acad Sci USA. 2001;98:13838–43. doi: 10.1073/pnas.251531798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sharif S, Arreaza GA, Zucker P, et al. Activation of natural killer T cells by α-galactosylceramide treatment prevents the onset and recurrence of autoimmune Type 1 diabetes. Nature Med. 2001;7:1057–62. doi: 10.1038/nm0901-1057. [DOI] [PubMed] [Google Scholar]
- 106.Wang B, Geng YB, Wang CR. CD1-restricted NK T cells protect nonobese diabetic mice from developing diabetes. J Exp Med. 2001;194:313–20. doi: 10.1084/jem.194.3.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Jahng AW, Maricic I, Pedersen B, Burdin N, Naidenko O, Kronenberg M, Koezuka Y, Kumar V. Activation of natural killer T cells potentiates or prevents experimental autoimmune encephalomyelitis. J Exp Med. 2001;194:1789–99. doi: 10.1084/jem.194.12.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Beaudoin L, Laloux V, Novak J, Lucas B, Lehuen A. NKT cells inhibit the onset of diabetes by impairing the development of pathogenic T cells specific for pancreatic beta cells. Immunity. 2002;17:725–36. doi: 10.1016/s1074-7613(02)00473-9. [DOI] [PubMed] [Google Scholar]
- 109.Chen H, Huang H, Paul WE. NK1.1+ CD4+ T cells lose NK1.1 expression upon in vitro activation. J Immunol. 1997;158:5112–9. [PubMed] [Google Scholar]
- 110.Eberl G, MacDonald HR. Rapid death and regeneration of NKT cells in anti-CD3ε- or IL-12-treated mice: a major role for bone marrow in NKT cell homeostasis. Immunity. 1998;9:345–53. doi: 10.1016/s1074-7613(00)80617-2. [DOI] [PubMed] [Google Scholar]
- 111.Bendelac A. Positive selection of mouse NK1+ T cells by CD1 expressing cortical thymocytes. J Exp Med. 1995;182:2091–6. doi: 10.1084/jem.182.6.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Coles MC, Raulet DH. NK1.1+ T cells in the liver arise in the thymus and are selected by interactions with class I molecules on CD4+CD8+ cells. J Immunol. 2000;164:2412–8. doi: 10.4049/jimmunol.164.5.2412. [DOI] [PubMed] [Google Scholar]
- 113.Tilloy F, Di Santo JP, Bendelac A, Lantz O. Thymic dependence of invariant Vα14+ natural killer-T cell development. Eur J Immunol. 1999;29:3313–8. doi: 10.1002/(SICI)1521-4141(199910)29:10<3313::AID-IMMU3313>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 114.Makino Y, Kanno R, Koseki H, Taniguchi M. Development of Vα14+ NK T cells in the early stages of embryogenesis. Proc Natl Acad Sci USA. 1996;93:6516–20. doi: 10.1073/pnas.93.13.6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shimamura M, Huang YY, Suda Y, et al. Positive selection of NKT cells by CD1+, CD11c+ non-lymphoid cells residing in the extrathymic organs. Eur J Immunol. 1999;29:3962–70. doi: 10.1002/(SICI)1521-4141(199912)29:12<3962::AID-IMMU3962>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 116.Shimamura M, Ohteki T, Launois P, Garcia AM, MacDonald HR. Thymus-independent generation of NK1+ T cells in vitro from fetal liver precursors. J Immunol. 1997;158:3682–9. [PubMed] [Google Scholar]
- 117.Gapin L, Matsuda JL, Surh CD, Kronenberg M. NKT cells derive from double-positive thymocytes that are positively selected by CD1d. Nature Immunol. 2001;2:971–8. doi: 10.1038/ni710. [DOI] [PubMed] [Google Scholar]
- 118.Chun T, Page MJ, Gapin L, et al. CD1d-expressing dendritic cells but not thymic epithelial cells can mediate negative selection of NKT cells. J Exp Med. 2003;197:907–18. doi: 10.1084/jem.20021366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Bendelac A, Matzinger P, Seder RA, Paul WE, Schwartz RH. Activation events during thymic selection. J Exp Med. 1992;175:731–42. doi: 10.1084/jem.175.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bix M, Coles MC, Raulet DH. Positive selection of Vb8+ CD4−8− thymocytes by class I molecules expressed by hematopoietic cells. J Exp Med. 1993;178:901–8. doi: 10.1084/jem.178.3.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Coles MC, Raulet DH. Class I dependence of the development of CD4+CD8− NK1.1+ thymocytes. J Exp Med. 1994;180:395–9. doi: 10.1084/jem.180.1.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Walunas TL, Wang B, Wang CR, Leiden JM. The Ets1 transcription factor is required for the development of NK T cells in mice. J Immunol. 2000;164:2857–60. doi: 10.4049/jimmunol.164.6.2857. [DOI] [PubMed] [Google Scholar]
- 123.Lacorazza HD, Miyazaki Y, Di Cristofano A, et al. The ETS protein MEF plays a critical role in perforin gene expression and the development of natural killer and NK-T cells. Immunity. 2002;17:437–49. doi: 10.1016/s1074-7613(02)00422-3. [DOI] [PubMed] [Google Scholar]
- 124.Ohteki T, Yoshida H, Matsuyama T, Duncan GS, Mak TW, Ohashi PS. The transcription factor interferon regulatory factor 1 (IRF-1) is important during the maturation of natural killer 1.1+ T cell receptor-αβ+ (NK1+ T) cells, natural killer cells, and intestinal intraepithelial T cells. J Exp Med. 1998;187:967–72. doi: 10.1084/jem.187.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Elewaut D, Brossay L, Santee SM, et al. Membrane lymphotoxin is required for the development of different subpopulations of NK T cells. J Immunol. 2000;165:671–9. doi: 10.4049/jimmunol.165.2.671. [DOI] [PubMed] [Google Scholar]
- 126.Eberl G, Lowin-Kropf B, MacDonald HR. NKT cell development is selectively impaired in Fyn-deficient mice. J Immunol. 1999;163:4091–4. [PubMed] [Google Scholar]
- 127.Gadue P, Morton N, Stein PL. The src family tyrosine kinase Fyn regulates natural killer T cell development. J Exp Med. 1999;190:1189–95. doi: 10.1084/jem.190.8.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Muthusamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995;377:639–42. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- 129.Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–63. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 130.Stein PL, Lee HM, Rich S, Soriano P. pp59fyn mutant mice display differential signaling in thymocytes and peripheral T cells. Cell. 1992;70:741–50. doi: 10.1016/0092-8674(92)90308-y. [DOI] [PubMed] [Google Scholar]
- 131.Binstadt BA, Brumbaugh KM, Dick CJ, et al. Sequential involvement of Lck and SHP-1 with MHC-recognizing receptors on NK cells inhibits FcR-initiated tyrosine kinase activation. Immunity. 1996;5:629–38. doi: 10.1016/s1074-7613(00)80276-9. [DOI] [PubMed] [Google Scholar]
- 132.Held W, Coudert JD, Zimmer J. The NK cell receptor repertoire: formation, adaptation and exploitation. Curr Opin Immunol. 2003;15:233–7. doi: 10.1016/s0952-7915(02)00031-6. [DOI] [PubMed] [Google Scholar]
- 133.Groves T, Smiley P, Cooke MP, Forbush K, Perlmutter RM, Guidos CJ. Fyn can partially substitute for Lck in T lymphocyte development. Immunity. 1996;5:417–28. doi: 10.1016/s1074-7613(00)80498-7. [DOI] [PubMed] [Google Scholar]
- 134.Campbell KS, Buder A, Deuschle U. Interactions between the amino-terminal domain of p56lck and cytoplasmic domains of CD4 and CD8 alpha in yeast. Eur J Immunol. 1995;25:2408–12. doi: 10.1002/eji.1830250842. [DOI] [PubMed] [Google Scholar]
- 135.Zamoyska R, Derham P, Gorman SD, von Hoegen P, Bolen JB, Veillette A, Parnes JR. Inability of CD8 alpha polypeptides to associate with p56lck correlates with impaired function in vitro and lack of expression in vivo. Nature. 1989;342:278–81. doi: 10.1038/342278a0. [DOI] [PubMed] [Google Scholar]
- 136.Salmon P, Mong M, Kang XJ, Cado D, Robey E. The role of CD8α′ in the CD4 versus CD8 lineage choice. J Immunol. 1999;163:5312–8. [PubMed] [Google Scholar]
- 137.Boesteanu A, De Silva AD, Nakajima H, Leonard WJ, Peschon JJ, Joyce S. Distinct roles for signals relayed through the common cytokine receptor γ chain and interleukin 7 receptor α chain in natural T cell development. J Exp Med. 1997;186:331–6. doi: 10.1084/jem.186.2.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Peschon JJ, Morrissey PJ, Grabstein KH, et al. Early lymphocyte expansion is severly impaired in interleukin 7 receptor deficient mice. J Exp Med. 1994;180:1955–60. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kennedy MK, Glaccum M, Brown SN, et al. Reversible defects in natural killer and memory CD8 T cell lineages in interleukin 15-deficient mice. J Exp Med. 2000;191:771–80. doi: 10.1084/jem.191.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lodolce JP, Boone DL, Chai S, Swain RE, Dassopoulos T, Trettin S, Ma A. IL-15 receptor maintains lymphoid homeostasis by supporting lymphocyte homing and proliferation. Immunity. 1998;9:669–76. doi: 10.1016/s1074-7613(00)80664-0. [DOI] [PubMed] [Google Scholar]
- 141.Ohteki T, Ho S, Suzuki H, Mak TW, Ohashi PS. Role for IL-15/IL-15 receptor β-chain in natural killer 1.1+ T cell receptor-αβ+ cell development. J Immunol. 1997;159:5931–5. [PubMed] [Google Scholar]
- 142.Lantz O, Sharara LI, Tilloy F, Anderson A, DiSanto JP. Lineage relationships and differentiation of natural killer (NK) T cells: intrathymic selection and interleukin (IL)-4 production in the absence of NKR-P1 and Ly49 molecules. J Exp Med. 1997;185:1395–401. doi: 10.1084/jem.185.8.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Pandey A, Ozaki K, Baumann H, et al. Cloning of a receptor subunit required for signaling by thymic stromal lymphopoietin. Nature Immunol. 2000;1:59–64. doi: 10.1038/76923. [DOI] [PubMed] [Google Scholar]
- 144.Park SH, Kyin T, Bendelac A, Carnaud C. The contribution of NKT cells, NK cells, and other gamma-chain-dependent non-T non-B cells to IL-12-mediated rejection of tumors. J Immunol. 2003;170:1197–1201. doi: 10.4049/jimmunol.170.3.1197. [DOI] [PubMed] [Google Scholar]
- 145.Falcone M, Yeung B, Tucker L, Rodriguez E, Searvetnick N. A defect in IL-12-induced activation and IFN-γ secretion of peripheral natural killer T cells in nonobese diabetic mice suggests new pathogenic mechanisms for insulin-dependent diabetes mellitus (IDDM) J Exp Med. 1999;190:963–72. doi: 10.1084/jem.190.7.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Oya H, Kawamura T, Shimizu T, et al. The differential effect of stress on natural killer T (NKT) and NK cell function. Clin Exp Immunol. 2000;121:384–90. doi: 10.1046/j.1365-2249.2000.01310.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Tamada K, Harada M, Abe K, Li T, Nomoto K. IL-4-producing NK1.1+ T cells are resistant to glucocorticoid-induced apoptosis: implications for the Th1/Th2 balance. J Immunol. 1998;161:1239–47. [PubMed] [Google Scholar]
- 148.Emoto M, Mittrucker HW, Schmits R, Mak TW, Kaufmann SH. Critical role of leukocyte function-associated antigen-1 in liver accumulation of CD4+NKT cells. J Immunol. 1999;162:5094–8. [PubMed] [Google Scholar]
- 149.Miyamoto M, Emoto M, Brinkmann V, van Rooijen N, Schmits R, Kita E, Kaufmann SH. Contribution of NK cells to the homing of thymic CD4+NKT cells to the liver. J Immunol. 2000;165:1729–32. doi: 10.4049/jimmunol.165.4.1729. [DOI] [PubMed] [Google Scholar]
- 150.Kobayashi E, Motoki K, Uchida T, Fukushima H, Koezuka Y. KRN7000, a novel immunomodulator, and its antitumor activities. Oncol Res. 1995;7:529–34. [PubMed] [Google Scholar]
- 151.Chen H, Paul WE. Cultured NK1.1+CD4+ T cells produce large amounts of IL-4 and IFN-γ upon activation by anti-CD3 or CD1. J Immunol. 1997;159:2240–9. [PubMed] [Google Scholar]
- 152.Park S-H, Benlagha K, Lee D, Balish E, Bendelac A. Unaltered phenotype, tissue distribution and function of Vα14+ NKT cells in germ-free mice. Eur J Immunol. 2000;30:620–5. doi: 10.1002/1521-4141(200002)30:2<620::AID-IMMU620>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 153.Shinkai K, Locksley RM. CD1, tuberculosis, and the evolution of major histocompatibility complex molecules. J Exp Med. 2000;191:907–14. doi: 10.1084/jem.191.6.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chapman R, Sidrauski C, Walter P. Intracellular signalling from the endoplasmic reticulum to the nucleus. Annu Rev Cell Biol. 1998;14:459–85. doi: 10.1146/annurev.cellbio.14.1.459. [DOI] [PubMed] [Google Scholar]
- 155.Carman GM, Henry SA. Phospholipid biosynthesis in yeast. Annu Rev Biochem. 1989;58:635–69. doi: 10.1146/annurev.bi.58.070189.003223. [DOI] [PubMed] [Google Scholar]
- 156.Nunnari J, Walter P. Regulation of organelle biogenesis. Cell. 1996;84:389–94. doi: 10.1016/s0092-8674(00)81283-0. [DOI] [PubMed] [Google Scholar]